Performances Evaluation of a Low-Cost Platform for High-Resolution Plant Phenotyping
Abstract
1. Introduction
2. Materials and Methods
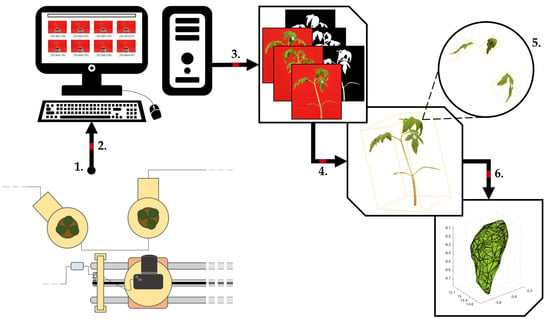
2.1. Phenotyping Platform Setup
2.2. Experimental Setup and Observed Data
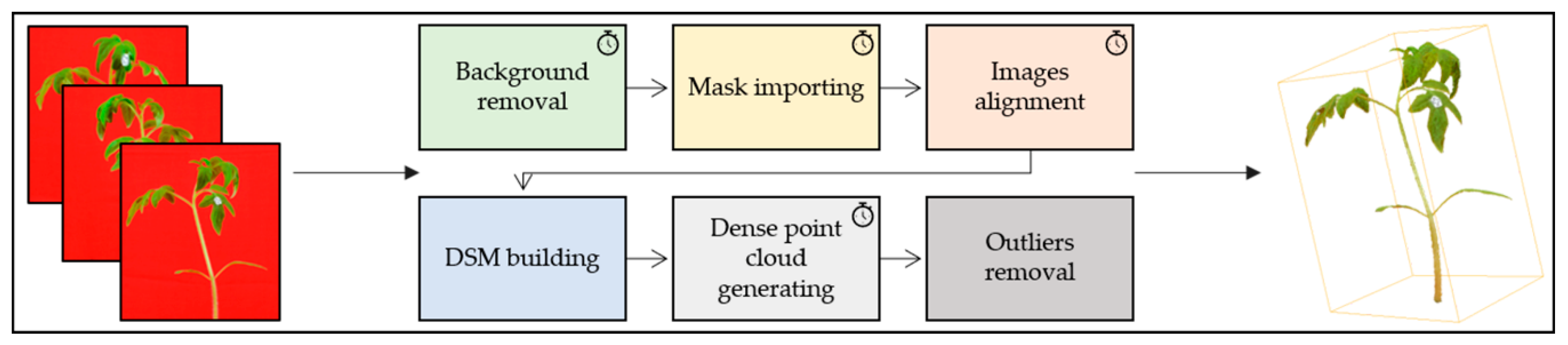
2.3. Image Processing
2.4. Architecture Segmentation
2.5. Statistical Analysis
3. Results
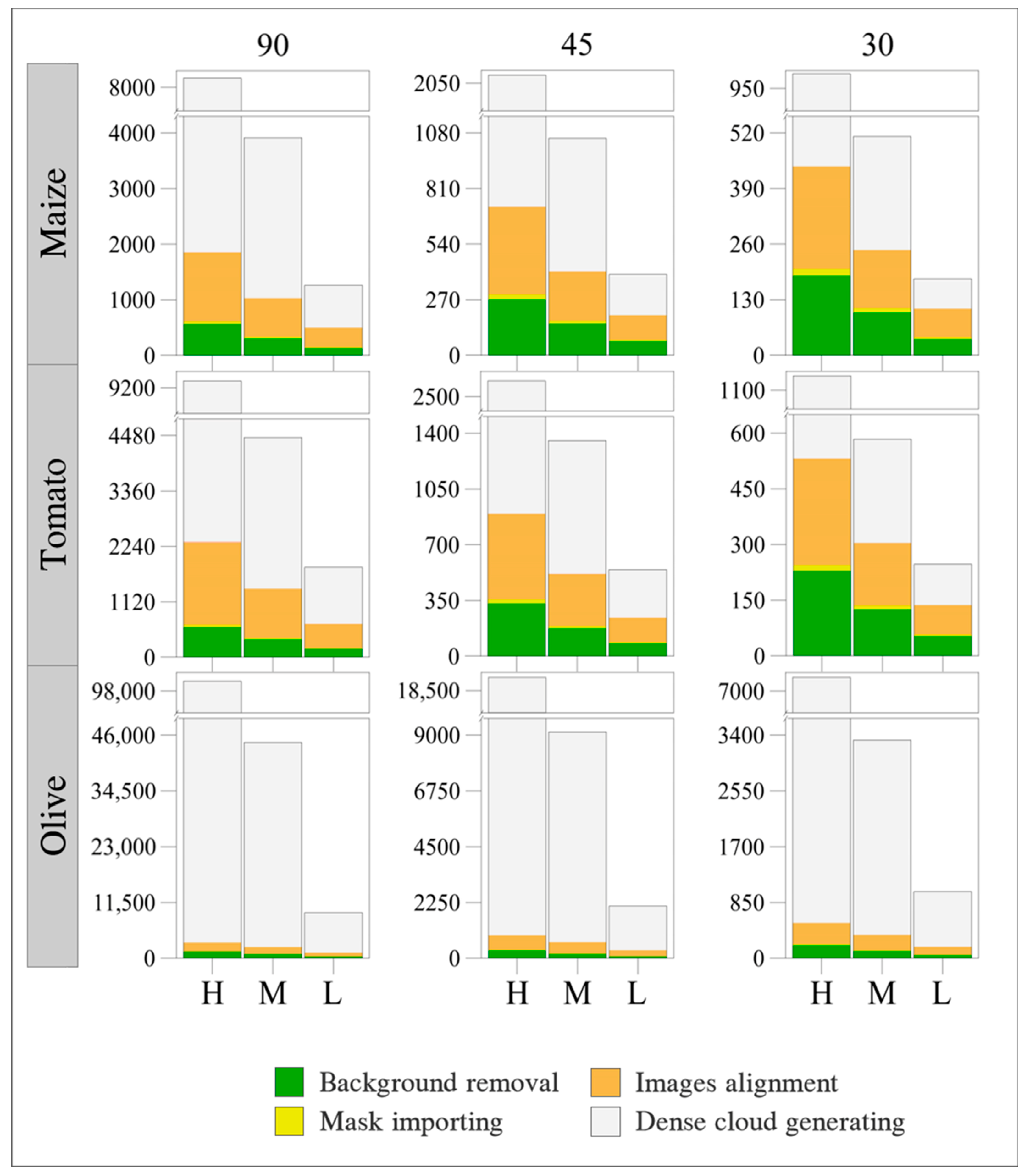
3.1. Time Processing
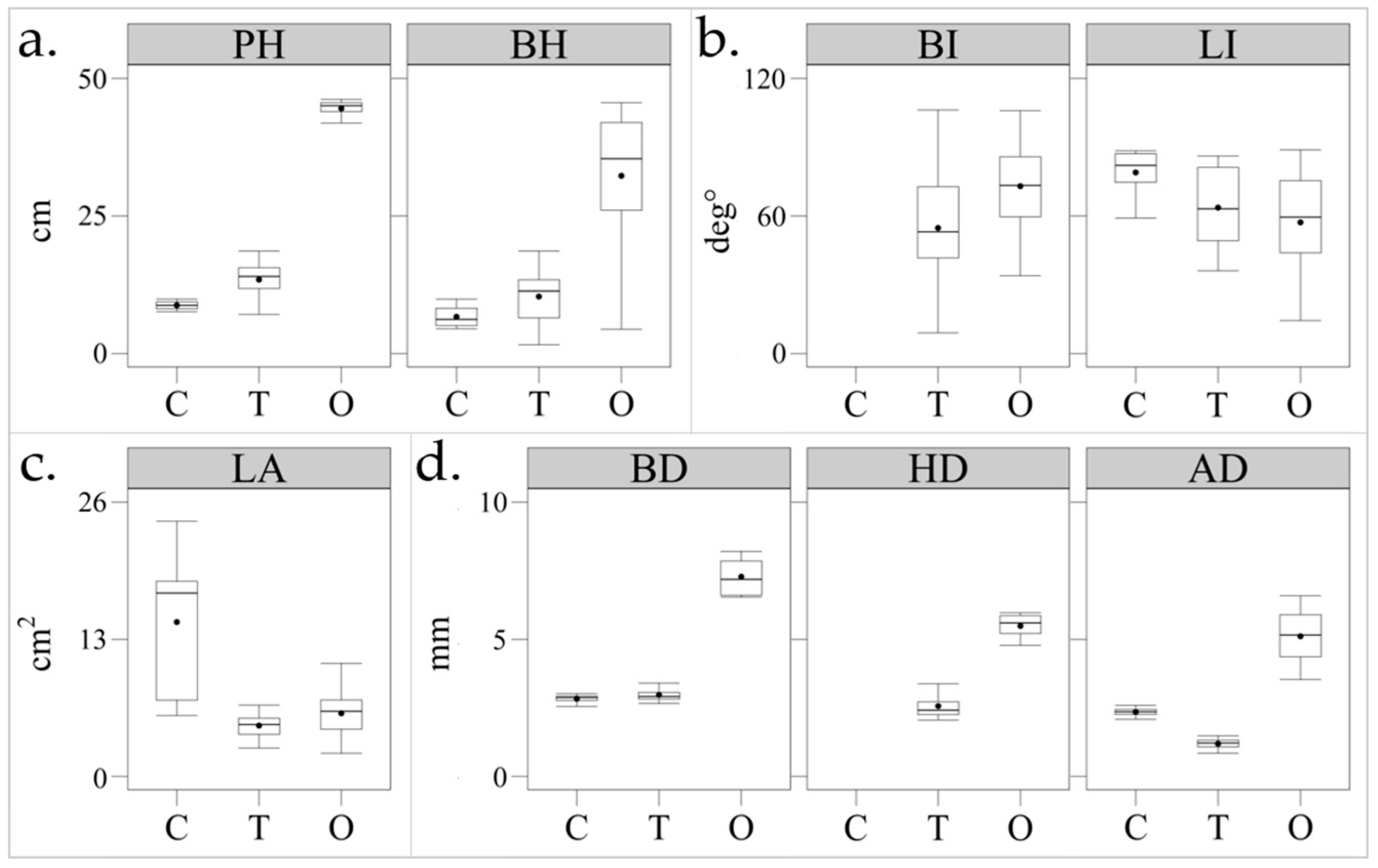
3.2. Morphometric Traits Extraction
3.2.1. Plant Heights
3.2.2. Stem Diameters
3.2.3. Petiole/Branch Inclination
3.2.4. Leaf Inclination
3.2.5. Leaf Area
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, L.; Zhang, Q.; Huang, D. A review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef] [PubMed]
- Araus, J.L.; Kefauver, S.C.; Zaman-Allah, M.; Olsen, M.S.; Cairns, J.E. Phenotyping: New Crop Breeding Frontier. In Encyclopedia of Sustainability Science and Technology; Meyers, R., Ed.; Springer: New York, NY, USA, 2018. [Google Scholar]
- Wyatt, J. Grain and plant morphology of cereals and how characters can be used to identify varieties. In Encyclopedia of Food Grains; Colin, W., Harold, C., Koushik, S., Eds.; Elsevier: Oxford, UK, 2016; Volume 1, pp. 51–72. [Google Scholar]
- Pangga, I.B.; Hanan, J.; Chakraborty, S. Pathogen dynamics in a crop canopy and their evolution under changing climate. Plant Pathol. 2011, 60, 70–81. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wen, W.; Wu, S.; Wang, C.; Yu, Z.; Guo, X.; Zhao, C. Maize plant phenotyping: Comparing 3D laser scanning, multi-view stereo reconstruction, and 3D digitizing estimates. Remote Sens. 2019, 11, 63. [Google Scholar] [CrossRef]
- Laxman, R.H.; Hemamalini, P.; Bhatt, R.M.; Sadashiva, A.T. Non-invasive quantification of tomato (Solanum lycopersicum L.) plant biomass through digital imaging using phenomics platform. Indian J. Plant Physiol. 2018, 23, 369–375. [Google Scholar] [CrossRef]
