The Implications of Fragmented Genomic DNA Size Range on the Hybridization Efficiency in NanoGene Assay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Cell Culture and gDNA Extraction
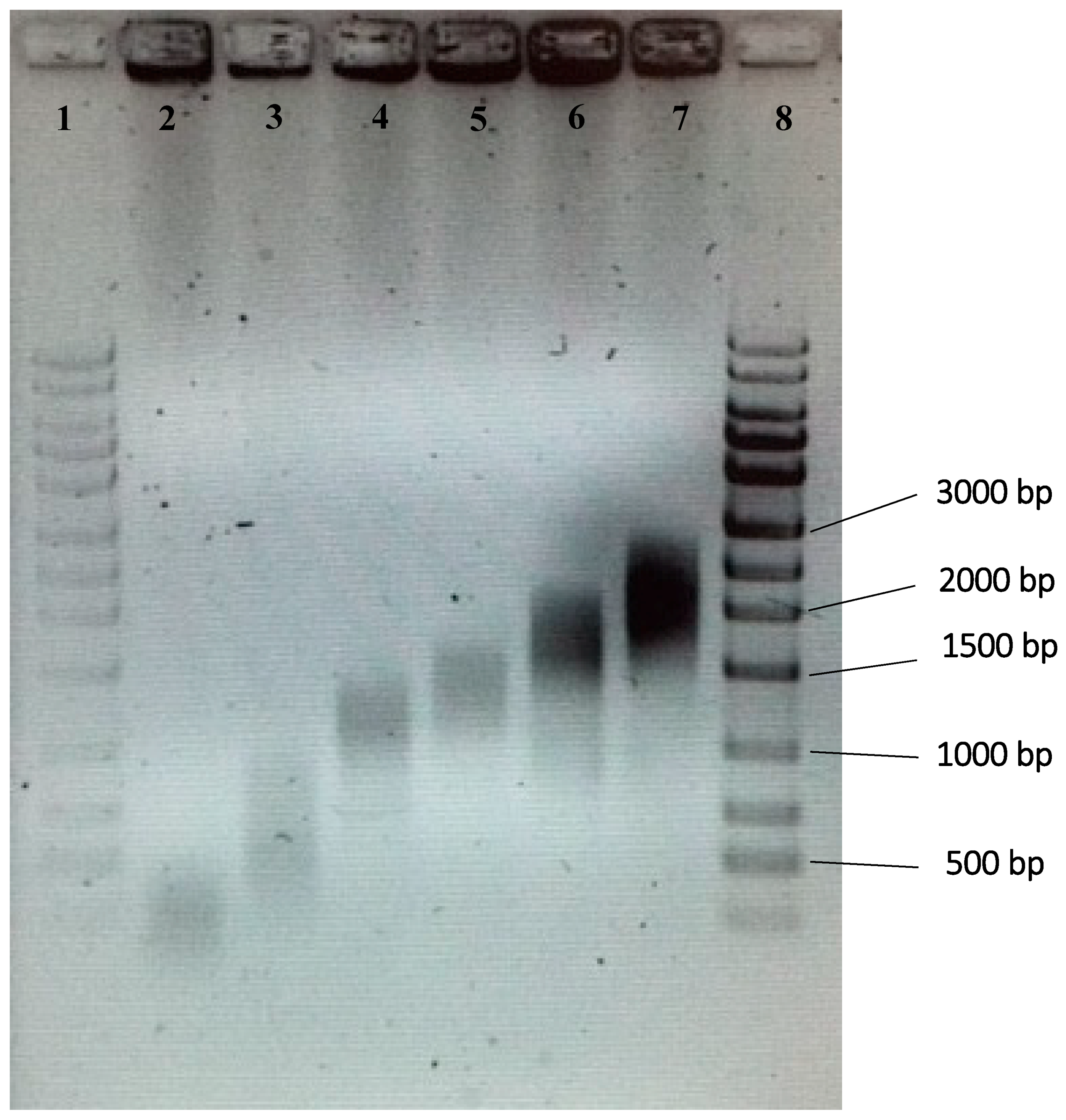
2.2. Preparation and Sizing of Fragmented gDNA Samples
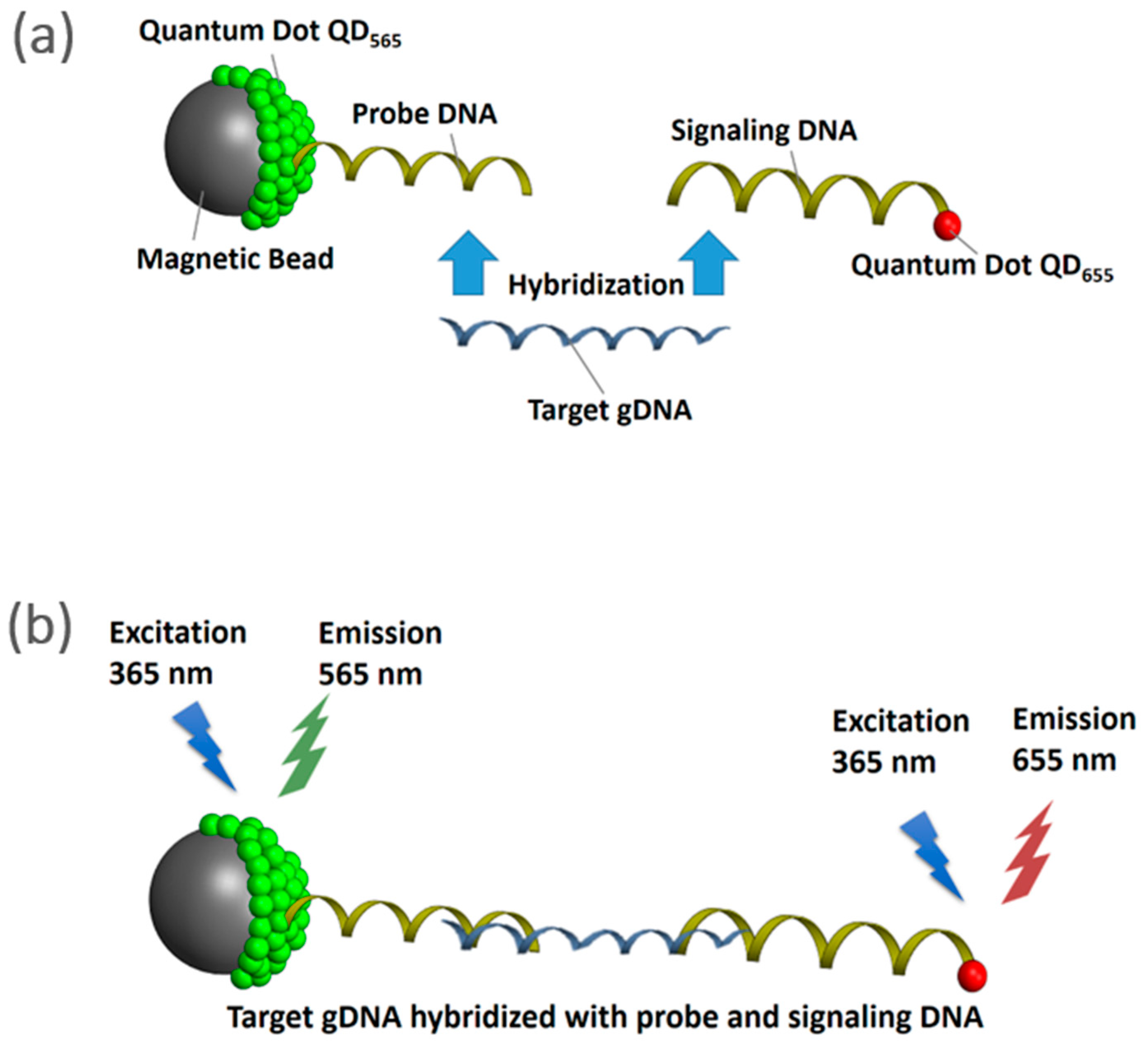
2.3. Quantification Capability of NanoGene Assay Using Fragmented gDNA of Different Size Ranges
- (i)
- Preparation of probe complex: 8 µL of 2 µmol/L Qdot® 565 ITK™ carboxyl quantum dot nanoparticles (QD565, Invitrogen, Carlsbad, CA, USA) were immobilized on the surface of 100 µL of 107 beads/mL aminated Dynabeads® M-270 magnetic beads (MB, Invitrogen, Carlsbad, CA, USA). Dynabeads® M-270 MB with 2.8 µm in diameter are superparamagnetic, and have a monolayer of amine functional group on their surface. This was followed by conjugating 5 µL of 100 nmol/L aminated probe DNA (5′-NH2-C6-AAG CTG TTG TTC GGG AAG ARW GTG C-3′, IDT, Coralville, IA, USA) on