Development of CdSe–ZnO Flower-Rod Core-Shell Structure Based Photoelectrochemical Biosensor for Detection of Norovirous RNA
Abstract
1. Introduction
2. Experimental
2.1. Materials and Reagents
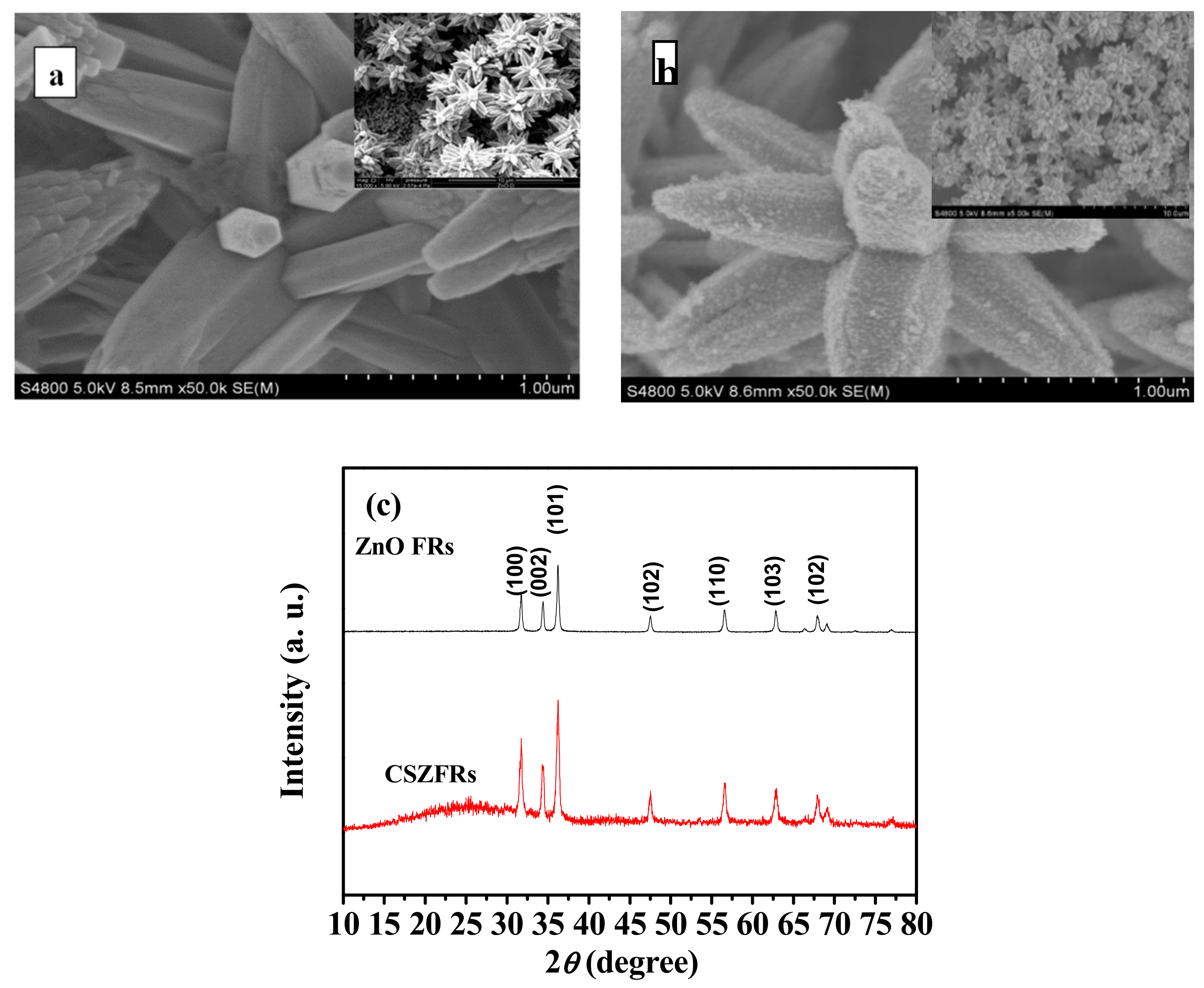
2.2. Preparation of CdSe Sensitized ZnO Flower-Rods
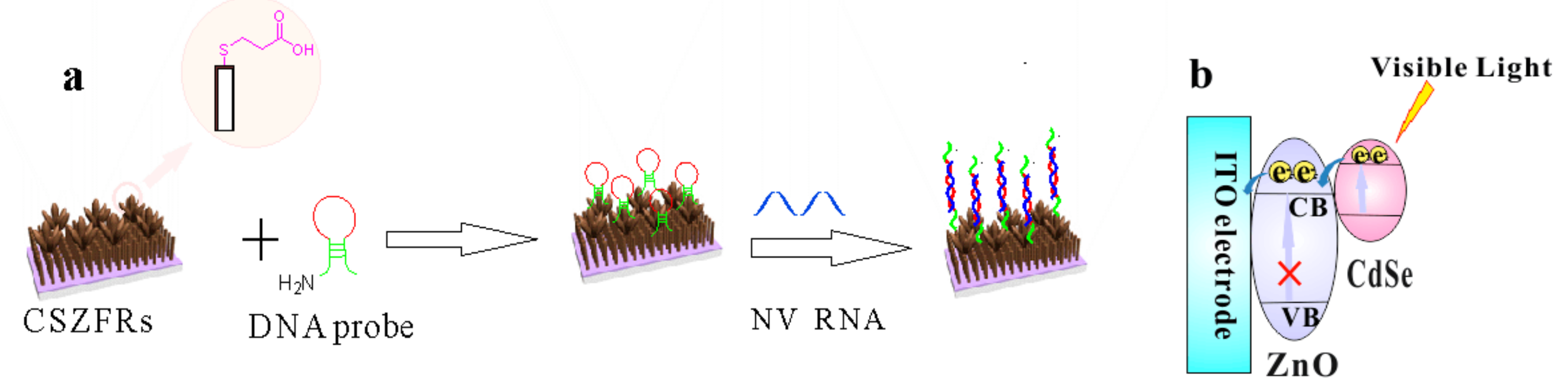
2.3. Construction of CdSe–ZnO Flower-Rods Based PEC Biosensor
2.4. Material Characterizations
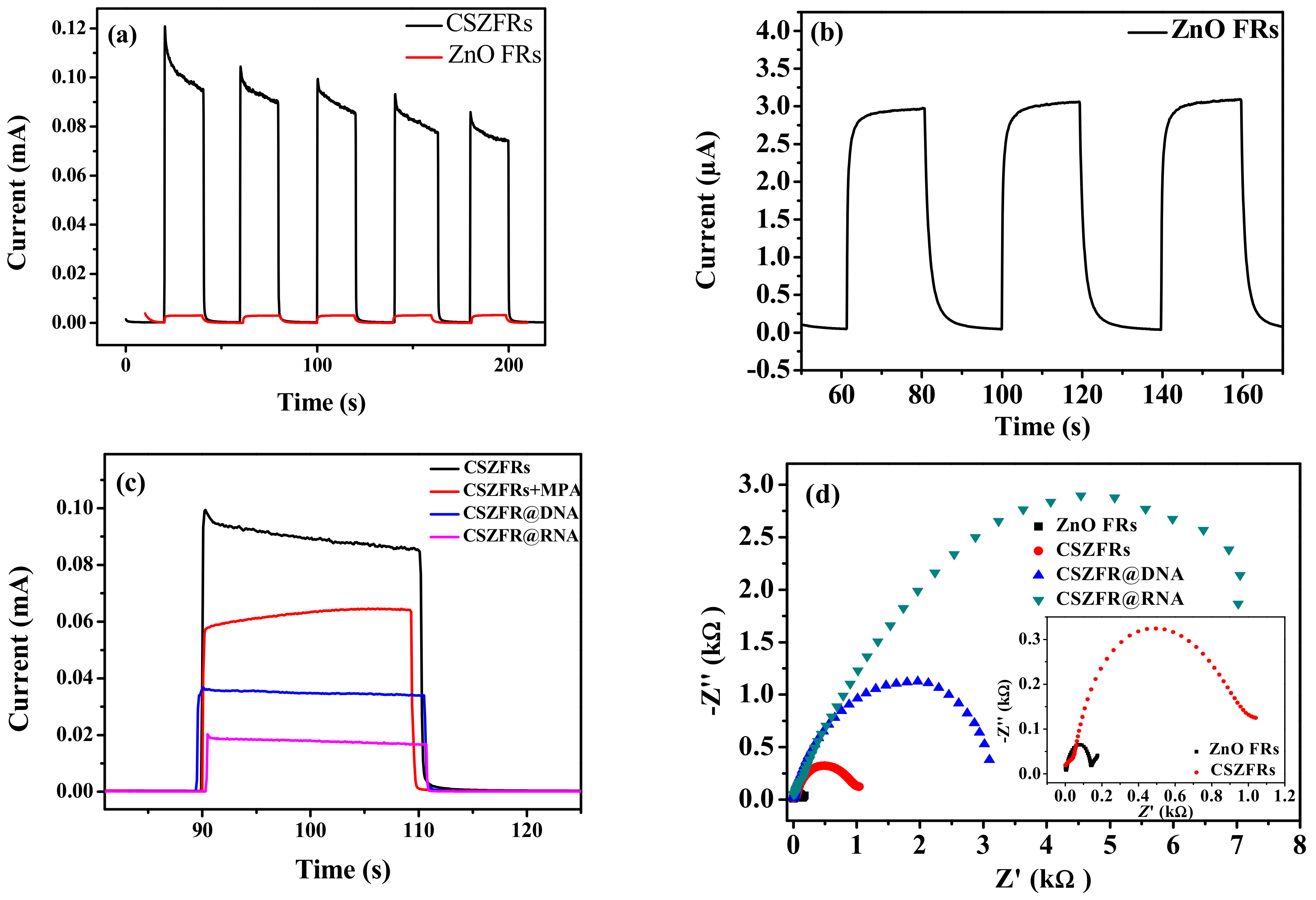
2.5. Photoelectrochemical Measurements
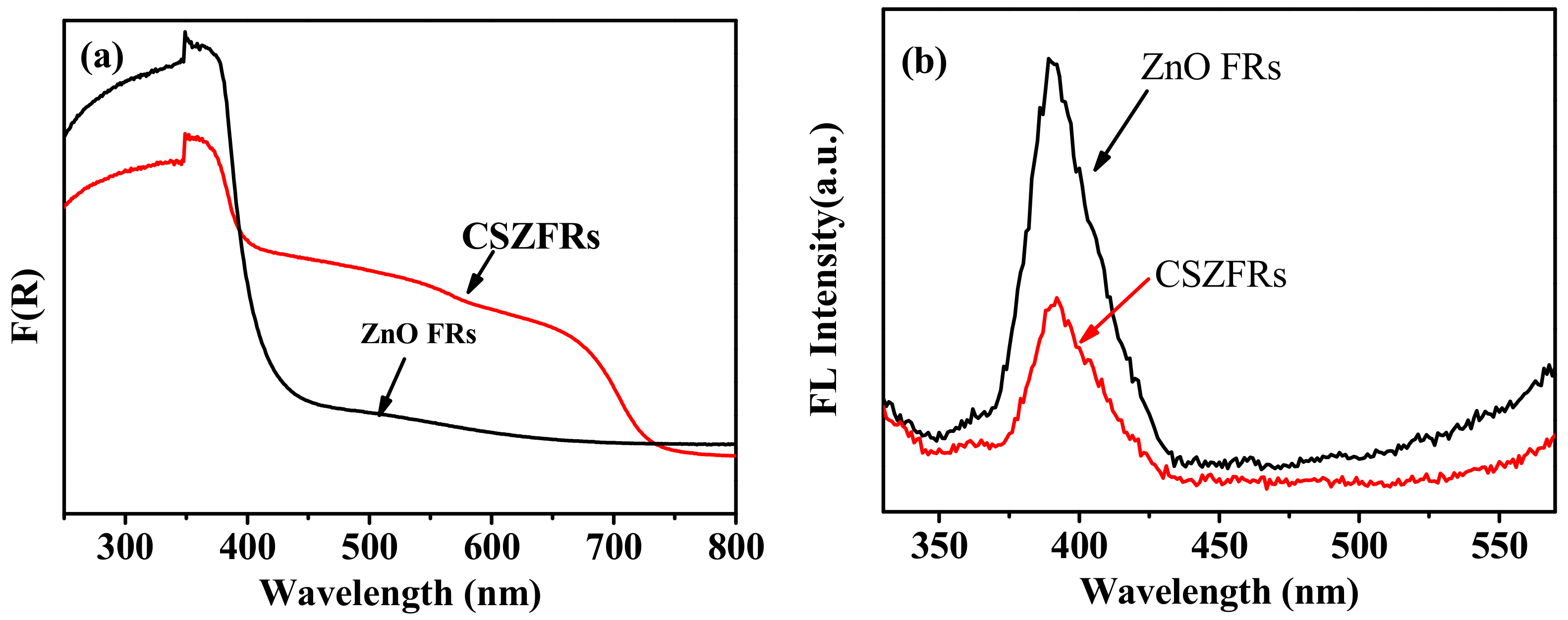
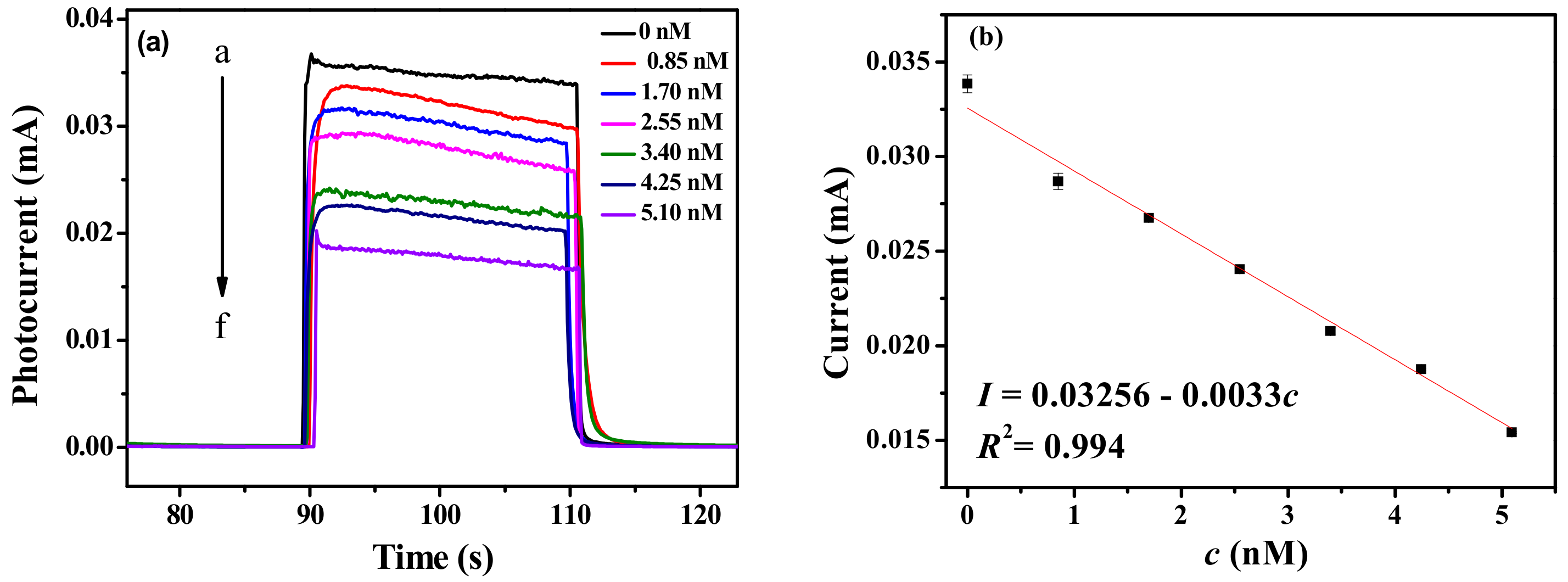
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Patel, M.M.; Widdowson, M.A.; Glass, R.I.; Akazawa, K.; Vinje, J.; Parashar, U.D. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 2008, 14, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.J.; Vinjé, J.; Lopman, B.; Park, G.W.; Yen, C.; Gregoricus, N.; Parashar, U. Updated norovirus outbreak management and disease prevention guidelines. MMWR 2011, 60, 1–15. [Google Scholar] [PubMed]
- Thongprachum, A.; Khamrin, P.; Tran, D.N.; Okitsu, S.; Mizuguchi, M.; Hayakawa, S.; Maneekarn, N.; Ushijima, H. Evaluation and comparison of the efficiency of immunochromatography methods for norovirus detection. Clin. Lab. 2012, 58, 489–493. [Google Scholar] [PubMed]