- Qiu, R.; Wei, S.; Zhang, M.; Li, H.; Sun, H.; Liu, G.; Li, M. Sensors for measuring plant phenotyping: A review. Int. J. Agric. Biol. Eng. 2018, 11, 1–17. [Google Scholar] [CrossRef]
- Zhang, Y.; Teng, P.; Shimizu, Y.; Hosoi, F.; Omasa, K. Estimating 3D leaf and stem shape of nursery paprika plants by a novel multi-camera photography system. Sensors 2016, 16, 874. [Google Scholar] [CrossRef]
- Guo, Q.; Wu, F.; Pang, S.; Zhao, X.; Chen, L.; Liu, J.; Xue, B.; Xu, G.; Li, L.; Jing, H.; et al. Crop 3D—A LiDAR based platform for 3D high-throughput crop phenotyping. Sci. China Life Sci. 2018, 61, 328–339. [Google Scholar] [CrossRef]
- Chawade, A.; Van Ham, J.; Blomquist, H.; Bagge, O.; Alexandersson, E.; Ortiz, R. High-throughput field-phenotyping tools for plant breeding and precision agriculture. Agronomy 2019, 9, 258. [Google Scholar] [CrossRef]
- Pratap, A.; Tomar, R.; Kumar, J.; Vankat, R.P.; Mehandi, S.; Katiyar, P.K. High-throughput plant phenotyping platforms. Phenomics Crop Plants: Trends, Options Limit. 2015, 285–296, ISBN 9788132222262. [Google Scholar]
- Roitsch, T.; Cabrera-Bosquet, L.; Fournier, A.; Ghamkhar, K.; Jiménez-Berni, J.; Pinto, F.; Ober, E.S. Review: New sensors and data-driven approaches—A path to next generation phenomics. Plant Sci. 2019, 282, 2–10. [Google Scholar] [CrossRef] [PubMed]
- De Diego, N.; Fürst, T.; Humplík, J.F.; Ugena, L.; Podlešáková, K.; Spíchal, L. An automated method for high-throughput screening of Arabidopsis rosette growth in multi-well llates and its validation in stress conditions. Front. Plant Sci. 2017, 8, 1702. [Google Scholar] [CrossRef] [PubMed]
- Granier, C.; Aguirrezabal, L.; Chenu, K.; Cookson, S.J.; Dauzat, M.; Hamard, P.; Thioux, J.-J.; Rolland, G.; Bouchier-Combaud, S.; Lebaudy, A.; et al. PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 2006, 169, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.; Scharr, H.; Gilmer, F.; Zierer, R.; Nagel, K.A.; Ernst, M.; Wiese, A.; Virnich, O.; Christ, M.M.; Uhlig, B.; et al. Dynamics of seedling growth acclimation towards altered light conditions can be quantified via GROWSCREEN: A setup and procedure designed for rapid optical phenotyping of different plant species. New Phytol. 