the surface of QD565 by forming an amide bond via the reaction of ethylcarbodiimide hydrochloride (EDC, Sigma-Aldrich, St. Louis, MO, USA) and N-hydroxysuccinimide (NHS, Sigma-Aldrich, St. Louis, MO, USA).
- (ii)
- Preparation of signaling complex: Aliquots (1.6 µL) of 100 nmol/L aminated signaling probe DNA (5′-CAA CCM ACG TGG TAT GCA TCT CAT C-NH2-3′) were attached covalently to the surface of 8 µL of 2 µmol/L carboxyl quantum dots (Qdot® 655 ITK™, QD655, Invitrogen, Carlsbad, CA, USA) via EDC-NHS reaction. Qdot® ITK™ carboxyl quantum dots are made from nanometer-scale crystals of a semiconductor material (CdSe), which are shelled with an additional semiconductor layer (ZnS). The size of QDs that were used in this study were within 15–20 nm.
- (iii)
- gDNA hybridization: Five µL of the target gDNA in six size ranges (200–500 bp, 500–1000 bp, 1000–1500 bp, 1500–2000 bp, 2000–2500 bp, and 2500–3000 bp) with a negative control (water) and a positive control (unfragmented gDNA of P. putida, ~4 Mbp) were mixed with the probe and signaling complexes. Subsequently, a slow vertical rotation in a hybridization oven (UVP HB-500 Minidizer Hybridization, Fisher Scientific) was used to enable DNA hybridization overnight at 37 °C.
- (iv)
- Fluorescence measurement: Following DNA hybridization, the hybridized complexes were washed three times using a phosphate buffer (pH = 7.4) to remove the untethered complexes. The endpoint fluorescence of QD565 and QD655 were measured using a SpectraMax M2 spectrofluorometer (Molecular Devices, Sunnyvale, CA, USA) at 570 and 660 nm, respectively, under excitation at 360 nm. All experiments were performed in triplicate, unless otherwise stated. The output of quantification was expressed by the ratio of the fluorescence intensity between QD655 and QD565 (QD655/QD565), as the signal (QD655) was normalized by the internal standard (QD565) to comprehend the different numbers of nanoparticles in each reaction.
