Use of Discrete Wavelet Transform to Assess Impedance Fluctuations Obtained from Cellular Micromotion
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Measurement of Impedance Time Course by ECIS
2.3. Numerical Analysis of Impedance Fluctuations
2.3.1. Shannon Entropy
2.3.2. Discrete Wavelet Transform
2.4. Statistical Analysis
3. Results and Discussion
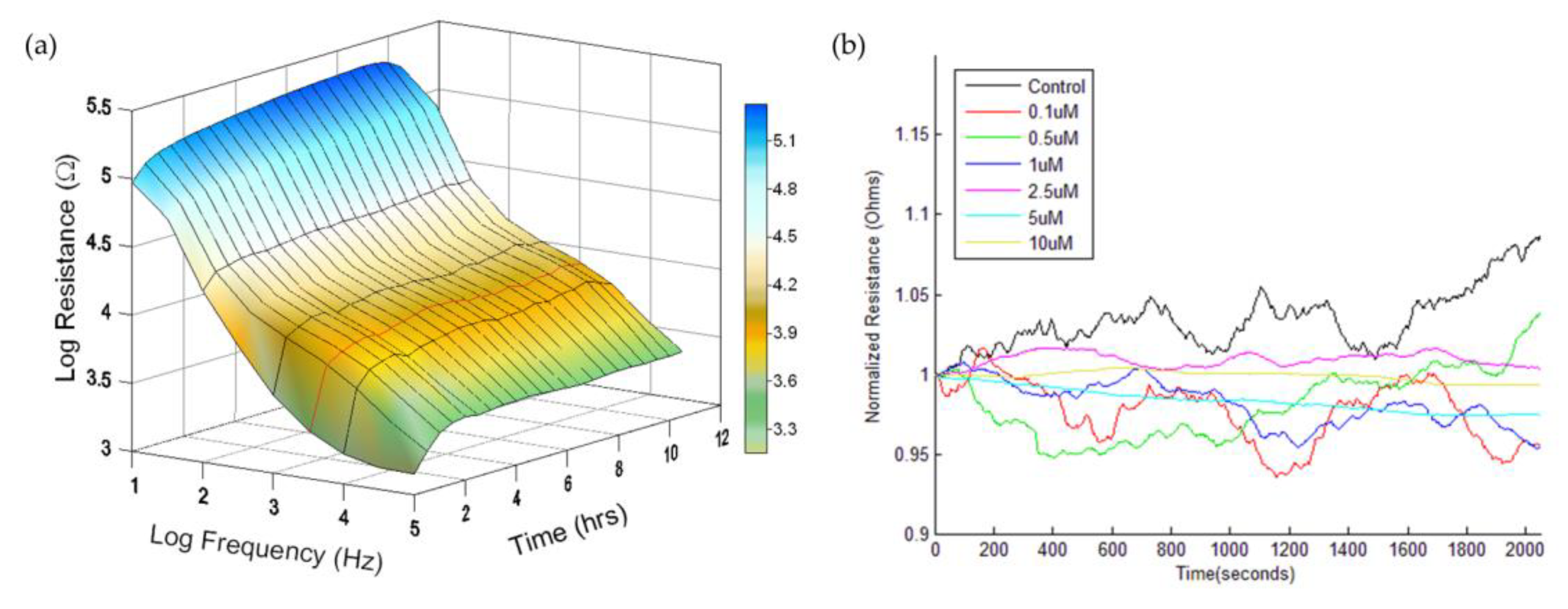
3.1. Real-Time Monitoring of HUVEC Attachment and Micromotions
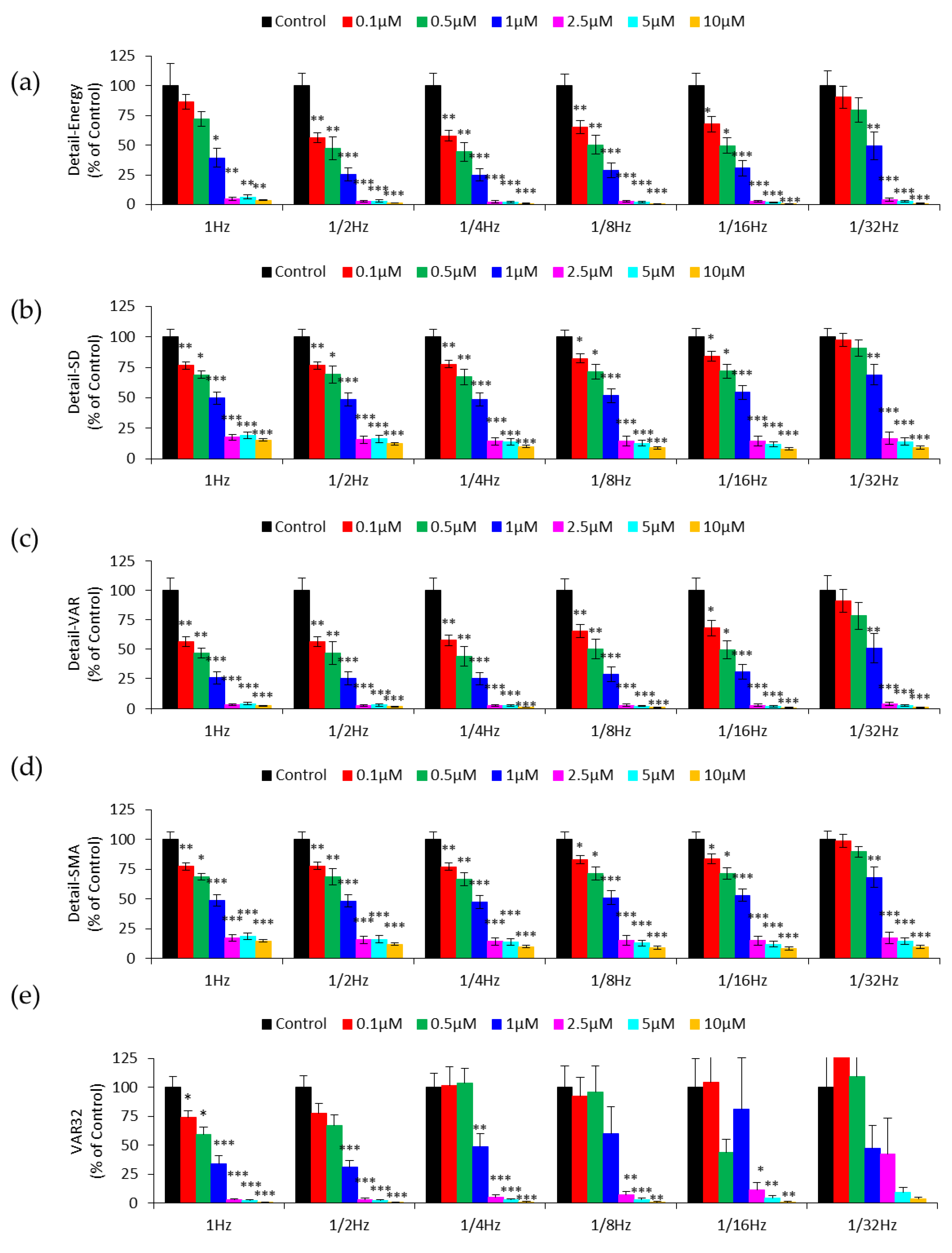
3.2. DWT Analysis of the Effect of Cytochalasin B on HUVEC Micromotion
3.3. Effect of Downsampling on Micromotion Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Giaever, I.; Keese, C.R. Monitoring fibroblast behavior in tissue culture with an applied electric field. Proc. Natl. Acad. Sci. USA 1984, 81, 3761–3764. [Google Scholar] [CrossRef]
- Giaever, I.; Keese, C.R. Micromotion of mammalian cells measured electrically. Proc. Natl. Acad. Sci. USA 1991, 88, 7896–7900. [Google Scholar] [CrossRef]
- Giaever, I.; Keese, C.R. A morphological biosensor for mammalian cells. Nature 1993, 366, 591–592. [Google Scholar] [CrossRef]
- Stolwijk, J.A.; Matrougui, K.; Renken, C.W.; Trebak, M. Impedance analysis of GPCR-mediated changes in endothelial barrier function: Overview and fundamental considerations for stable and reproducible measurements. Pflug. Arch. 2015, 467, 2193–2218. [Google Scholar] [CrossRef]
- Stolwijk, J.A.; Wegener, J. Impedance-Based Assays along the Life Span of Adherent Mammalian Cells in Vitro: From Initial Adhesion to Cell Death. In Label-Free Monitoring of Cells In Vitro; Wegener, J., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–75. [Google Scholar] [CrossRef]
- Lang, O.; Kohidai, L.; Wegener, J. Label-free profiling of cell dynamics: A sequence of impedance-based assays to estimate tumor cell invasiveness in vitro. Exp. Cell Res. 2017, 359, 243–250. [Google Scholar] [CrossRef]
- Szulcek, R.; Beckers, C.M.; Hodzic, J.; de Wit, J.; Chen, Z.; Grob, T.; Musters, R.J.; Minshall, R.D.; van Hinsbergh, V.W.; van Nieuw Amerongen, G.P. Localized RhoA GTPase activity regulates dynamics of endothelial monolayer integrity. Cardiovasc. Res. 2013, 99, 471–482. [Google Scholar] [CrossRef]