2007, 174, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, H.; Zhou, J.; Fu, X.; Ye, H.; Nguyen, H.T. Development of an automated phenotyping platform for quantifying soybean dynamic responses to salinity stress in greenhouse environment. Comput. Electron. Agric. 2018, 151, 319–330. [Google Scholar] [CrossRef]
- Kolukisaoglu, Ü.; Thurow, K. Future and frontiers of automated screening in plant sciences. Plant Sci. 2010, 178, 476–484. [Google Scholar] [CrossRef]
- Zhang, C.; Pumphrey, M.O.; Zhou, J.; Zhang, Q.; Sankaran, S. Development of an automated high-throughput phenotyping system for wheat evaluation in a controlled environment. Trans. ASABE 2019, 62, 61–74. [Google Scholar] [CrossRef]
- Huang, P.; Luo, X.; Jin, J.; Wang, L.; Zhang, L.; Liu, J.; Zhang, Z. Improving high-throughput phenotyping using fusion of close-range hyperspectral camera and low-cost depth sensor. Sensors 2018, 18, 2711. [Google Scholar] [CrossRef]
- Gibbs, J.A.; Pound, M.; French, A.P.; Wells, D.M.; Murchie, E.; Pridmore, T. Approaches to three-dimensional reconstruction of plant shoot topology and geometry. Funct. Plant Biol. 2017, 44, 62–75. [Google Scholar] [CrossRef]
- Nguyen, C.V.; Fripp, J.; Lovell, D.R.; Furbank, R.; Kuffner, P.; Daily, H.; Sirault, X. 3D scanning system for automatic high-resolution plant phenotyping. In Proceedings of the 2016 International Conference on Digital Image Computing: Techniques and Applications (DICTA), Gold Coast, Australia, 30 November–2 December 2016; pp. 148–155. [Google Scholar]
- Paulus, S.; Behmann, J.; Mahlein, A.-K.; Plümer, L.; Kuhlmann, H. Low-cost 3D systems: Suitable tools for plant phenotyping. Sensors 2014, 14, 3001–3018. [Google Scholar] [CrossRef]
- Müller-Linow, M.; Wilhelm, J.; Briese, C.; Wojciechowski, T.; Schurr, U.; Fiorani, F. Plant Screen Mobile: An open-source mobile device app for plant trait analysis. Plant Methods 2019, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.; Paulus, S.; Kuhlmann, H. Accuracy analysis of a multi-view stereo approach for phenotyping of tomato plants at the organ level. Sensors 2015, 15, 9651–9665. [Google Scholar] [CrossRef] [PubMed]
