Non-Local Based Denoising Framework for In Vivo Contrast-Free Ultrasound Microvessel Imaging
Abstract
1. Introduction
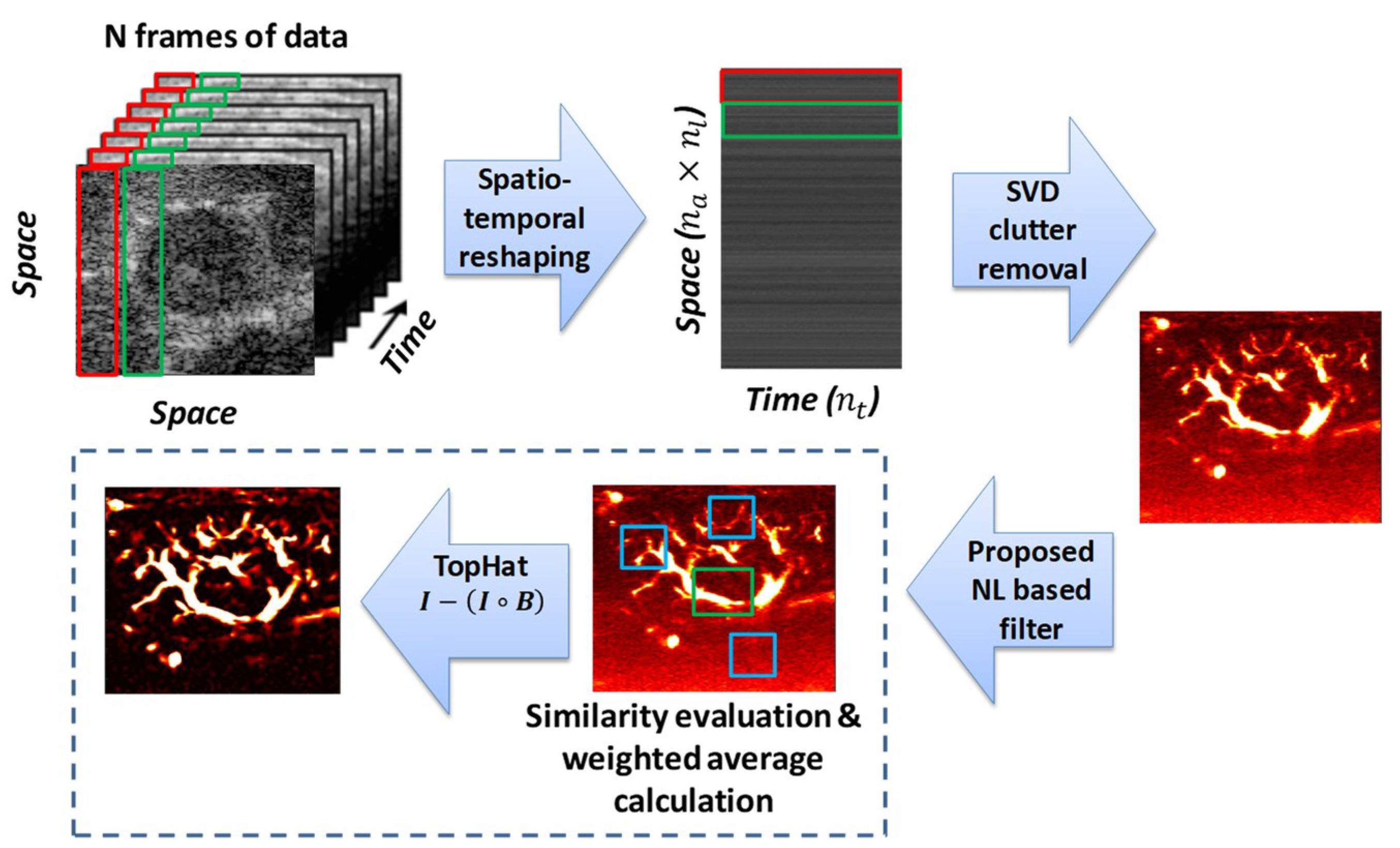
2. Materials and Methods
2.1. SVD Filtering and Clutter Rank Selection
2.2. Background Removal
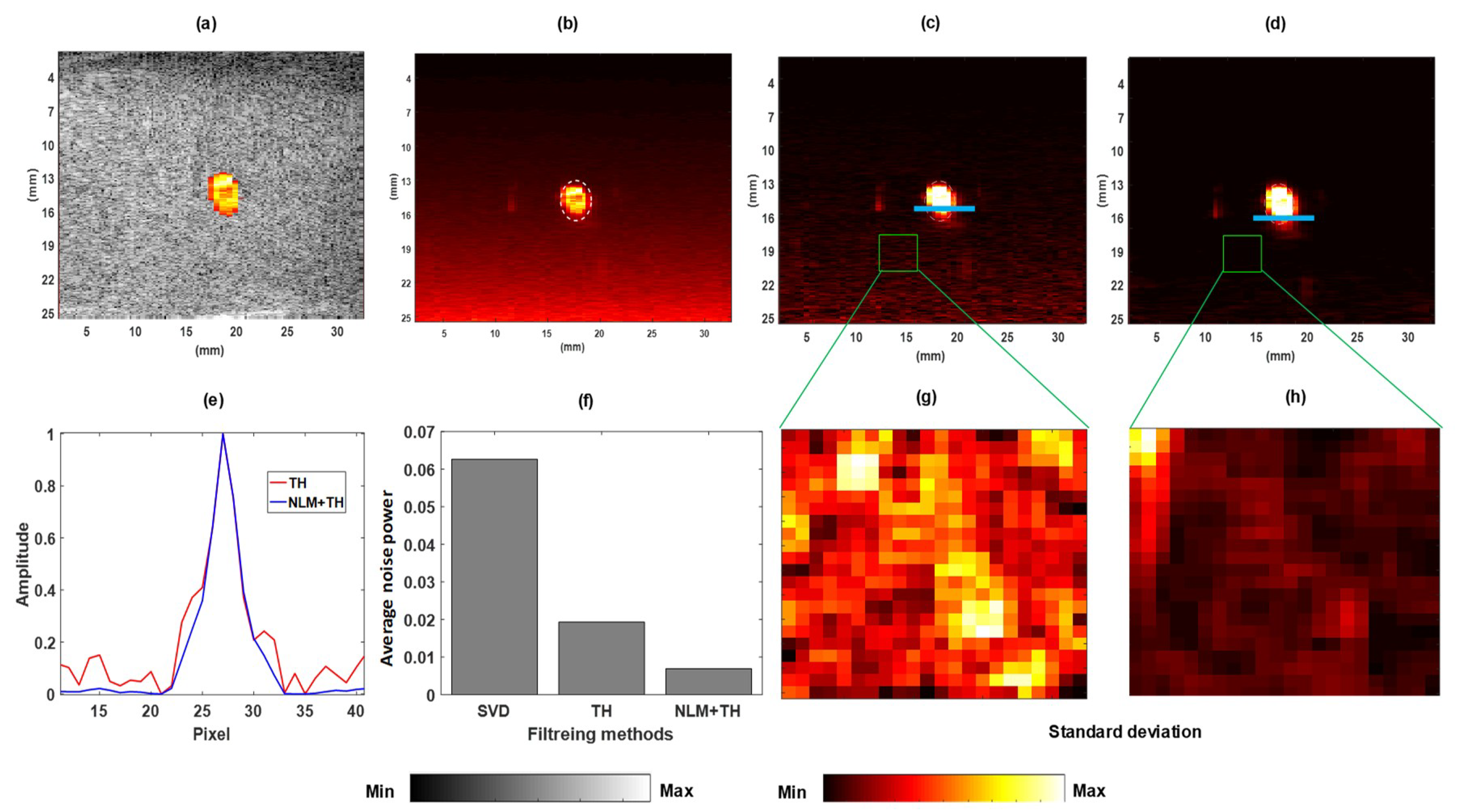
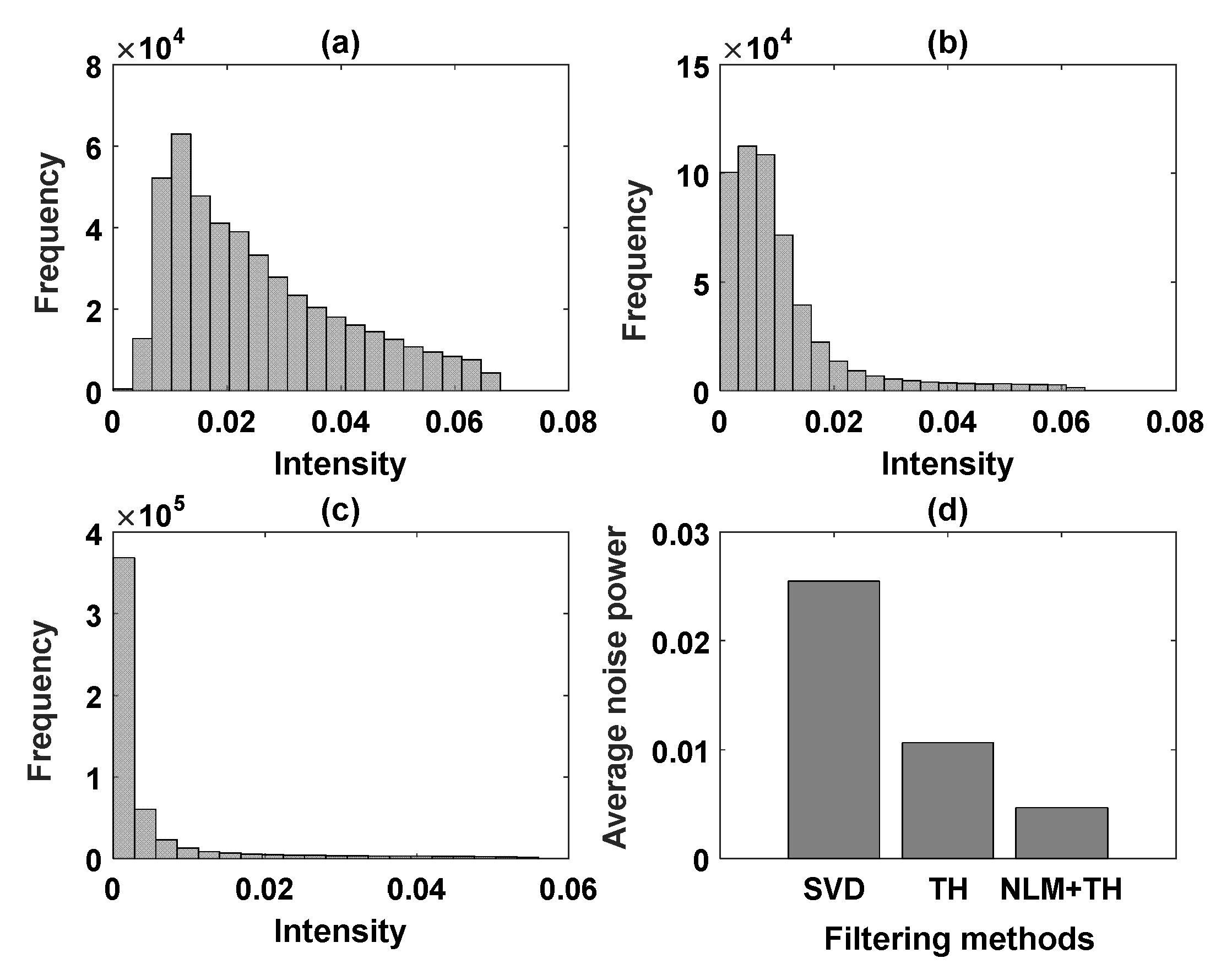
2.3. Quantitative Assessment Measures
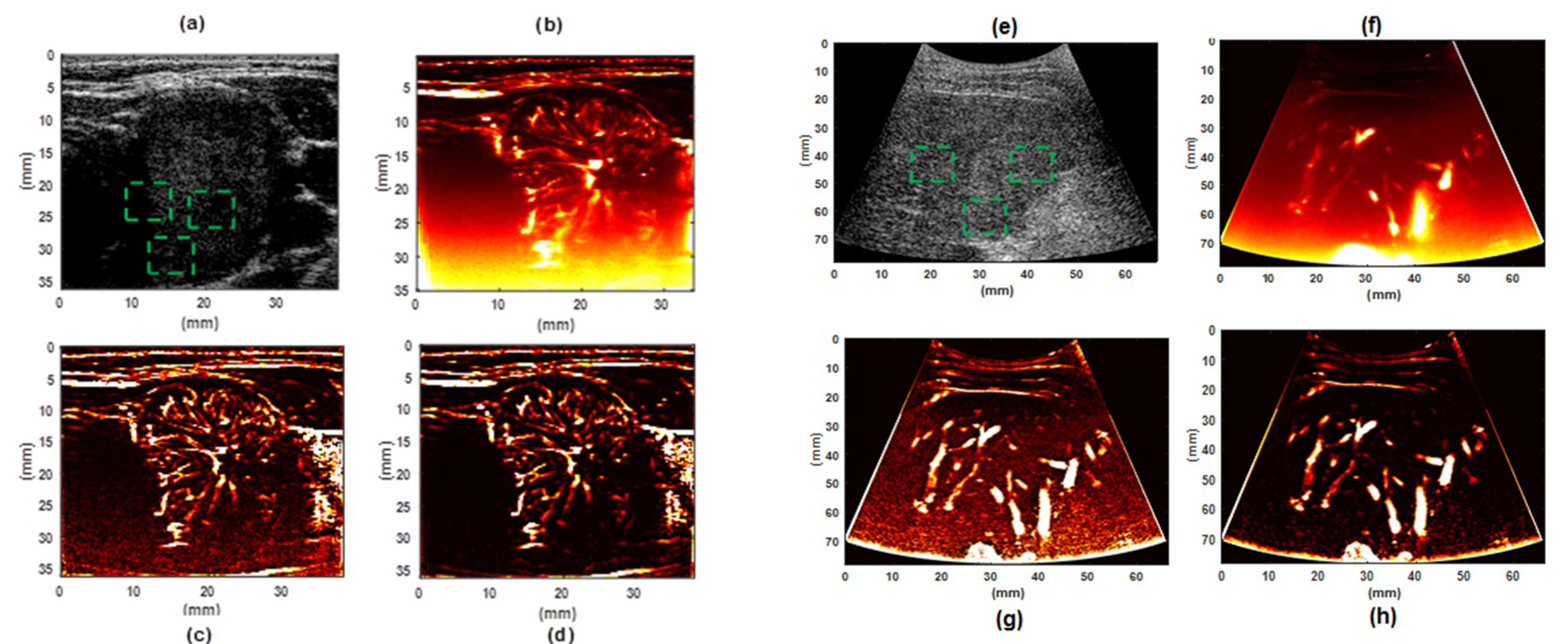
3. Results
3.1. Validation of Flow Phantom Data
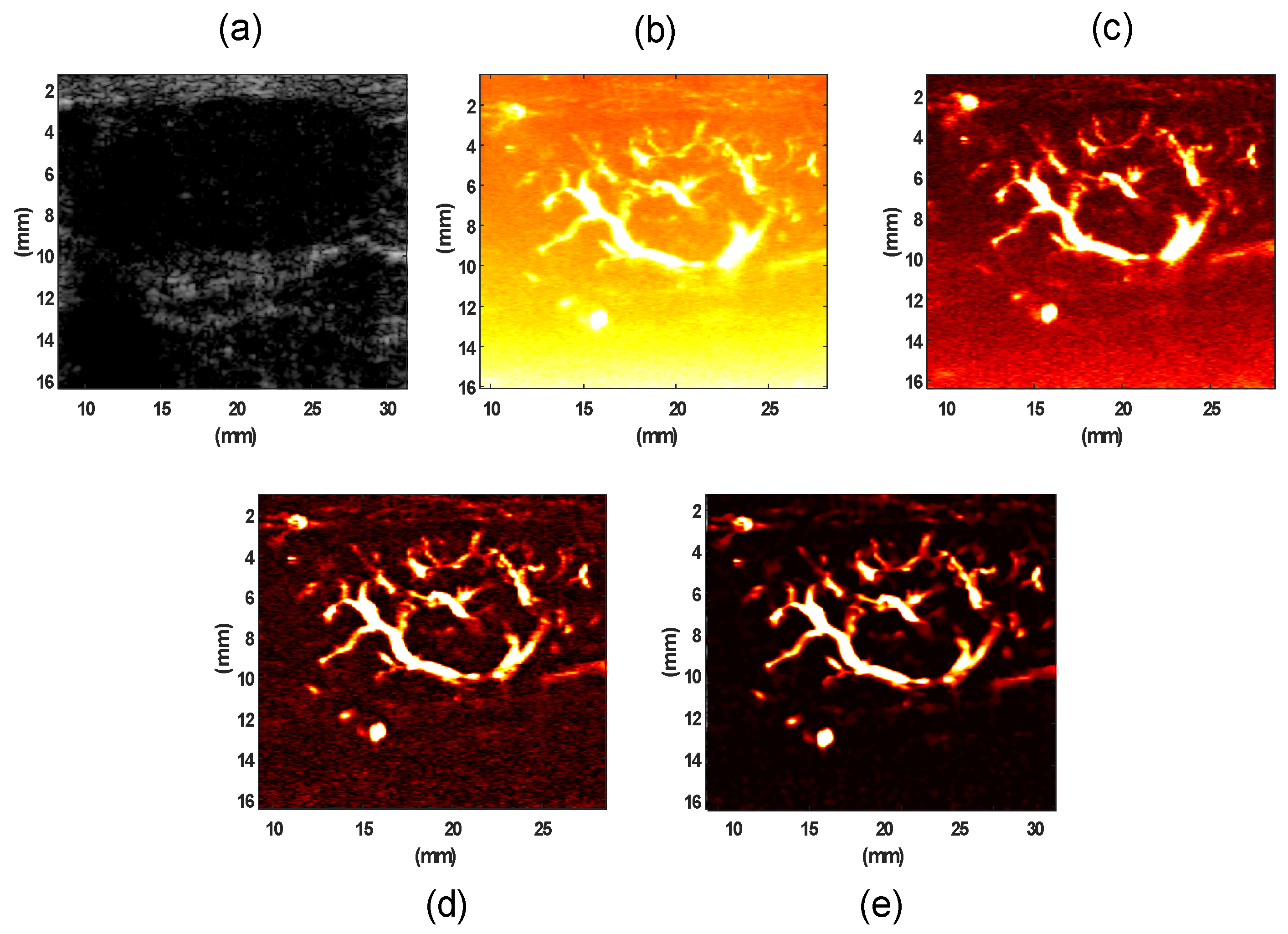
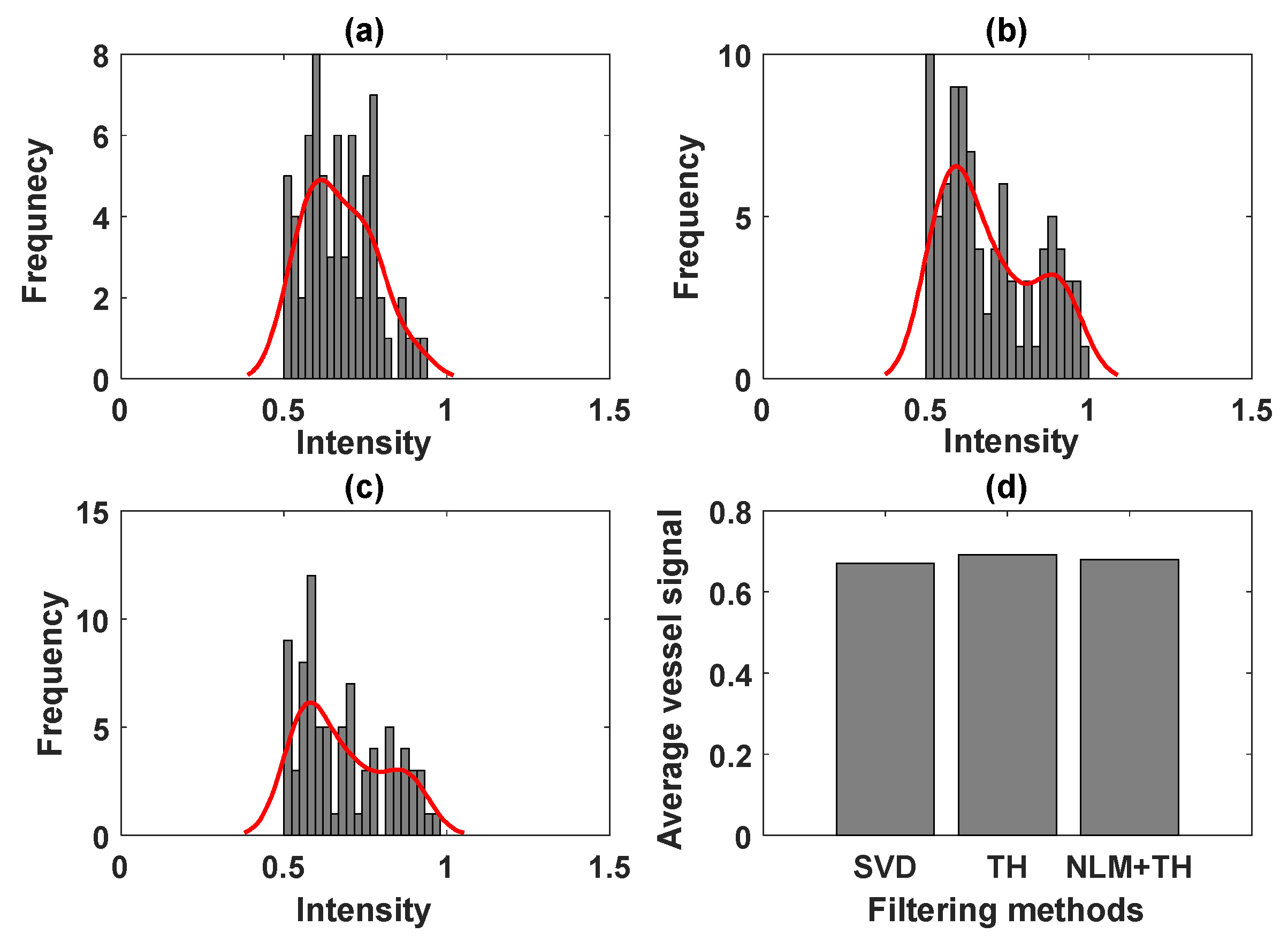
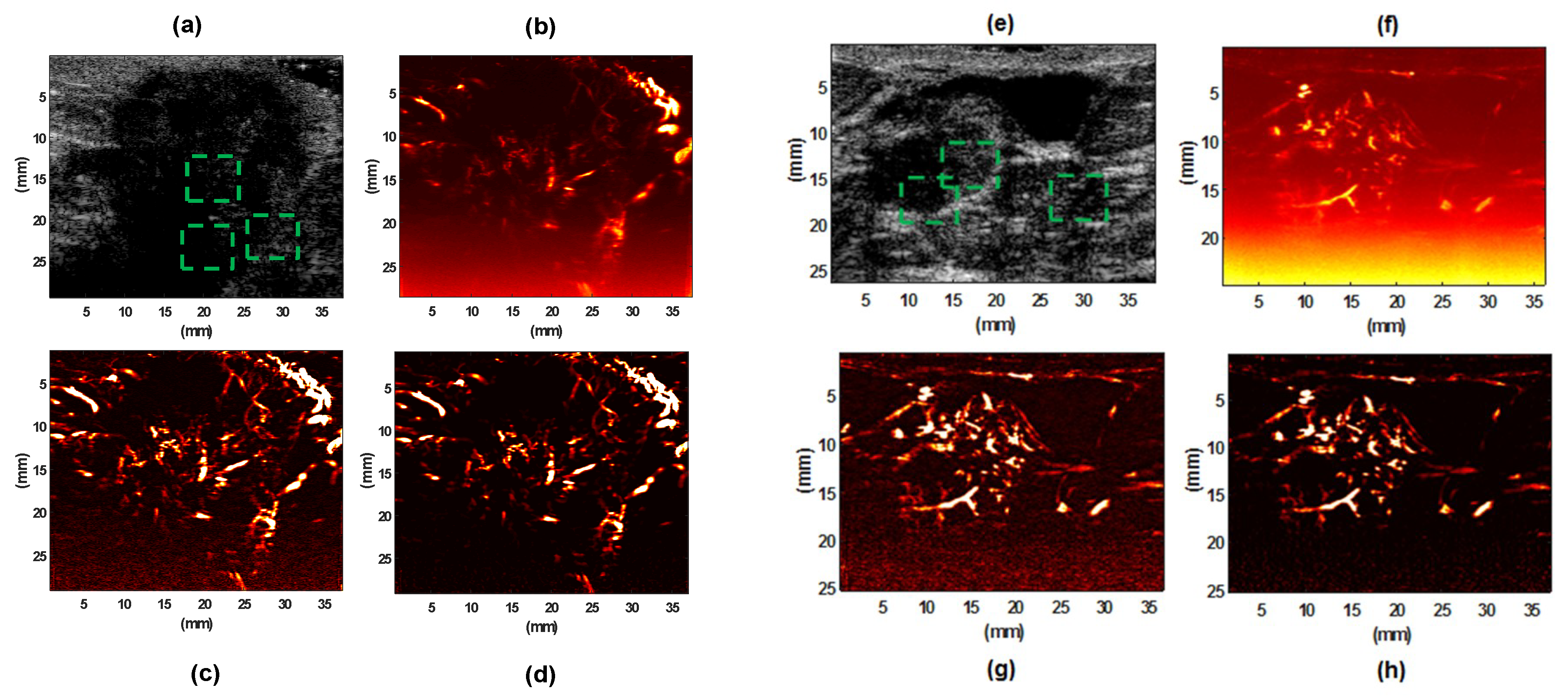
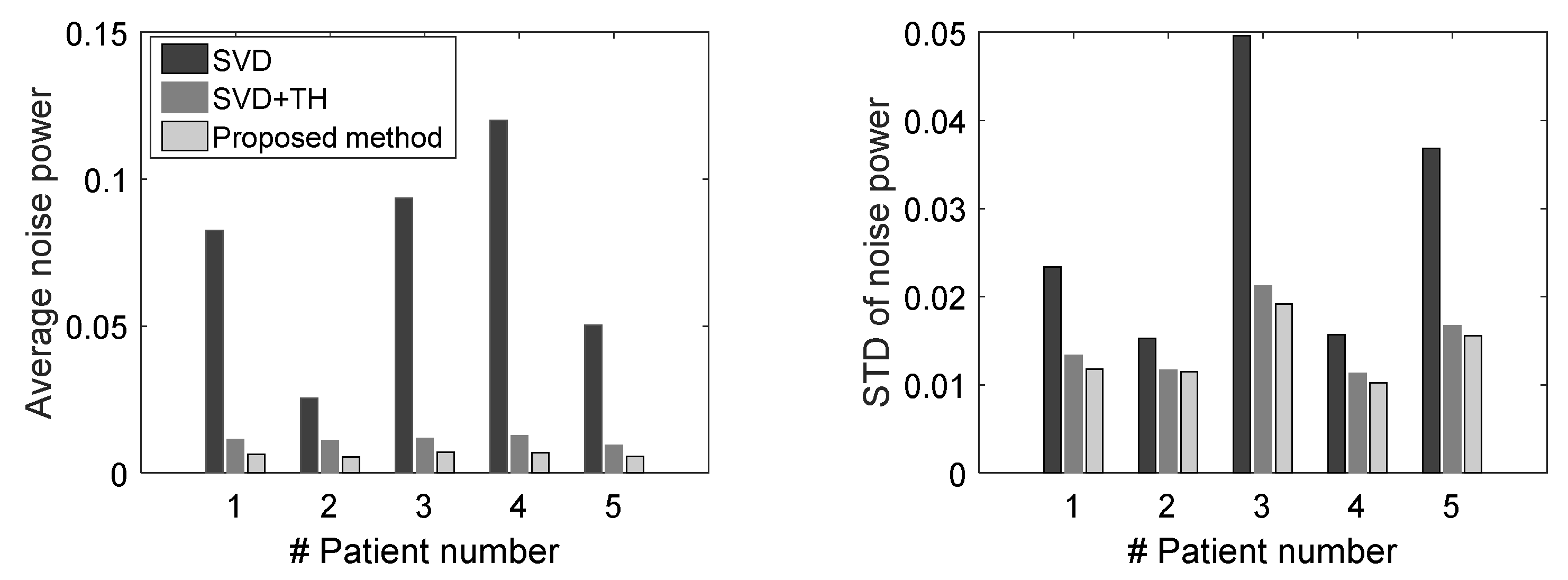
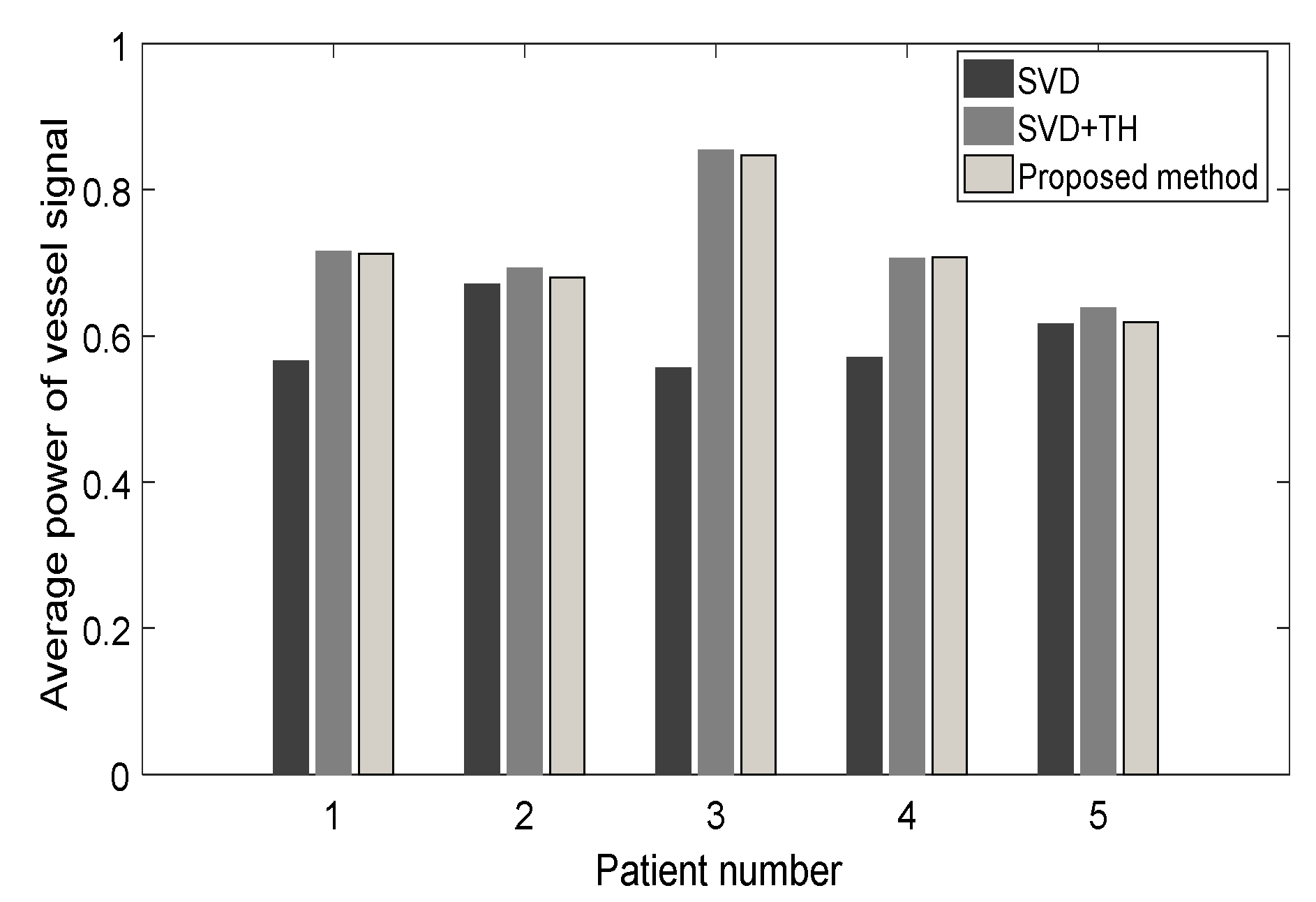
3.2. Evaluation Performance on In Vivo Clinical Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fukumura, D.; Duda, D.G.; Munn, L.L.; Jain, R.K. Tumor microvasculature and microenvironment: Novel insights through intravital imaging in pre-clinical models. Microcirculation 2010, 17, 206–225. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Dromain, C.; Balleyguier, C. Contrast-enhanced digital mammography. In Digital Mammography; Springer: Berlin, Germany, 2010; pp. 187–198. [Google Scholar]