- Morillo, S.G.; Luchs, A.; Cilli, A.; Ribeiro, C.D.; Calux, S.J.; Carmona, R.C.; Timenersky, M.C. Norovirus 3rd generation kit: An improvement for rapid diagnosis of sporadic gastroenteritis cases and valuable for outbreak detection. J. Virolo. Methods 2011, 173, 13–16. [Google Scholar] [CrossRef]
- Reymão, T.K.A.; Fumian, T.M.; Justino, M.C.A.; Hernandez, J.M.; Bandeira, R.S.; Lucena, M.S.S.; Teixeira, M.D.; Farias, F.P.; Silva, L.D.; Linhares, A.C.; et al. Norovirus RNA in serum associated with increased fecal viral load in children: Detection, quantification and molecular analysis. PLoS ONE 2018, 13, e0199763. [Google Scholar] [CrossRef] [PubMed]
- Robilotti, E.; Deresinski, S.; Pinsky, B.A. Norovirus. Clin. Microbiol. Rev. 2015, 28, 134–164. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Qiu, Z.L.; Zhuang, J.Y.; Xu, M.D.; Tang, D.P. In situ generation of electron donor to assist signal amplification on porphyrin-sensitized titanium dioxide nanostructures for ultrasensitive photoelectrochemical immunoassay. ACS Appl. Mater. Interfaces 2015, 7, 23812–23818. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. ; Liu. Z.; Hu, C.; Hu, S. Ultrasensitive photoelectrochemical biosensing of multiple biomarkers on a single electrode by a light addressing strategy. Anal. Chem. 2015, 87, 9368–9375. [Google Scholar] [CrossRef] [PubMed]