- Moriondo, M.; Leolini, L.; Staglianò, N.; Argenti, G.; Trombi, G.; Brilli, L.; Dibari, C.; Leolini, C.; Bindi, M. Use of digital images to disclose canopy architecture in olive tree. Sci. Hortic. 2016, 209, 1–13. [Google Scholar] [CrossRef]
- Caruso, G.; Zarco-Tejada, P.J.; González-Dugo, V.; Moriondo, M.; Tozzini, L.; Palai, G.; Rallo, G.; Hornero, A.; Primicerio, J.; Gucci, R. High-resolution imagery acquired from an unmanned platform to estimate biophysical and geometrical parameters of olive trees under different irrigation regimes. PLoS ONE 2019, 14, e0210804. [Google Scholar] [CrossRef]
- Caruso, G.; Tozzini, L.; Rallo, G.; Primicerio, J.; Moriondo, M.; Palai, G.; Gucci, R. Estimating biophysical and geometrical parameters of grapevine canopies (‘Sangiovese’) by an unmanned aerial vehicle (UAV) and VIS-NIR cameras. Vitis J. Grapevine Res. 2017, 56, 63–70. [Google Scholar]
- Smith, M.W.; Carrivick, J.L.; Quincey, D.J. Structure from motion photogrammetry in physical geography. Prog. Phys. Geogr. 2015, 40, 247–275. [Google Scholar] [CrossRef]
- Pagán, J.I.; López, I.; Bañón, L.; Aragonés, L. 3D modelling of dune ecosystems using photogrammetry from remotely piloted air systems surveys. WIT Trans. Eng. Sci. 2019, 125, 163–171. [Google Scholar]
- Micheletti, N.; Chandler, J.H.; Lane, S.N. Structure from Motion (SfM) Photogrammetry. In Geomorphological Techniques; Clarke, L.E., Nield, J.M., Eds.; British Society for Geomorphology: London, UK, 2015; Volume 2, pp. 1–12. [Google Scholar]
- Cao, W.; Zhou, J.; Yuan, Y.; Ye, H.; Nguyen, H.T.; Chen, J.; Zhou, J. Quantifying variation in soybean due to flood using a low-cost 3D imaging system. Sensors 2019, 19, 2682. [Google Scholar] [CrossRef]
- Liu, S.; Acosta-Gamboa, L.; Huang, X.; Lorence, A. Novel low cost 3D surface model reconstruction system for plant phenotyping. J. Imaging 2017, 3, 39. [Google Scholar] [CrossRef]
