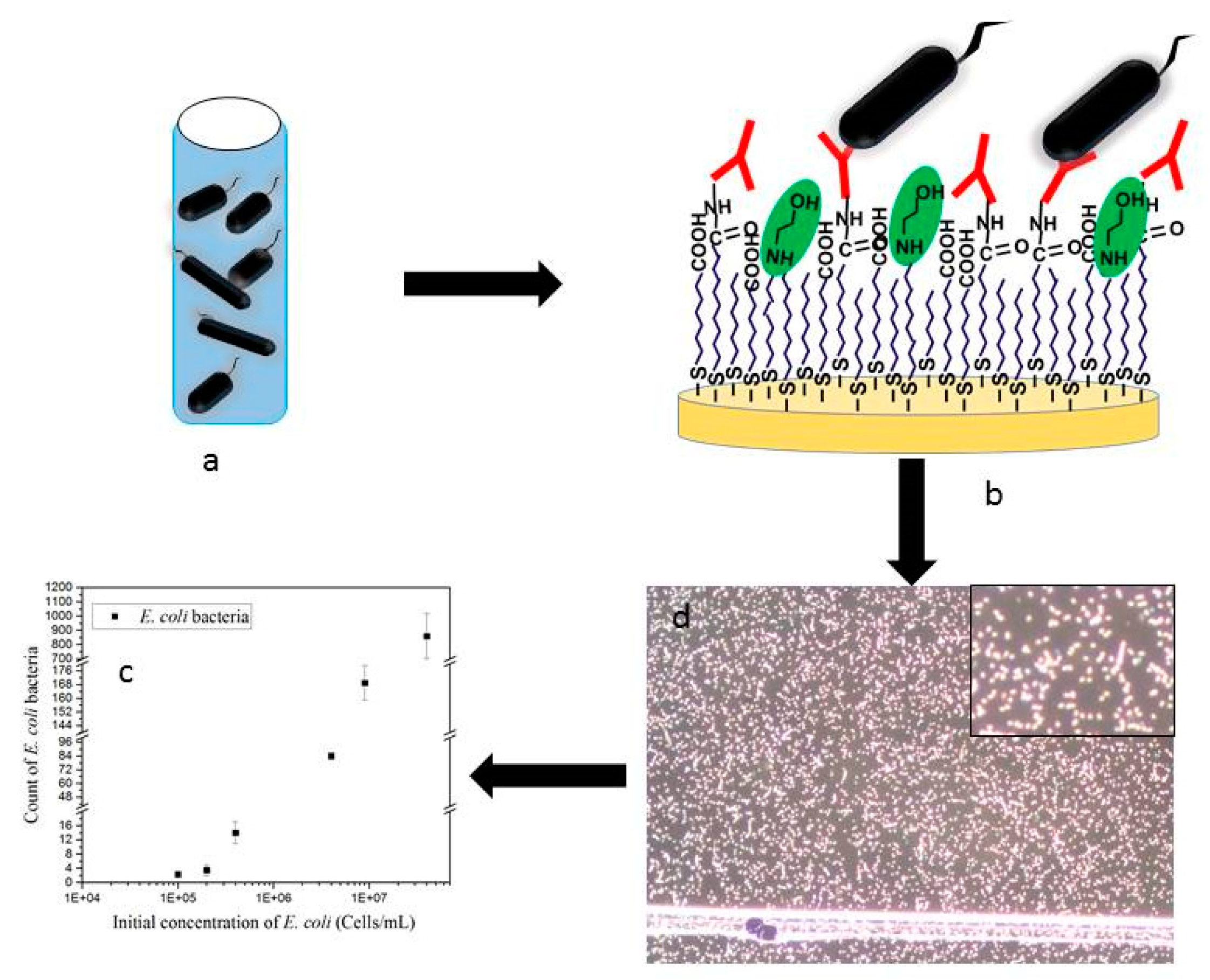
The different antibodies used in these experiments were immobilized on the gold coated surface using carbodiimide chemistry as described above. Each channel was then connected to a peristaltic pump to run the sample containing bacteria over the antibody-functionalized surface. The flow was stopped and the sample was incubated for 75 min on the sensing platform. The bacteria captured on the surface were then detected by dark field microscopy and statistical analysis was performed on the recorded image for the counting of the bacteria captured by antibodies.