- Aryaa, S.K.; Ramirez-Vick, J.E.; Singhe, S.P.; Bhansali, S.; Gupta, V. Recent advances in ZnO nanostructures and thin films for biosensor applications: Review. Anal. Chim. Acta 2012, 737, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Vuong, N.M.; Lee, Y.I.; Conte, E.; Reynolds, J.L. H: ZnO nanorod-based photoanode sensitized by CdS and carbon quantum dots for photoelectrochemical water splitting. J. Phys. Chem. C 2015, 119, 24323–24331. [Google Scholar] [CrossRef]
- Bu, Y.; Chen, Z. Effect of oxygen-doped C3N4 on the separation capability of the photoinduced electron-hole pairs generated by O-C3N4@TiO2 with quasi-shell-core nanostructure. Electrochim. Acta 2014, 144, 42–49. [Google Scholar] [CrossRef]
- Jindal, K.; Tomar, M.; Gupta, V. Nitrogen-doped zinc oxide thin films biosensor for determination of uric acid. Analyst 2013, 138, 4353–4362. [Google Scholar] [CrossRef] [PubMed]
- Anta, J.A.; Guillén, E.; Tenazaera, R. ZnO-Based dye-sensitized solar cells. J. Phys. Chem. C 2012, 116, 11413–11425. [Google Scholar] [CrossRef]
- Chen, H.M.; Chen, C.K.; Lin, C.C.; Liu, R.S.; Yang, H.; Chang, W.S.; Chen, K.H.; Chan, T.S.; Lee, J.F.; Tsai, D.P. Multi-bandgap-sensitized ZnO nanorod photoelectrode arrays for water splitting: An X-ray absorption spectroscopy approach for the electronic evolution under solar illumination. J. Phys. Chem. C 2011, 115, 21971–21980. [Google Scholar] [CrossRef]