- Santos, T.T.; de Oliveira, A.A. Image-based 3D digitizing for plant architecture analysis and phenotyping. In Proceedings of the Workshop on Industry Applications (WGARI) in SIBGRAPI 2012 (XXV Conference on Graphics, Patterns and Images), Ouro Preto, Brazil, 22–25 August 2012. [Google Scholar]
- Torres-Sánchez, J.; López-Granados, F.; Borra-Serrano, I.; Manuel Peña, J. Assessing UAV-collected image overlap influence on computation time and digital surface model accuracy in olive orchards. Precis. Agric. 2018, 19, 115–133. [Google Scholar] [CrossRef]
- Zhou, J.; Fu, X.; Schumacher, L.; Zhou, J. Evaluating geometric measurement accuracy based on 3d reconstruction of automated imagery in a greenhouse. Sensors 2018, 18, 2270. [Google Scholar] [CrossRef] [PubMed]
- Paulus, S. Measuring crops in 3D: Using geometry for plant phenotyping. Plant Methods 2019, 15, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Biasi, N.; Setti, F.; Tavernini, M.; Fornaser, A.; Lunardelli, M.; Da Lio, M.; De Cecco, M. Low-Cost Garment-Based 3D Body Scanner. In Proceedings of the 3rd International Conference on 3D Body Scanning Technologies, Lugano, Switzerland, 16–17 October 2012; pp. 106–114. [Google Scholar]
- Remondino, F.; El-Hakim, S. Image-based 3D modelling: A review. Photogramm. Rec. 2006, 21, 269–291. [Google Scholar] [CrossRef]
- Martinez-Guanter, J.; Ribeiro, Á.; Peteinatos, G.G.; Pérez-Ruiz, M.; Gerhards, R.; Bengochea-Guevara, J.M.; Machleb, J.; Andújar, D. Low-cost three-dimensional modeling of crop plants. Sensors 2019, 19, 2883. [Google Scholar] [CrossRef]
- Pound, M.P.; French, A.P.; Murchie, E.H.; Pridmore, T.P. Breakthrough technologies automated recovery of three-dimensional models of plant shoots from multiple color images. Plant Physiol. 2014, 166, 1688–1698. [Google Scholar] [CrossRef]
- Verma, A.K.; Bourke, M.C. A method based on structure-from-motion photogrammetry to generate sub-millimetre-resolution digital elevation models for investigating rock breakdown features. Earth Surf. Dyn. 2019, 7, 45–66. [Google Scholar] [CrossRef]