3.1. Optimization of the Binding Assay in Surface Plasmon Resonance Measurements
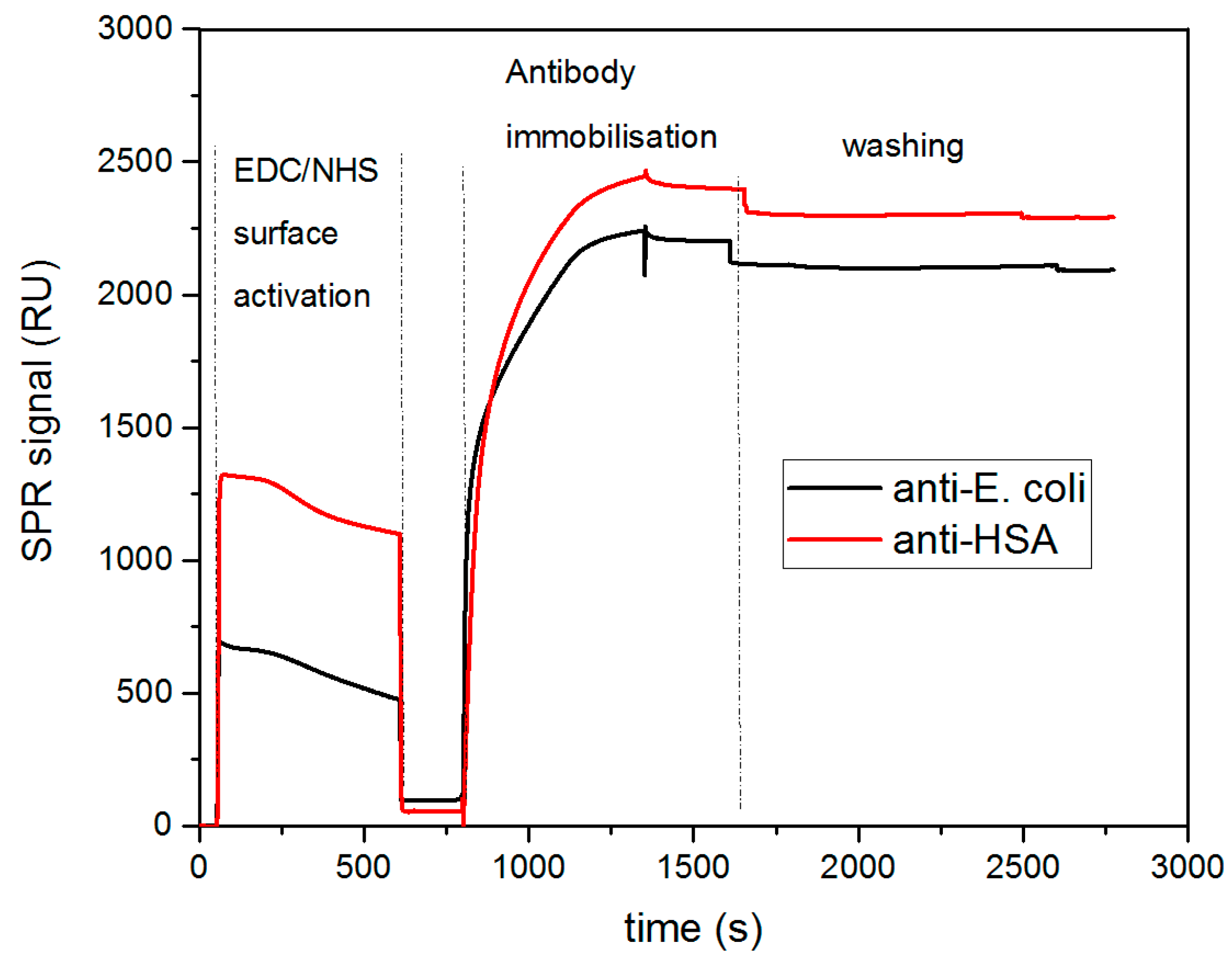
SPR experiments were first carried out to characterize and optimize the immobilization of the antibodies on the gold surfaces as well as the capture of bacteria. In this study, the antibodies were covalently immobilized on the gold surface functionalized with a self-assembled monolayer (SAM) terminated with carboxyl functional groups. To enable antibody covalent coupling, the –COOH groups were activated using standard EDC/NHS activation chemistry. The carbodiimide activation (EDC) along with the addition of NHS created amine-reactive NHS-esters able to form stable amide bonds with primary amines exposed by antibody molecules (mainly Lysine moieties). SPR measurements confirmed the antibody immobilization, as shown in
Figure 2. Two antibodies (anti-HSA and anti-
E. coli) were immobilized onto two separate channels and the SPR signal was recorded, being referenced to the signal from a reference channel, which was not functionalized with any antibody. The first step of EDC/NHS activation was observed as expected with a small increase in the SPR signal and was followed by a considerable signal rise upon the injection of antibody solutions. In this step, the immobilization of anti-
E. coli and anti-HSA antibodies is obtained, with an increase in signal of near 2300 RU and 2100 RU, respectively. Loading the antibodies at a concentration of 50 µg mL
−1 ensured the stable immobilization of the bioreceptors, with a similar surface coverage observed for both antibodies. After deactivation and BSA blocking of the surface (data not shown), the biosensor was washed in PBS and the resulting SPR baseline remained stable for further measurements.
Once the SPR biochips were functionalized with the antibodies, the detection specificity was assessed. The different channels were exposed to
E. coli suspensions at increasing concentration.
Figure 3a shows the SPR sensograms obtained during incubation of the bacteria onto anti-
E. coli and anti-has antibodies functionalized surfaces.
Figure 3b compares the SPR signal for each bacteria concentration, registered after each PBS washing step. The observed SPR curves demonstrated specific recognition of