- Lo, C.M.; Keese, C.R.; Giaever, I. Monitoring motion of confluent cells in tissue culture. Exp. Cell Res. 1993, 204, 102–109. [Google Scholar] [CrossRef]
- Lai, Y.T.; Chu, Y.S.; Lo, J.C.; Hung, Y.H.; Lo, C.M. Effects of electrode diameter on the detection sensitivity and frequency characteristics of electric cell-substrate impedance sensing. Sens. Actuators B Chem. 2019, 288, 707–715. [Google Scholar] [CrossRef]
- Tarantola, M.; Schneider, D.; Sunnick, E.; Adam, H.; Pierrat, S.; Rosman, C.; Breus, V.; Sonnichsen, C.; Basche, T.; Wegener, J.; et al. Cytotoxicity of metal and semiconductor nanoparticles indicated by cellular micromotility. ACS Nano 2009, 3, 213–222. [Google Scholar] [CrossRef]
- Sapper, A.; Wegener, J.; Janshoff, A. Cell motility probed by noise analysis of thickness shear mode resonators. Anal. Chem. 2006, 78, 5184–5191. [Google Scholar] [CrossRef]
- Yang, C.T.; Mejard, R.; Griesser, H.J.; Bagnaninchi, P.O.; Thierry, B. Cellular micromotion monitored by long-range surface plasmon resonance with optical fluctuation analysis. Anal. Chem. 2015, 87, 1456–1461. [Google Scholar] [CrossRef]
- Opp, D.; Wafula, B.; Lim, J.; Huang, E.; Lo, J.C.; Lo, C.M. Use of electric cell-substrate impedance sensing to assess in vitro cytotoxicity. Biosens. Bioelectron. 2009, 24, 2625–2629. [Google Scholar] [CrossRef]
- Lovelady, D.C.; Friedman, J.; Patel, S.; Rabson, D.A.; Lo, C.M. Detecting effects of low levels of cytochalasin B in 3T3 fibroblast cultures by analysis of electrical noise obtained from cellular micromotion. Biosens. Bioelectron. 2009, 24, 2250–2254. [Google Scholar] [CrossRef]
- Chiu, S.P.; Lee, Y.W.; Wu, L.Y.; Tung, T.H.; Gomez, S.; Lo, C.M.; Wang, J.Y. Application of ECIS to Assess FCCP-Induced Changes of MSC Micromotion and Wound Healing Migration. Sensors 2019, 19, 3210. [Google Scholar] [CrossRef]
- Hlawatsch, F.; Boudreaux-Bartels, G.F. Linear and quadratic time-frequency signal representations. IEEE Signal Process. Mag. 1992, 9, 21–67. [Google Scholar] [CrossRef]