- Li, J.H.; Li, J.; Fang, X.; Wang, X.H.; Wei, Z.P. Fabrication and characteristic of ZnO glucose oxidase enzyme electrode based on flexible substrate. J. Acta Phys. Chim. Sin. 2012, 28, 1393–1397. [Google Scholar]
- Lee, K.; Park, S.W.; Min, J.K.; Kim, K.; Park, N.G. Selective positioning of organic dyes in a mesoporous inorganic oxide film. Nat. Mater. 2009, 8, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.B.; Jiang, Z.F.; Wei, L.; Chen, Y.X.; Jiao, J.; Eastman, M.; Liu, H. Photosensitization of TiO2, nanorods with CdS quantum dots for photovoltaic applications: A wet-chemical approach. Nano Energy. 2012, 1, 440–447. [Google Scholar] [CrossRef]
- Bora, T.; Kyaw, H.H.; Dutta, J. Zinc oxide-zinc stannate core-shell nanorod arrays for CdS quantum dot sensitized solar cells. Electrochim. Acta 2012, 68, 141–145. [Google Scholar] [CrossRef]
- Xu, C.; Wu, J.; Desai, U.V.; Gao, D. Multilayer assembly of nanowire arrays for dye-sensitized solar cell. J. Am. Chem. Soc. 2011, 133, 8122–8125. [Google Scholar] [CrossRef] [PubMed]