E. coli by the corresponding antibodies, with a signal increase proportional to the concentration of bacteria loaded on the channel. The response from the anti-HSA functionalized channel was found to be substantially lower when compared to the specific antibody. Interestingly, we observed a signal drop at the end of each injection, which was probably due to shear forces exercised on bacteria while washing in PBS. Nevertheless, the SPR signal remained still much higher for anti-
E. coli than for the negative control (anti-HSA) as can be seen in
Figure 3b. The described signal drop was certainly due to the particular behavior of bacterial cells in the confinement of SPR instrument microfluidic system. Indeed, the dimensions of
E.coli were comparable to the tiny diameter of microfluidic channels available in Biacore T200 system. The bacteria, which are around 2 µm of diameter, were therefore subjected to shear stress, especially upon modification of the flow conditions (PBS washing after injection), leading to their desorption from the surface. Furthermore, the SPR responses to bacteria might not appear proportional but it is related to method limitation. SPR measurement enables the detection of changes in the refractive indexes at a distance of up to 150 nm from the sensing surface while the size of bacteria largely exceeds this scale, so the SPR signal results principally from the lower part of the bacterial wall in contact with the sensing surface. Moreover, the limited penetration of bacteria by the electromagnetic field and the similarity in refractive index between the bacterial cytoplasm and the aqueous medium is a drawback for the sensitivity of SPR in this application [
13]. As such, SPR is probably not an optimal method for bacteria quantification. Nevertheless, the described SPR tests confirmed the correct immobilization of the antibodies onto the biochip surface and the subsequent specific recognition of
E. coli bacteria. The SPR experimental conditions for the antibody immobilization were applied for the development of the biosensing platform using in DF detection methods.