- Lai, Y.T.; Lo, C.M. Assessing in vitro cytotoxicity of cell micromotion by Hilbert-Huang transform. In Proceedings of the 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 3200–3203. [Google Scholar] [CrossRef]
- Akay, M. Time Frequency and Wavelets in Biomedical Signal Processing. In IEEE Press Series in Biomedical Engineering; Wiley-IEEE Press: Hoboken, NJ, USA, 1998. [Google Scholar]
- Percival, D.B.; Walden, A.T. Wavelet Methods for Time Series Analysis; Cambridge University Press: Cambridge, UK, 2000; Volume 4. [Google Scholar]
- Chen, S.W.; Yang, J.M.; Yang, J.H.; Yang, S.J.; Wang, J.S. A computational modeling and analysis in cell biological dynamics using electric cell-substrate impedance sensing (ECIS). Biosens. Bioelectron. 2012, 33, 196–203. [Google Scholar] [CrossRef]
- Yang, J.M.; Chen, S.W.; Yang, J.H.; Hsu, C.C.; Wang, J.S. A quantitative cell modeling and wound-healing analysis based on the Electric Cell-substrate Impedance Sensing (ECIS) method. Comput. Biol. Med. 2016, 69, 134–143. [Google Scholar] [CrossRef]
- Das, D.; Shiladitya, K.; Biswas, K.; Dutta, P.K.; Parekh, A.; Mandal, M.; Das, S. Wavelet-based multiscale analysis of bioimpedance data measured by electric cell-substrate impedance sensing for classification of cancerous and normal cells. Phys. Rev. E 2015, 92, 062702. [Google Scholar] [CrossRef]
- Parekh, A.; Das, D.; Das, S.; Dhara, S.; Biswas, K.; Mandal, M.; Das, S. Bioimpedimetric analysis in conjunction with growth dynamics to differentiate aggressiveness of cancer cells. Sci. Rep. 2018, 8, 783. [Google Scholar] [CrossRef]
- Costa, M.; Goldberger, A.L.; Peng, C.K. Multiscale entropy analysis of biological signals. Phys. Rev. E 2005, 71, 021906. [Google Scholar] [CrossRef]
- Nordberg, R.C.; Zhang, J.; Griffith, E.H.; Frank, M.W.; Starly, B.; Loboa, E.G. Electrical Cell-Substrate Impedance Spectroscopy Can Monitor Age-Grouped Human Adipose Stem Cell Variability During Osteogenic Differentiation. Stem Cells Transl. Med. 2017, 6, 502–511. [Google Scholar] [CrossRef]
- Shannon, C.E. Communication in the Presence of Noise. Proc. IRE 1949, 37, 10–21. [Google Scholar] [CrossRef]
- Bouten, C.V.; Westerterp, K.R.; Verduin, M.; Janssen, J.D. Assessment of energy expenditure for physical activity using a triaxial accelerometer. Med. Sci. Sports Exerc. 1994, 26, 1516–1523. [Google Scholar] [CrossRef]