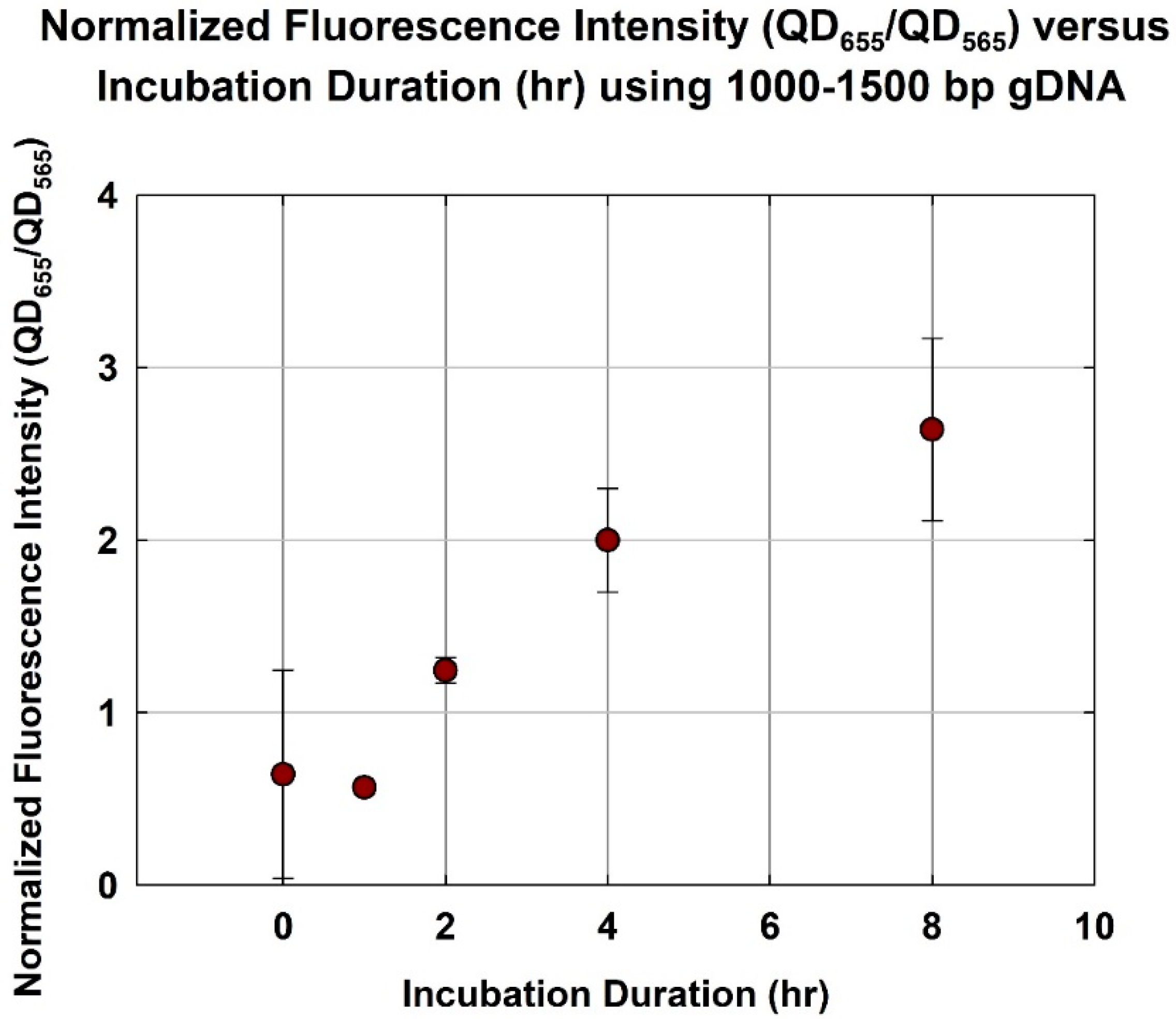
2.4. Different Hybridization Duration Using Fragmented gDNA at Optimum Size Range
2.5. Comparison of Fragmented (at Optimum Size Range) and Unfragmented gDNA via Quantification Capability of NanoGene Assay
3. Results and Discussion
3.1. Size Ranges of Fragmented gDNA
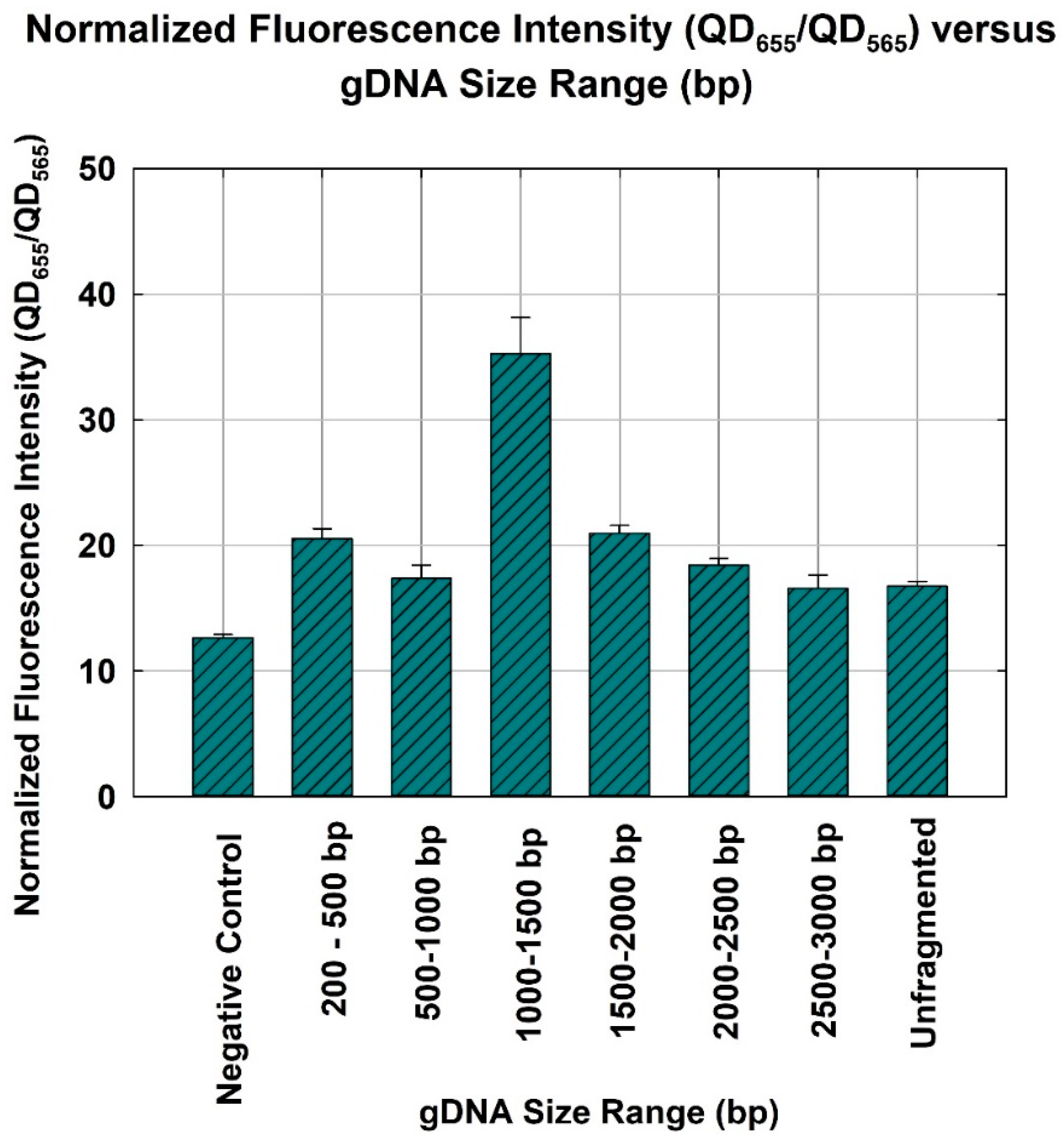
3.2. Quantification Capability of NanoGene Assay Using Fragmented gDNA of Different Size Ranges
3.3. Different Incubation Duration Using Fragmented gDNA at Optimum Size Range
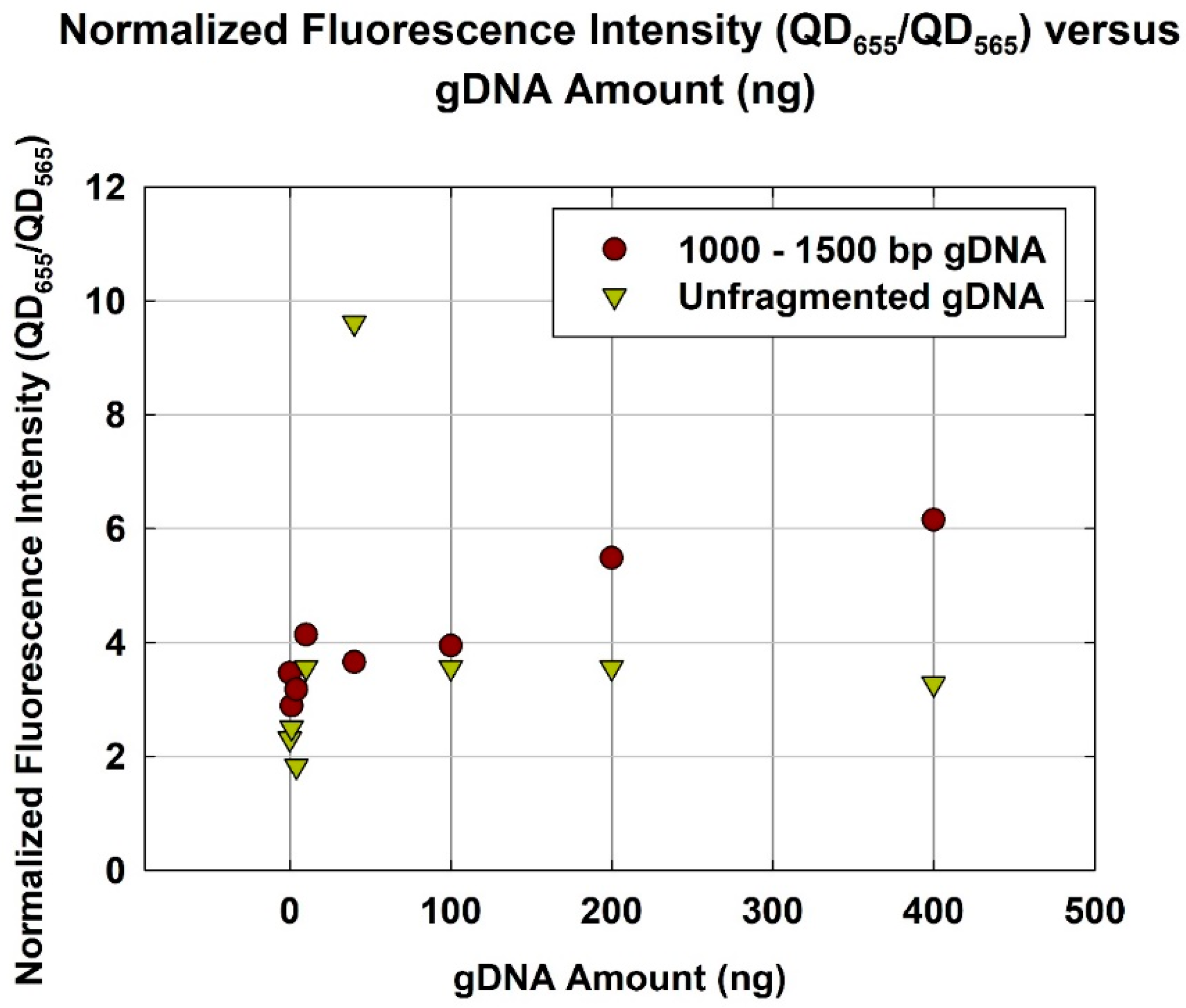
3.4. Comparison of Quantification Capability NanoGene Assay with Optimum gDNA Size Range and Unfragmented gDNA
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khandjian, E.W. Optimized hybridization of DNA blotted and fixed to nitrocellulose and nylon membranes. Nat. Biotechnol. 1987, 5, 165–167. [Google Scholar] [CrossRef]
- Poltronieri, P.; D’Urso, O.F.; Blaiotta, G.; Morea, M. DNA arrays and membrane hybridization methods for screening of six Lactobacillus species common in food products. Food Anal. Method 2008, 1, 171–180. [Google Scholar] [CrossRef]
- Su, W.; Song, S.; Long, M.; Liu, G. Multiplex polymerase chain reaction/membrane hybridization assay for detection of genetically modified organisms. J. Biotechnol. 2003, 105, 227–233. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.F.; Wickham, G.S.; Pace, N.R. Phylogenetic strains: Ribosomal RNA-based probes for identification of single cells. Science 1989, 243, 1360–1363. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Horny, M.; Daimsz, H. Fluorescence in situ hybridisation for the identification and characterization of prokaryotes. Curr. Opin. Microbiol. 2003, 6, 302–309. [Google Scholar] [CrossRef]