3.3. Dark Field Data Treatment
The detection of bacteria immobilized on the surface of the sensor was performed by dark field microscopy. Images were recorded after 75 min of sample incubation and 30 min of rinsing to remove loosely bound bacteria. ImageJ software was used to analyze the images and count the bound bacteria.
The average area and circularity distribution of the
E. coli bacteria was determined by using all bacteria detected by ImageJ without geometric restrictions (
Figure 1c is an example of the original microscopic image). These characteristics were further used to screen and quantify the amount of
E. coli bacteria in a field sample.
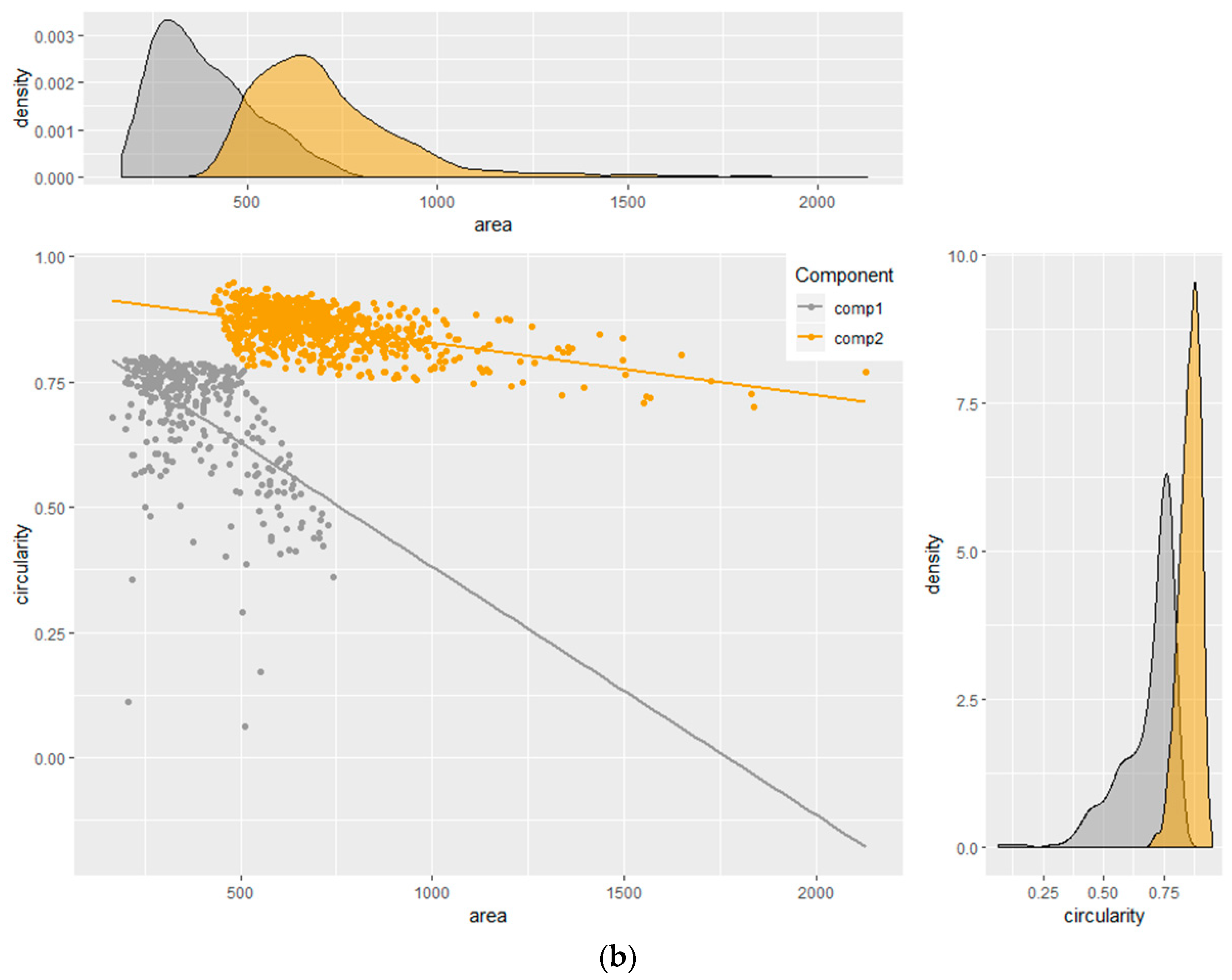
Figure 4 illustrates the measurements of area versus the circularity of a total sample of
2459
E. coli bacteria obtained from five independent experiments. The bacteria measurements were analyzed using mixtures of regressions models, which then enabled us to discriminate the shape and consider its binding orientation to the functionalized surface. Mixture regression models are applied when there are several mixed groups in the population that the sample represents. In this case, the
E. coli population on the surface could assume several shapes versus orientation such as a round-shape and rod like shape. Round shapes can be observed when single bacteria assume an up-right position or several bacteria are associated together. In both scenarios, a circularity value close to 1 was measured. Furthermore, rod like shapes can be observed if a single bacteria is side-on to the surface (circularity lower than 1) or assume any intermediate position (circularity between 0 and 1). By modeling the conditional distribution of the response given for the covariate as a mixture, the sample can be clustered into groups and the regression models for the groups can be estimated at the same time. Mixtures of linear regression models were first studied in the econometrics literature and were introduced by Quandt [
14] and Quandt and Ramsey [
15] as the “switching regression” problem. There are several applications of mixture distributions in agriculture, biology, economics, medicine, and genetics (see e.g., [
16] and [
17] for reviews).
Figure 4a illustrates the presence of two, although not well separated, clusters in the sample. In the first instance, the measurements of circularity were modeled as the response of the area covariates. Suppose that we have
n independent observations of circularity,
, each with a corresponding predictor measurement of area,
. Furthermore, each observation (
belongs to either one of two main groups of
E. coli bacteria distribution. Conditional on the membership in the