- Heywang-Köbrunner, S.H.; Katzberg, R.W. Contrast-enhanced magnetic resonance imaging of the breast. Investig. Radiol. 1994, 29, 94–104. [Google Scholar] [CrossRef]
- Tromberg, B.J.; Pogue, B.W.; Paulsen, K.D.; Yodh, A.G.; Boas, D.A.; Cerussi, A.E. Assessing the future of diffuse optical imaging technologies for breast cancer management. Med. Phys. 2008, 35, 2443–2451. [Google Scholar] [CrossRef] [PubMed]
- Kinsella, J.M.; Jimenez, R.E.; Karmali, P.P.; Rush, A.M.; Kotamraju, V.R.; Gianneschi, N.C.; Ruoslahti, E.; Stupack, D.; Sailor, M.J. X-ray computed tomography imaging of breast cancer by using targeted peptide-labeled bismuth sulfide nanoparticles. Angew. Chem. Int. Ed. 2011, 50, 12308–12311. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, M.; Ku, G.; Li, C.; Wang, L.V. Design and evaluation of a novel breast cancer detection system combining both thermoacoustic (TA) and photoacoustic (PA) tomography. Med. Phys. 2008, 35, 2218–2223. [Google Scholar] [CrossRef]
- Zhao, H.; Xu, R.; Ouyang, Q.; Chen, L.; Dong, B.; Huihua, Y. Contrast-enhanced ultrasound is helpful in the differentiation of malignant and benign breast lesions. Eur. J. Radiol. 2010, 73, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Renaud, G.; Bosch, J.; Ten Kate, G.; Shamdasani, V.; Entrekin, R.; de Jong, N.; Van der Steen, A. Counter-propagating wave interaction for contrast-enhanced ultrasound imaging. Phys. Med. Biol. 2012, 57, L9. [Google Scholar] [CrossRef]
- Yildiz, Y.O.; Eckersley, R.J.; Senior, R.; Lim, A.K.; Cosgrove, D.; Tang, M.-X. Correction of non-linear propagation artifact in contrast-enhanced ultrasound imaging of carotid arteries: Methods and in vitro evaluation. Ultrasound Med. Biol. 2015, 41, 1938–1947. [Google Scholar] [CrossRef]
- Demi, L.; Van Sloun, R.J.; Wijkstra, H.; Mischi, M. Cumulative phase delay imaging for contrast-enhanced ultrasound tomography. Phys. Med. Biol. 2015, 60, L23. [Google Scholar] [CrossRef]
- Tanter, M.; Fink, M. Ultrafast imaging in biomedical ultrasound. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2014, 61, 102–119. [Google Scholar] [CrossRef] [PubMed]
- Bjaerum, S.; Torp, H.; Kristoffersen, K. Clutter filter design for ultrasound color flow imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2002, 49, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Kadi, A.P.; Loupas, T. On the performance of regression and step-initialized IIR clutter filters for color Doppler systems in diagnostic medical ultrasound. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 1995, 42, 927–937. [Google Scholar] [CrossRef]
- Gallippi, C.M.; Trahey, G.E. Adaptive clutter filtering via blind source separation for two-dimensional ultrasonic blood velocity measurement. Ultrason. Imaging 2002, 24, 193–214. [Google Scholar] [CrossRef] [PubMed]
- Demené, C.; Deffieux, T.; Pernot, M.; Osmanski, B.-F.; Biran, V.; Gennisson, J.-L.; Sieu, L.-A.; Bergel, A.; Franqui, S.; Correas, J.-M. Spatiotemporal clutter filtering of ultrafast ultrasound data highly increases Doppler and fUltrasound sensitivity. IEEE Trans. Med. Imaging 2015, 34, 2271–2285. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.C.; Lovstakken, L. Eigen-based clutter filter design for ultrasound color flow imaging: A review. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2010, 57, 1096–1111. [Google Scholar] [CrossRef]
- Alberti, G.S.; Ammari, H.; Romero, F.; Wintz, T. Mathematical analysis of ultrafast ultrasound imaging. SIAM J. Appl. Math. 2017, 77, 1–25. [Google Scholar] [CrossRef]
- Song, P.; Manduca, A.; Trzasko, J.D.; Chen, S. Ultrasound small vessel imaging with block-wise adaptive local clutter filtering. IEEE Trans. Med. Imaging 2017, 36, 251–262. [Google Scholar] [CrossRef]
- Bayat, M.; Fatemi, M.; Alizad, A. Background Removal and Vessel Filtering of Non-Contrast Ultrasound Images of Microvasculature. IEEE Trans. Biomed. Eng. 2018. [Google Scholar] [CrossRef]
- Song, P.; Manduca, A.; Trzasko, J.D.; Chen, S. Noise Equalization for Ultrafast Plane Wave Microvessel Imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2017, 64, 1776–1781. [Google Scholar] [CrossRef]
- Vincent, L.M.; Masters, B.R. Morphological image processing and network analysis of cornea endothelial cell images. In Proceedings of the Image Algebra and Morphological Image Processing III, San Diego, CA, USA, 20–22 July 1992; pp. 212–227. [Google Scholar]
- Buades, A.; Coll, B.; Morel, J.-M. A non-local algorithm for image denoising. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Diego, CA, USA, 20–25 June 2005; pp. 60–65. [Google Scholar]