- Chianucci, F.; Pisek, J.; Raabe, K.; Marchino, L.; Ferrara, C.; Corona, P. A dataset of leaf inclination angles for temperate and boreal broadleaf woody species. Ann. For. Sci. 2018, 75, 50–57. [Google Scholar] [CrossRef]
- Pisek, J.; Ryu, Y.; Alikas, K. Estimating leaf inclination and G-function from leveled digital camera photography in broadleaf canopies. Trees Struct. Funct. 2011, 25, 919–924. [Google Scholar] [CrossRef]
- Jianchang, G.; Guangjun, G.; Yanmei, G.; Xiaoxuan, W.; YongChen, D. Measuring plant leaf area by scanner and ImageJ software. China Veg. 2011, 1, 73–77. [Google Scholar]
- Cosmulescu, S.; Scrieciu, F.; Manda, M. Determination of leaf characteristics in different medlar genotypes using the ImageJ program. Hortic. Sci. 2019. [Google Scholar]
- Wu, J.; Xue, X.; Zhang, S.; Qin, W.; Chen, C.; Sun, T. Plant 3D reconstruction based on LiDAR and multi-view sequence images. Int. J. Precis. Agric. Aviat. 2018, 1, 37–43. [Google Scholar] [CrossRef]
- Gélard, W.; Herbulot, A.; Devy, M.; Debaeke, P.; McCormick, R.F.; Truong, S.K.; Mullet, J. Leaves Segmentation in 3D Point Cloud. Lect. Notes Comput. Sci. 2017, 10617, 664–674. [Google Scholar]
- Wright, S. Correlation and causation. J. Agric. Res. 1921, 20, 557–585. [Google Scholar]
- Loague, K.; Green, R.E. Statistical and graphical methods for evaluating solute transport models: Overview and application. J. Contam. Hydrol. 1991, 7, 51–73. [Google Scholar] [CrossRef]
- Akaike, H. Information Theory and an Extension of the Maximum Likelihood Principle. In Selected Papers of Hirotugu Akaike. Springer Series in Statistics (Perspectives in Statistics); Parzen, E., Tanabe, K., Kitagawa, G., Eds.; Springer: New York, NY, USA, 1998; pp. 199–213. [Google Scholar]
- Jamieson, P.D.; Porter, J.R.; Wilson, D.R. A test of the computer simulation model ARCWHEAT1 on wheat crops grown in New Zealand. Field Crops Res. 1991, 27, 337–350. [Google Scholar] [CrossRef]