jth group (component), the relationship between
and
is the normal regression model and the mixture of regression models is given by:
where the
are independent,
1, 2,
1, …,
n.
The problem is to estimate the seven parameters that specify the proportion of points to each cluster (belonging to each regression line), the slope and the intercept of each line, and the variance around each line. The parameter vector is denoted as
and accounting for the mixture structure, conditional on
,
has the density
where
is the normal density with a mean of
and variance of
,
1, 2. We assume that
are such that
is positive definite. The expectation maximization (EM) algorithm, originally introduced by Dempster, Laird, and Rubin [
18], has been used to compute the maximum likelihood estimators (MLE). The EM estimates are iteratively reweighted MLE with weights equal to the current conditional probability of the component membership. Second, the measurements of the area were modeled as the response of circularity with a mixture of regression models. The two mixtures of regression models were subsequently matched (i.e., when the calculated posterior probabilities of belonging to one component coincide). Through the matching, the clusters were further separated into two main components, which represented the average area and the circularity of
E. coli bacteria. Specifically, the clusters could be separated in Component 1 and Component 2, and the estimated parameter vector was
= (0.17, 0.56, 0.78, 0.00012, 0.00010, 0.14
2, 0.05
2). The results of the average area ±σ and average circularity ±σ of
E. coli bacteria are listed in
Table 1 and represented in
Figure 4. Data analysis was carried out in the R environment for statistical computing and visualization [
19] using the following libraries: dplyr [
20], mixtools [
21], and mixture [
22].
Bacteria immobilized on the surface can be divided in two populations as a function of their arrangement and orientation. The first population, represented by Component 2, is characterized by a circularity value close to 1 (i.e., 0.86) and the largest area, representing most objects grouping two or more bacteria. The second population (Component 1), characterized by a lower circularity (<1) and an area lower than Component 2, might indicate single E. coli bacteria with side-on or intermediate orientations onto the functionalized surface. Therefore, Component 1 seems to statistically represent the average area and circularity of single E. coli bacteria. In further experiments, Component 2 was not used for the statistical analysis of E. coli in the field water sample because of the lack of specific information regarding the average area and circularity of single bacteria.