- Sudhagar, P.; Song, T.; Dong, H.L.; Moraseró, I.; Bisquert, J.; Laudenslager, M.; Wolfgang, M.S.; Won, I.P.; Ungyu, P.; Yong, S.K. High open circuit voltage quantum dot sensitized solar cells manufactured with ZnO nanowire arrays and Si/ZnO branched hierarchical structures. J. Phys. Chem. Lett. 2011, 2, 1984–1990. [Google Scholar] [CrossRef]
- Mokari, T.; Rothenberg, E.; Popov, I.; Costi, R.; Banin, U. Selective growth of metal tips onto semiconductor quantum rods and tetrapods. Science 2004, 304, 1787–1790. [Google Scholar] [CrossRef] [PubMed]
- Hodes, G. Comparison of dye- and semiconductor-sensitized porous nanocrystalline liquid junction solar cells. J. Mod. Opt. 2008, 112, 191–195. [Google Scholar] [CrossRef]
- Anusorn, K.; Kevin, T.; Kensuke, T.; Masaru, K.; Prashant, V.K. Quantum dot solar cells. Tuning photoresponse through size and shape control of CdSe-TiO2 architecture. J. Phys. Chem. C 2008, 130, 4007–4015. [Google Scholar]
- Mali, S.S.; Desai, S.K.; Kalagi, S.S.; Betty, C.A.; Bhosale, P.N.; Devan, R.S.; Ma, Y.R.; Patil, P.S. PbS quantum dot sensitized anatase TiO2 nanocorals for quantum dot-sensitized solar cell applications. Dalton Trans. 2012, 41, 6130–6136. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.W.; Wang, J.; Zhu, Y.C.; Xu, J.J.; Chen, H.Y. Quantum dots: Electrochemiluminescent and photoelectrochemical bioanalysis. Anal. Chem. 2015, 87, 9520–9531. [Google Scholar] [CrossRef] [PubMed]