- Gilboa, G.; Osher, S. Nonlocal linear image regularization and supervised segmentation. Multiscale Model. Simul. 2007, 6, 595–630. [Google Scholar] [CrossRef]
- Singer, A.; Shkolnisky, Y.; Nadler, B. Diffusion interpretation of nonlocal neighborhood filters for signal denoising. Siam. J. Imaging Sci. 2009, 2, 118–139. [Google Scholar] [CrossRef]
- Coupé, P.; Hellier, P.; Kervrann, C.; Barillot, C. Nonlocal means-based speckle filtering for ultrasound images. IEEE Trans. Image Process. 2009, 18, 2221–2229. [Google Scholar] [CrossRef] [PubMed]
- Coupé, P.; Yger, P.; Prima, S.; Hellier, P.; Kervrann, C.; Barillot, C. An optimized blockwise nonlocal means denoising filter for 3-D magnetic resonance images. IEEE Trans. Med. Imaging 2008, 27, 425–441. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, E.R. An Introduction to Morphological Image Processing (Tutorial Texts in Optical Engineering); DC O’Shea, SPIE Optical Engineering Press: Bellingham, WA, USA, 1992. [Google Scholar]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, L.; Zuo, W.; Zhang, D.; Feng, X. Patch group based nonlocal self-similarity prior learning for image denoising. In Proceedings of the IEEE International Conference on Computer Vision, Washington, DC, USA, 7–13 December 2015; pp. 244–252. [Google Scholar]