In order to assess the reliability of the described counting method, Component 1 values were applied and compared to manual counting using an antibody functionalized surface exposed to a known concentration of cultured
E. coli bacteria. The analysis of known samples suggests that using Component 1 values with one standard deviation (area 386 ± 127 and circularity 0.68 ± 0.12) is very restrictive. Indeed, for a field of view of 400 × 400 μm
2 and incubation time of 75 min, 259 bacteria were manually counted for a known concentration of 4 × 10
6 cells mL
−1. The application of the statistical parameters (area 386 ± 127 and circularity 0.68 ± 0.12) generated 44 counts of bacteria, which represented only 17% of the total count of bacteria. By using Component 1 values with 1.5 and 2 standard deviations, the number of detected bacteria increased to 74 (28% of the total count of bacteria) and 214 bacteria (82% of the total count of bacteria), respectively. However, the 2σ results overlapped between Components 1 and 2 (
Figure 4), so were not retained as suitable for counting single bacteria population. Therefore, the counting of bacteria on the functionalized surface was carried out by considering 1.5σ as the statistical error.
3.5. Biosensor Calibration
After optimization of the assay, the detection limit of the system was determined by spiking cultured E. coli bacteria in PBS solution. The flow cell was filled with 30 μL of PBS solution with an increasing concentration of bacteria (from 104 to 107 cells mL−1). The experiments were performed in triplicate. The bacteria solutions were flowed over the sensor surface and left to sediment toward the surface for 75 min. The surface was then washed with PBS for 20 min and the number of bacteria immobilized on the surface was counted by ImageJ after DF image recording. The field of view used for the counting was 400 × 400 μm2.
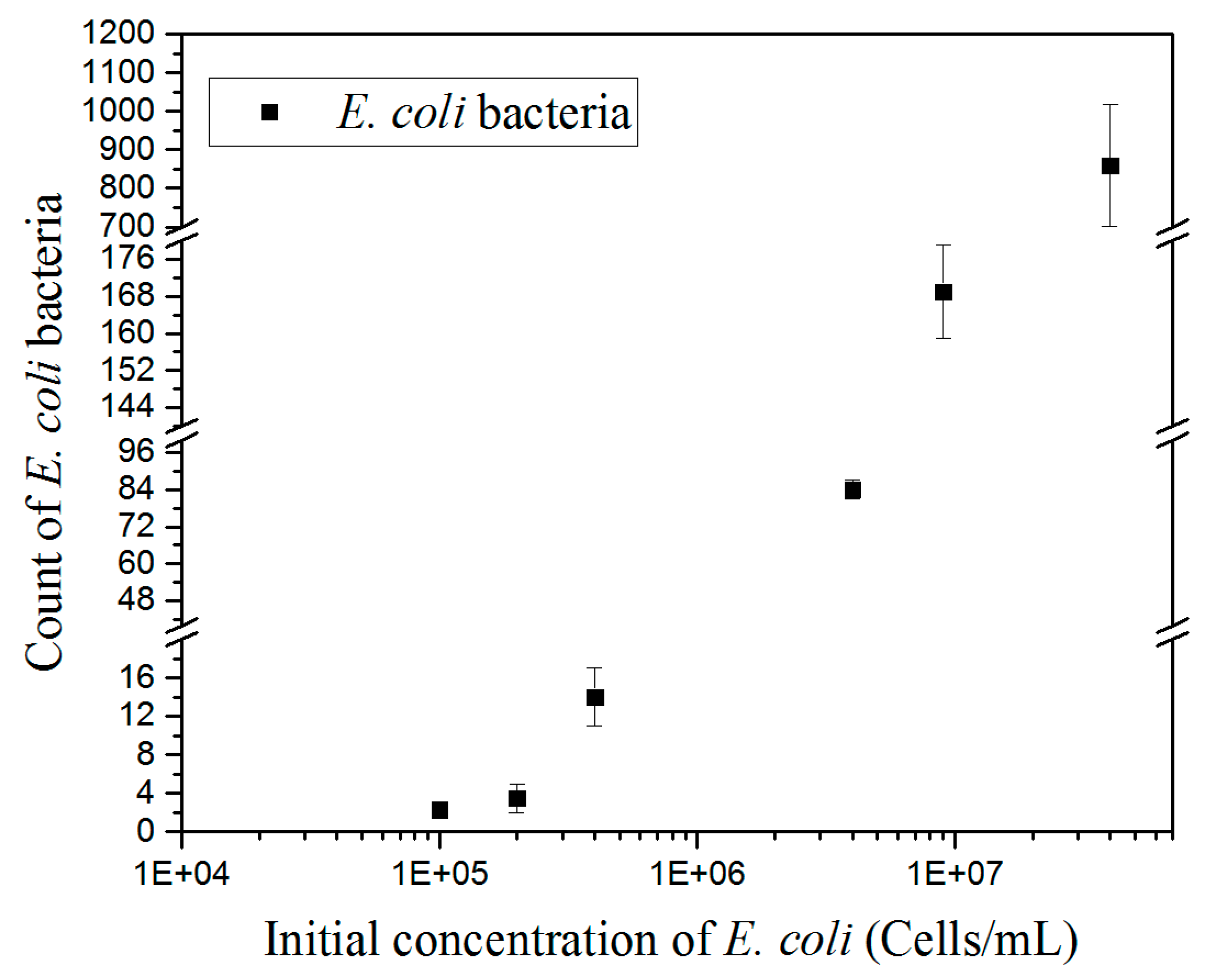
The counting results of immobilized bacteria at different concentrations are shown in
Figure 6. Increasing concentrations of
E. coli generated a proportional increase of the number of bacteria immobilized on the surface. Quantitative detection of bacteria was carried out by considering a threshold of at least five bacteria for ensuring reliable counting [
7], which is defined here as the detection limit. This reached a concentration of 2.10
5 cells mL
−1. Initial concentrations of
E. coli bacteria of 4 × 10
5, 4 × 10
6, and 4 × 10
7 cells mL
−1 generated a number of immobilized
E. coli bacteria of 15, 80, and 800, respectively, within an error of 4% and 18% for the highest concentration. The calibration curve could be fitted with a linear regression with R
2 of 0.9. Nevertheless, increasing the DF sampling area from 0.16 mm
2 to 1 mm
2 would improve the quantitative detection of bacteria by a factor of 5–6 and reach a detection limit in the order of 5 × 10
4 cells mL
−1, which are values comparable to other conventional methods.