- Daims, H.; Nielsen, J.L.; Nielsen, P.H.; Schleifer, K.-H.; Wagner, M. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 2001, 67, 5273–5284. [Google Scholar] [CrossRef] [PubMed]
- Pernthaler, A.; Amann, R. Simultaneous fluorescence in situ hybridization of mRNA and rRNA in environmental bacteria. Appl. Environ. Microbiol. 2004, 70, 5426–5433. [Google Scholar] [CrossRef] [PubMed]
- Shalon, D.; Smith, S.J.; Brown, P.O. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996, 6, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, M.; Yoshiura, K.-I.; Kondo, H.; Miura, S.; Nagayasu, T.; Nakashima, M. Significance of genomic instability in breast cancer in atomic bomb survivors: analysis of microarray-comparative genomic hybridization. Radiat. Oncol. 2011, 6, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laassri, M.; Bidzhieva, B.; Speicher, J.; Pletnev, A.G.; Chumakov, K. Microarray hybridization for assessment of the genetic stability of chimeric west nile/dengue 4 virus. J. Med. Virol. 2011, 83, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Spellman, P.T.; Sherlock, G.; Zhang, M.Q.; Iyer, V.R.; Anders, K.; Eisen, M.B.; Brown, P.O.; Botstein, D.; Futcher, B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 1998, 9, 3273–3297. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Li, X.-B.; Lopa, N.S.; Ahn, S.J.; Lee, J.-J. Electrochemical DNA hybridization sensors based on conducting polymers. Sensors 2015, 15, 3801–3829. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Kim, Y.J.; Lee, J.-J. Sensitivity control of label-free DNA hybridization detection based on poly(thionine)-modified glassy carbon and gold electrodes. J. Electrochem. Soc. 2016, 163, B153–B157. [Google Scholar] [CrossRef]
- Corrigan, D.K.; Schulze, H.; McDermott, R.A.; Schmuser, I.; Henihan, G.; Henry, J.B.; Bachmann, T.T.; Mount, A.R. Improving electrochemical biosensor performance by understanding the influence of target DNA length on assay sensitivity. J. Electroanal. Chem. 2014, 732, 25–29. [Google Scholar] [CrossRef]
- Corrigan, D.K.; Schulze, H.; Henihan, G.; Hardie, A.; Ciani, I.; Giraud, G.; Terry, J.G.; Walton, A.J.; Pethig, R.; Ghazal, P.; et al. Development of a PCR-free electrochemical point of care test for clinical detection of methicillin resistant Staphylococcus aureus (MRSA). Analyst 2013, 138, 6997–7005. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.-Y.; Son, A. Development and characterization of a magnetic bead-quantum dot nanoparticles based assay capable of Escherichia coli O157:H7 quantification. Anal. Chim. Acta 2010, 677, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-H.; Chua, B.; Son, A. Detection of cyanobacteria in eutrophic water using a portable electrocoagulator and NanoGene assay. Environ. Sci. Technol. 2018, 52, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.-Y.; Wang, X.; Son, A. Inhibitor resistance and in situ capability of nanoparticle based gene quantification. J. Environ. Monitor. 2011, 13, 1344–1350. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.A.; Chua, B.; Son, A. Development of first generation in-situ pathogen detection system (Gen1-IPDS) based on NanoGene assay for near real time E. coli O157:H7 detection. Biosens. Bioelectron. 2014, 54, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-H.; Chua, B.; Son, A. Detection of airborne bacteria with disposable bio-precipitator and NanoGene assay. Biosens. Bioelectron. 2016, 83, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Son, A. Effects of pretreatment on the denaturation and fragmentation of genomic DNA for DNA hybridization. Environ. Sci. Process. Impacts 2013, 15, 2204–2212. [Google Scholar] [CrossRef] [PubMed]
- Cébron, A.; Norini, M.-P.; Beguiristain, T.; Leyval, C. Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J. Microbiol. Methods 2008, 73, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cho, K.-S.; Son, A. Ultrasonication as a rapid and high yield DNA extraction method for bacterial gene quantification by NanoGene assay. Biotech. Bioproc. Eng. 2015, 20, 1133–1140. [Google Scholar] [CrossRef]
- Wang, X.; Liles, M.R.; Son, A. Quantification of Escherichia coli O157:H7 in soils using an inhibitor-resistant NanoGene assay. Soil Biol. Biochem. 2013, 58, 9–15. [Google Scholar] [CrossRef]
- Lee, E.-H.; Cho, K.-S.; Son, A. Detection and quantification of toxin-producing Microcystis aeruginosa strain in water by NanoGene assay. J. Microbiol. Biotechnol. 2017, 27, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Son, A.; Dosev, D.; Nichkova, M.; Kennedy, I.M.; Scow, K.M.; Hristova, K.R. Quantitative DNA hybridization in solution using magnetic/lumiscenct core-shell nanoparticles. Anal. Biochem. 