- Li, M.F.; Tang, X.P.; Wu, W.; Liu, H. Bin General models for estimating daily global solar radiation for different solar radiation zones in mainland China. Energy Convers. Manag. 2013, 70, 139–148. [Google Scholar] [CrossRef]
- Itakura, K.; Hosoi, F. Automatic leaf segmentation for estimating leaf area and leaf inclination angle in 3D plant images. Sensors 2018, 18, 3576. [Google Scholar] [CrossRef]
- Teixeira Santos, T.; Vieira Koenigkan, L.; Garcia Arnal Barbedo, J.; Costa Rodrigues, G. 3D Plant Modeling: Localization, Mapping and Segmentation for Plant Phenotyping Using a Single Hand-held Camera. In Computer Vision-ECCV 2014 Workshops; Springer: New York, NY, USA, 2015; pp. 247–263. ISBN 1916220118. [Google Scholar]
- Andújar, D.; Calle, M.; Fernández-Quintanilla, C.; Ribeiro, Á.; Dorado, J. Three-dimensional modeling of weed plants using low-cost photogrammetry. Sensors 2018, 18, 1077. [Google Scholar] [CrossRef]
- Andújar, D.; Dorado, J.; Bengochea-Guevara, J.; Conesa-Muñoz, J.; Fernández-Quintanilla, C.; Ribeiro, Á. Influence of wind speed on RGB-D images in tree plantations. Sensors 2017, 17, 914. [Google Scholar] [CrossRef]
- Jones, H.; Sirault, X. Scaling of thermal images at different spatial resolution: The mixed pixel problem. Agronomy 2014, 4, 380–396. [Google Scholar] [CrossRef]
- Paproki, A.; Sirault, X.; Berry, S.; Furbank, R.; Fripp, J. A novel mesh processing based technique for 3D plant analysis. BMC Plant Biol. 2012, 12, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Zeng, G.; Wang, J.; Kang, S.B.; Quan, L. Image-based tree modeling. ACM Trans. Graph. 2007, 26, 1–7. [Google Scholar] [CrossRef]
- Quan, L.; Tan, P.; Zeng, G.; Yuan, L.; Wang, J.; Kang, S.B. Image-based plant modeling. ACM Trans. Graph. 2006, 25, 599–604. [Google Scholar] [CrossRef]
- Bernotas, G.; Scorza, L.C.T.; Hansen, M.F.; Hales, I.J.; Halliday, K.J.; Smith, L.N.; Smith, M.L.; McCormick, A.J. A photometric stereo-based 3D imaging system using computer vision and deep learning for tracking plant growth. Gigascience 2019, 8. [Google Scholar] [CrossRef]