- Mahmoudi, M.; Sapiro, G. Fast image and video denoising via nonlocal means of similar neighborhoods. IEEE Signal Process. Lett. 2005, 12, 839–842. [Google Scholar] [CrossRef]
- Ghavami, S.; Bayat, M.; Fatemi, M.; Alizad, A. Quantification of Morphological Features in Non-Contrast Ultrasound Microvasculature Imaging. arXiv, 2018; arXiv:1812.03229. [Google Scholar]
| QAM (dB) | SVD | SVD+TH | Proposed |
|---|---|---|---|
| SNR | 30.80 | 32.21 | 42.07 |
| CNR | 27.23 | 31.12 | 41.52 |
| Breast Lesion# | SVD-Unbiased | SVD+TH | Proposed |
|---|---|---|---|
| 1 | 32.77 | 31.07 | 40.20 |
| 2 | 29.16 | 34.44 | 51.45 |
| 3 | 27.96 | 34.26 | 45.25 |
| 4 | 22.13 | 25.21 | 35.34 |
| 5 | 39.91 | 32.93 | 38.38 |
| Breast Lesion# | SVD-Unbiased | SVD+TH | Proposed |
|---|---|---|---|
| 1 | 27.88 | 30.57 | 38.72 |
| 2 | 27.49 | 32.90 | 42.69 |
| 3 | 32.71 | 31.60 | 42.20 |
| 4 | 23.49 | 29.20 | 38.49 |
| 5 | 29.98 | 33.49 | 41.01 |
| Organ | SVD-Unbiased | SVD+TH | Proposed |
|---|---|---|---|
| Thyroid 1 | 29.39 | 32.41 | 38.79 |
| Thyroid 2 | 34.75 | 40.77 | 45.94 |
| Liver 1 | 33.87 | 38.06 | 45.23 |