- Benshahar, Y.; Scotognella, F.; Kriegel, I.; Moretti, L.; Cerullo, G.; Rabani, E. Optimal metal domain size for photocatalysis with hybrid semiconductor-metal nanorods. Nat. Commun. 2016, 7, 10413. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Nanomaterial-based electrochemical biosensors. Analyst 2005, 130, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Xie, Y.B.; Liu, Z.; Rong, F.; Wang, Y.; Fu, D.G. Covalently immobilized biosensor based on gold nanoparticles modified TiO2 nanotube arrays. J. Electroanal. Chem. 2011, 650, 241–247. [Google Scholar] [CrossRef]
- Bruchez, M.; Moronne, M.; Gin, P.; Weiss, S. Alivisatos, A.P. Semiconductor nanocrystals as fluorescent biological labels. Science 1998, 281, 2013–2016. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.C.W.; Nie, S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 1998, 281, 2016–2018. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.Z.; Wei, L.Y.; Tang, L.L.; Chen, C.Q.; Pan, H.P.; Chen, J.Z. Aligned Cdse@ZnO flower-rod core-shell nanocable as photovoltaic application. J. Power Sources 2013, 239, 546–552. [Google Scholar] [CrossRef]
- Lu, W.; Wang, G.; Jin, Y.; Yao, X.; Hu, J.Q.; Li, J.H. Label-free photoelectrochemical strategy for hairpin DNA hybridization detection on titanium dioxide electrode. Appl. Phys. Lett. 2006, 89, 263902. [Google Scholar] [CrossRef]