- Zhou, R.; Yu, X.; Ottosen, C.O.; Rosenqvist, E.; Zhao, L.; Wang, Y.; Yu, W.; Zhao, T.; Wu, Z. Drought stress had a predominant effect over heat stress on three tomato cultivars subjected to combined stress. BMC Plant Biol. 2017, 17, 1–13. [Google Scholar] [CrossRef]

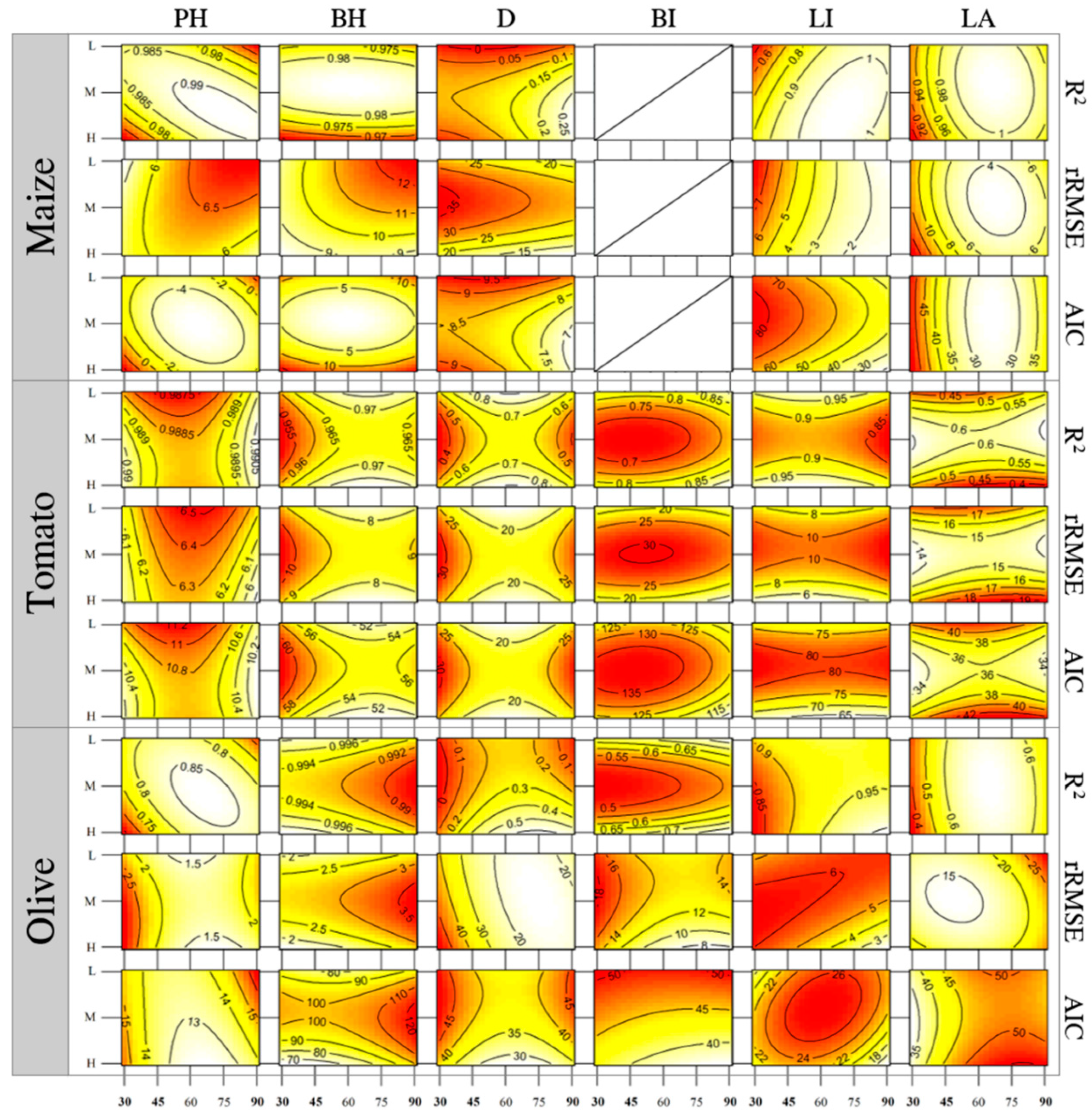
| Trait | Crop | R2 | rRMSE | AIC |
|---|---|---|---|---|
| HP | Maize | 0.99 | 5.03% | −2.74 |
| HP | Tomato | 0.99 | 5.69% | 10.00 |
| HP | Olive | 0.83 | 1.86% | 14.12 |
| BH | Maize | 0.97 | 8.37% | 12.39 |
| BH | Tomato | 0.97 | 7.74% | 52.30 |
| BH | Olive | 0.99 | 2.15% | 88.04 |
| D | Maize | 0.23 | 11.80% | 7.35 |
| D | Tomato | 0.67 | 21.86% | 22.51 |
| D | Olive | 0.56 | 16.39% | 35.61 |
| BI | Tomato | 0.95 | 11.19% | 104.55 |
| BI | Olive | 0.91 | 6.09% | 35.44 |
| LI | Maize | 0.99 | 0.58% | 13.17 |
| LI | Tomato | 0.96 | 6.26% | 63.58 |
| LI | Olive | 0.99 | 1.39% | 12.38 |
| LA | Maize | 0.97 | 6.89% | 40.22 |
| LA | Tomato | 0.36 | 19.69% | 42.95 |
| LA | Olive | 0.50 | 28.37% | 56.39 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, R.; Leolini, C.; Costafreda-Aumedes, S.; Leolini, L.; Bindi, M.; Zaldei, A.; Moriondo, M. Performances Evaluation of a Low-Cost Platform for High-Resolution Plant Phenotyping. Sensors 2020, 20, 3150. https://doi.org/10.3390/s20113150
Rossi R, Leolini C, Costafreda-Aumedes S, Leolini L, Bindi M, Zaldei A, Moriondo M. Performances Evaluation of a Low-Cost Platform for High-Resolution Plant Phenotyping. Sensors. 2020; 20(11):3150. https://doi.org/10.3390/s20113150
Chicago/Turabian StyleRossi, Riccardo, Claudio Leolini, Sergi Costafreda-Aumedes, Luisa Leolini, Marco Bindi, Alessandro Zaldei, and Marco Moriondo. 2020. "Performances Evaluation of a Low-Cost Platform for High-Resolution Plant Phenotyping" Sensors 20, no. 11: 3150. https://doi.org/10.3390/s20113150
APA StyleRossi, R., Leolini, C., Costafreda-Aumedes, S., Leolini, L., Bindi, M., Zaldei, A., & Moriondo, M. (2020). Performances Evaluation of a Low-Cost Platform for High-Resolution Plant Phenotyping. Sensors, 20(11), 3150. https://doi.org/10.3390/s20113150