3.6. Application to Field Samples and Comparison of Techniques
The presently developed method was compared to two conventional methods, namely Colilert-18 and qPCR. The Colilert-18 method is an ISO standard (ISO 9308-2:2012) for the detection of total coliforms and
E. coli. It has been demonstrated that the results are equivalent to traditional quantification methods [
23] and it is included in the Drinking Water Directive [
24]. The Colilert-18 method is very simple and does not require specific expertise and the results are obtained within 18 to 22 h. Quantitative PCR is another technique that is widely used for the quantification of known organisms. This technique has the potential to detect very low copy numbers of target genes. For quantification, a specific primer set is needed, which solely detects the region of interest of the target gene. Quantification may be done based on a non-specific dsDNA intercalating dye (SYBR Green) or by using a probe containing reporter and quencher molecules. The fact that different species, and even strains, may contain a different number of copies of a single gene impacts the extrapolation of the cell number based on qPCR analysis (which reports data as a gene copy number). For instance, it is of general agreement that
E. coli has a total of seven copies of the 16S rRNA gene, and therefore the cell quantification must take that into account. Moreover, the maintenance of extracellular DNA in field samples is a fact and may impact the quantification of the total cell number if it is not taken into consideration. Pre-digestion of extracellular DNA before cell disruption and successively intracellular-DNA extraction reduces the impact of non-viable cells/DNA on the quantification analysis. Other methods can be used to this end such as the relative quantification of the extracted RNA, however, this would increase the length of the analysis. The DNA extraction process may vary depending on the protocol used. Nevertheless, the correlation between qPCR and culturing methods for the detection of coliforms and
E. coli is known [
25]. In order to assess the robustness of the DF based bioassay, experiments were performed by using two water samples from the wastewater treatment plant (Samples A and B), which were subsequently filtered through a 45 µm metallic sieve and 5 µm (Sample A) and 20 µm (Sample B) filter membranes. In the field water sample, the expected level of
E. coli was estimated in the order of 10
4 cells mL
−1. Since our assay works in the order of 10
5 cells mL
−1 (i.e., the range of the calibration curve was from 5 × 10
4 to 5 × 10
7 cells mL
−1), the field water samples had to be concentrated. DF images of the bacteria bound to the surface were captured and processed using ImageJ software (
Figure 7). The screening of
E. coli bacteria was performed by using the Component 1 parameters with ±1.5σ. These parameters were previously discussed in the dark field data analysis treatment paragraph. The count of
E. coli bacteria was then converted in cells mL
−1 by using the calibration curve seen in
Figure 6.
3.7. Comparison of Techniques
Table 2 and
Figure 8 show the results obtained by the different methods and the method variations used in this work.
Overall, the results were consistent and comparable among the techniques. The results obtained specifically for E. coli by the Colilert-18 method exhibited the lowest quantification in both samples when compared to the other methods. This effect can be explained by qPCR and biosensor technique pitfalls in term of specificity. Indeed, the results obtained with these methods were close to the ones obtained with the Colilert-18 method for the total coliforms by relying on the monitoring of a color-based chemical transformation (i.e., the metabolic conversion of o-nitrophenyl into o-nitrophenol by the β-galactosidase enzyme), which detected the presence of all viable cells in the sample. We could then assume that the results of all techniques considered not only the E. coli strain detected by Colilert-18, but also other coliforms. The results of the Colilert-18 tests were considered as a reference since it is the most accepted standard.
Over-estimation observed from the DF measurements, in addition to the other coliforms, could be attributed to the detection of non-viable cells (which will not grow in Colilert-18 media, and therefore could not be detected). Furthermore, the attachment may also be partially impaired by the organic load present in wastewater samples, which may affect the specific interaction with the antibody.
Similarly, qPCR analysis also showed an over-estimation due to a lack of specificity. To investigate the reason, we used BLASTN 2.8.0+ software [
26] (NCBI) to analyze the qPCR target sequence
CCCTTACGACCAGGGCTACACA
CGTGCTACAATGGCGCAT
ACAAAGAGAAGCGACCTCGC, which was found by overlapping the primers/probe pair (primers underlined and probe in italic) with the complete genome of
E. coli O157:H7 str. Sakai DNA (NCBI code >gi|47118301 |dbj|BA000007.2). BLAST index results [
27] showed a high similarity with other bacteria species (even full identity) (data not shown). The lack of specificity of the primer set used in this study may explain the over-estimation of
E. coli by qPCR in Sample B and the non-digested samples. On the other hand, the contaminants expected on a DNA extract of the WWTP samples may interfere with the PCR reaction. Inhibition of the enzyme was expected, however, the increase may somehow be an impact of the contaminants, which was not further evaluated.
The observed over-quantification was roughly 10 times higher for qPCR and the biosensor when compared to the Colilert-18 test for E. coli. Considering the Colilert-18 test results for coliforms, all methods performed similarly, which supports the theory that the proposed method may unspecifically recognize other coliforms due to low specificity of the polyclonal antibodies, unspecific recognition due to the presence of organic compounds or other contaminants, or other interactions that were not explored.