| Samples | Addition (nM) | Found (nM, n = 5) | Recoveries (%) | Average Recovery (%) | RSD (%) |
|---|---|---|---|---|---|
| 1 | 1.70 | 1.73 | 101.80 | 101.97 | 8.58 |
| 2 | 3.40 | 3.43 | 100.88 | 5.88 | |
| 3 | 5.10 | 5.25 | 103.22 | 2.29 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Z.; Weng, Q.; Lin, C.; Yi, J.; Kang, J. Development of CdSe–ZnO Flower-Rod Core-Shell Structure Based Photoelectrochemical Biosensor for Detection of Norovirous RNA. Sensors 2018, 18, 2980. https://doi.org/10.3390/s18092980
Han Z, Weng Q, Lin C, Yi J, Kang J. Development of CdSe–ZnO Flower-Rod Core-Shell Structure Based Photoelectrochemical Biosensor for Detection of Norovirous RNA. Sensors. 2018; 18(9):2980. https://doi.org/10.3390/s18092980
Chicago/Turabian StyleHan, Zhizhong, Qinghua Weng, Chaofan Lin, Jinquan Yi, and Jie Kang. 2018. "Development of CdSe–ZnO Flower-Rod Core-Shell Structure Based Photoelectrochemical Biosensor for Detection of Norovirous RNA" Sensors 18, no. 9: 2980. https://doi.org/10.3390/s18092980
APA StyleHan, Z., Weng, Q., Lin, C., Yi, J., & Kang, J. (2018). Development of CdSe–ZnO Flower-Rod Core-Shell Structure Based Photoelectrochemical Biosensor for Detection of Norovirous RNA. Sensors, 18(9), 2980. https://doi.org/10.3390/s18092980