2007, 370, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.-Y.; Wang, X.; Ahn, H.; Son, A. Gene quantification by the NanoGene assay is resistant to inhibition by humic acids. Environ. Sci. Technol. 2011, 45, 8873–8880. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Son, A. Quantification of Salmonella Typhimurium in liquid food using NanoGene assay. Anal. Methods 2015, 7, 7674–7679. [Google Scholar] [CrossRef]
- Lee, E.-H.; Chua, B.; Son, A. Micro corona discharge based cell lysis method suitable for inhibitor resistant bacterial sensing systems. Sens. Actuators B Chem. 2015, 216, 17–23. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Chua, B.; Son, A. The Implications of Fragmented Genomic DNA Size Range on the Hybridization Efficiency in NanoGene Assay. Sensors 2018, 18, 2646. https://doi.org/10.3390/s18082646
Wang X, Chua B, Son A. The Implications of Fragmented Genomic DNA Size Range on the Hybridization Efficiency in NanoGene Assay. Sensors. 2018; 18(8):2646. https://doi.org/10.3390/s18082646
Chicago/Turabian StyleWang, Xiaofang, Beelee Chua, and Ahjeong Son. 2018. "The Implications of Fragmented Genomic DNA Size Range on the Hybridization Efficiency in NanoGene Assay" Sensors 18, no. 8: 2646. https://doi.org/10.3390/s18082646
APA StyleWang, X., Chua, B., & Son, A. (2018). The Implications of Fragmented Genomic DNA Size Range on the Hybridization Efficiency in NanoGene Assay. Sensors, 18(8), 2646. https://doi.org/10.3390/s18082646