| Liver 2 | 20.01 | 25.61 | 31.26 |
| Organ | SVD-Unbiased | SVD+TH | Proposed |
|---|---|---|---|
| Thyroid 1 | 24.39 | 29.74 | 32.70 |
| Thyroid 2 | 31.26 | 38.96 | 42.20 |
| Liver 1 | 28.49 | 37.61 | 45.26 |
| Liver 2 | 20.01 | 25.61 | 31.26 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adabi, S.; Ghavami, S.; Fatemi, M.; Alizad, A. Non-Local Based Denoising Framework for In Vivo Contrast-Free Ultrasound Microvessel Imaging. Sensors 2019, 19, 245. https://doi.org/10.3390/s19020245
Adabi S, Ghavami S, Fatemi M, Alizad A. Non-Local Based Denoising Framework for In Vivo Contrast-Free Ultrasound Microvessel Imaging. Sensors. 2019; 19(2):245. https://doi.org/10.3390/s19020245
Chicago/Turabian StyleAdabi, Saba, Siavash Ghavami, Mostafa Fatemi, and Azra Alizad. 2019. "Non-Local Based Denoising Framework for In Vivo Contrast-Free Ultrasound Microvessel Imaging" Sensors 19, no. 2: 245. https://doi.org/10.3390/s19020245
APA StyleAdabi, S., Ghavami, S., Fatemi, M., & Alizad, A. (2019). Non-Local Based Denoising Framework for In Vivo Contrast-Free Ultrasound Microvessel Imaging. Sensors, 19(2), 245. https://doi.org/10.3390/s19020245