- Mathie, M.J.; Coster, A.C.; Lovell, N.H.; Celler, B.G. Detection of daily physical activities using a triaxial accelerometer. Med. Biol. Eng. Comput. 2003, 41, 296–301. [Google Scholar] [CrossRef]
- Carter, S.B. Effects of cytochalasins on mammalian cells. Nature 1967, 213, 261–264. [Google Scholar] [CrossRef]
- Scherlach, K.; Boettger, D.; Remme, N.; Hertweck, C. The chemistry and biology of cytochalasans. Nat. Prod. Rep. 2010, 27, 869–886. [Google Scholar] [CrossRef]
- Chaovalit, P.; Gangopadhyay, A.; Karabatis, G.; Chen, Z.Y. Discrete Wavelet Transform-Based Time Series Analysis and Mining. ACM Comput. Surv. 2011, 43, 1–37. [Google Scholar] [CrossRef]
- Chan, K.-P.; Fu, A.W.-C. Efficient time series matching by wavelets. In Proceedings of the 15th International Conference on Data Engineering (Cat. No. 99CB36337), Sydney, Australia, 23–26 March 1999; pp. 126–133. [Google Scholar]
- Liu, S.; Cao, R.; Huang, Y.; Ouypornkochagorn, T.; Jia, J. Time Sequence Learning for Electrical Impedance Tomography Using Bayesian Spatiotemporal Priors. IEEE Trans. Instrum. Meas. 2020. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tung, T.-H.; Wang, S.-H.; Huang, C.-C.; Su, T.-Y.; Lo, C.-M. Use of Discrete Wavelet Transform to Assess Impedance Fluctuations Obtained from Cellular Micromotion. Sensors 2020, 20, 3250. https://doi.org/10.3390/s20113250
Tung T-H, Wang S-H, Huang C-C, Su T-Y, Lo C-M. Use of Discrete Wavelet Transform to Assess Impedance Fluctuations Obtained from Cellular Micromotion. Sensors. 2020; 20(11):3250. https://doi.org/10.3390/s20113250
Chicago/Turabian StyleTung, Tse-Hua, Si-Han Wang, Chun-Chung Huang, Tai-Yuan Su, and Chun-Min Lo. 2020. "Use of Discrete Wavelet Transform to Assess Impedance Fluctuations Obtained from Cellular Micromotion" Sensors 20, no. 11: 3250. https://doi.org/10.3390/s20113250
APA StyleTung, T.-H., Wang, S.-H., Huang, C.-C., Su, T.-Y., & Lo, C.-M. (2020). Use of Discrete Wavelet Transform to Assess Impedance Fluctuations Obtained from Cellular Micromotion. Sensors, 20(11), 3250. https://doi.org/10.3390/s20113250




