Detection of Salmonella Typhimurium on Spinach Using Phage-Based Magnetoelastic Biosensors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fabrication of ME Biosensor Platforms
2.2. E2 Phage Immobilization
2.3. Surface Blocking of the ME Biosensors
2.4. Salmonella Pre-Enrichment and Exposure to the Biosensors
2.5. Frequency Measurement and Student’s t-Test
2.6. Optical Microscopy Observation
2.7. Negative Control Test
3. Results and Discussion
3.1. Phage Immobilization and Surface Coverage
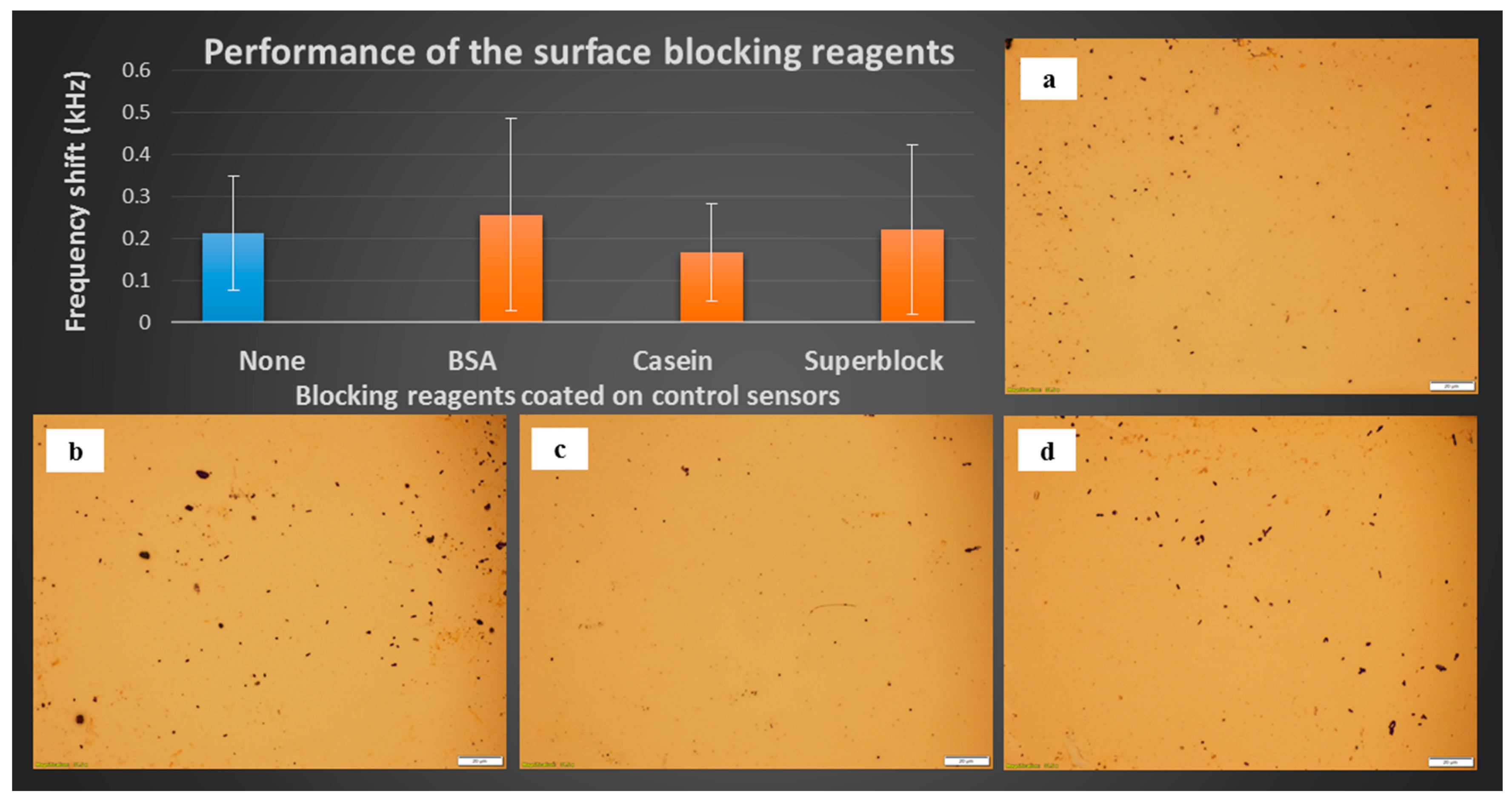
3.2. Performance of the Surface Blocking Reagents
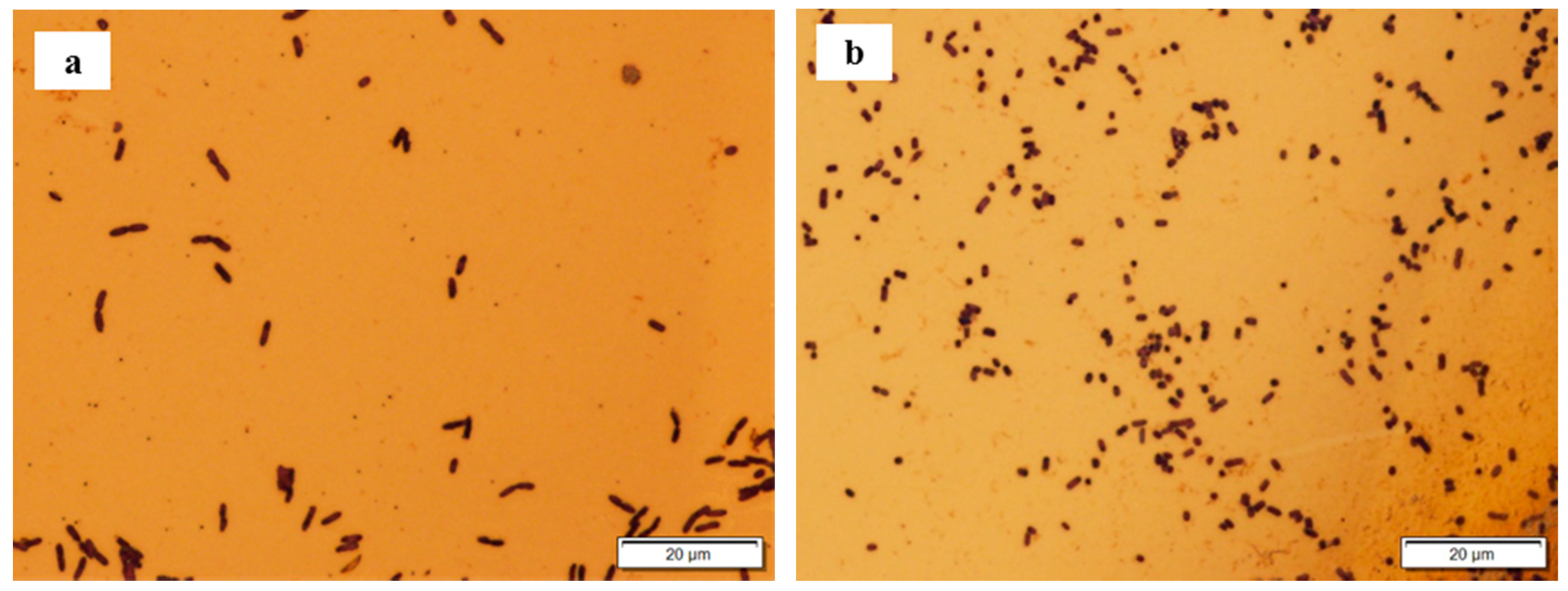
3.3. Lactose Broth vs. LB Broth
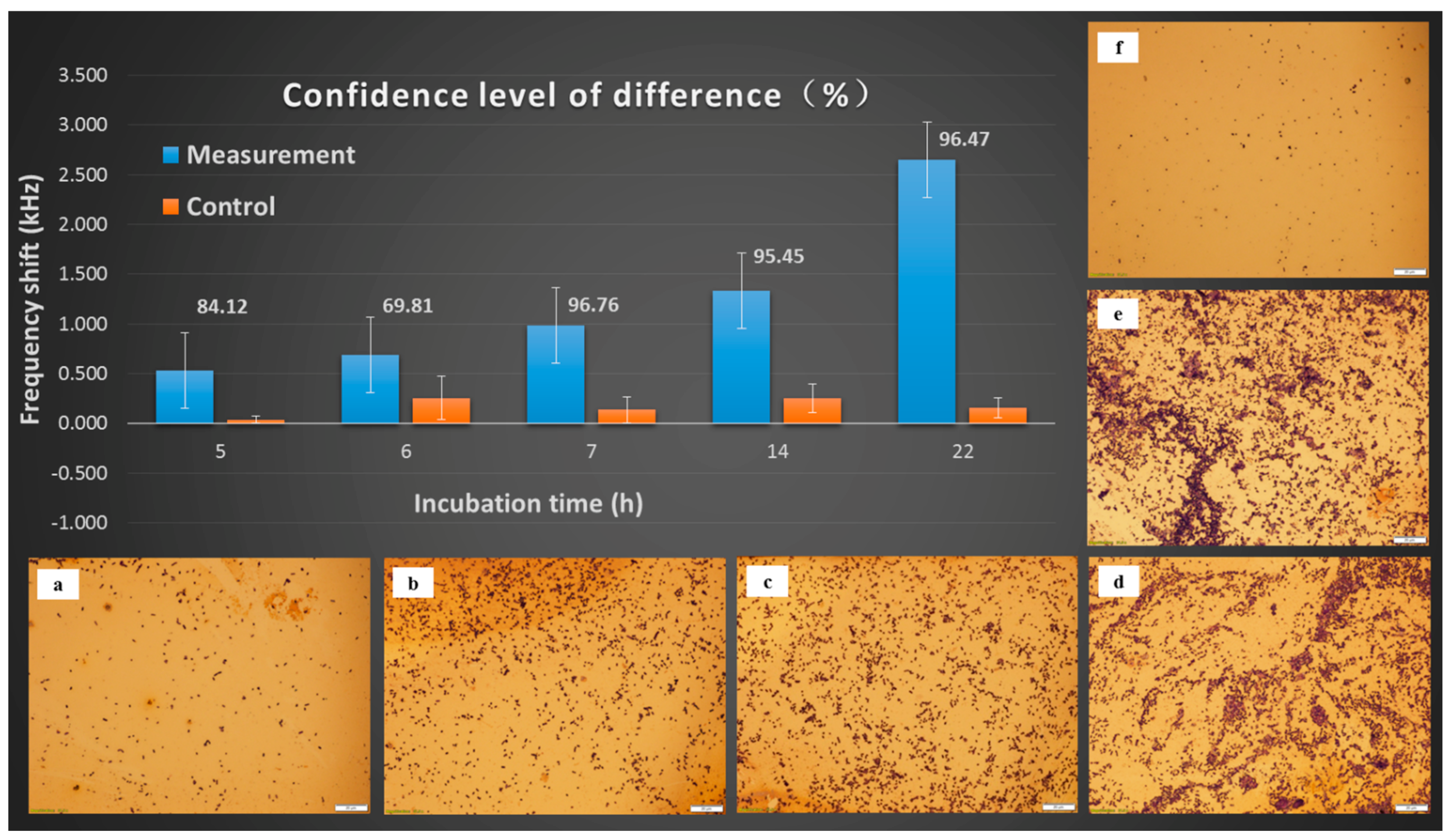
3.4. Frequency Shifts as a Function of Incubation Time
3.5. Negative Control
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Heaton, J.C.; Jones, K. Microbial contamination of fruit and vegetables and the behaviour of enteropathogens in the phyllosphere: A review. J. Appl. Microbiol. 2008, 104, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Park, M.K.; Oh, J.H. Rapid detection of E. coli O157:H7 on turnip greens using a modified gold nsor combined with light microscopic imaging system. J. Food Sci. 2012, 77, M127–M134. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.P.; Erickson, M.C. Summer meeting 2007-the problems with fresh produce: An iew. J. Appl. Microbiol. 2008, 105, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Hanning, I.B.; Nutt, J.D.; Ricke, S.C. Salmonellosis outbreaks in the United States due to fresh uce: Sources and potential intervention measures. Foodborne Pathog. Dis. 2009, 6, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne gen detection: In the perspective of biosensors. Biotech. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.; Moreira, R.G.; Castell-Perez, E. Radiosensitization of Salmonella spp. and Listeria spp. dy-to-eat baby spinach leaves. J. Food Sci. 2011, 76, E141–E148. [Google Scholar] [CrossRef] [PubMed]
- Buck, J.W.; Walcott, R.R.; Beuchat, L.R. Recent trends in microbiological safety of fruits and ables. Plant Health Prog. 2003, 10, 1094–1103. [Google Scholar]
- Guntupalli, R.; Hu, J.; Lakshmanan, R.S.; Huang, T.S.; Barbaree, J.M.; Chin, B.A. A magnetoelastic resonance biosensor immobilized with polyclonal antibody for the detection of Salmonella typhimurium. Biosens. Bioelectron. 2007, 22, 1474–1479. [Google Scholar] [CrossRef] [PubMed]
- Guntupalli, R.; Lakshmanan, R.S.; Hu, J.; Huang, T.S.; Barbaree, J.M.; Vodyanoy, V.; Chin, B.A. Rapid and sensitive magnetoelastic biosensors for the detection of Salmonella typhimurium in a mixed microbial population. J. Microbiol. Methods 2007, 70, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, R.S.; Guntupalli, R.; Hu, J.; Kim, D.J.; Petrenko, V.A.; Barbaree, J.M.; Chin, B.A. Phage immobilized magnetoelastic sensor for the detection of Salmonella typhimurium. J. Microbiol. Methods 2007, 71, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.L.; Wan, J.H.; Huang, S.C.; Cheng, Z.Y.; Petrenko, V.A.; Kim, D.J.; Chen, I.H.; Barbaree, J.M.; Hong, J.W.; Chin, B.A. A wireless biosensor using microfabricated phage-interfaced magnetoelastic particles. Sens. Actuators A 2008, 144, 38–47. [Google Scholar] [CrossRef]
- Horikawa, S.; Bedi, D.; Li, S.Q.; Shen, W.; Huang, S.C.; Chen, I.H.; Chai, Y.T.; Auad, M.L.; Bozack, M.; Barbaree, J.M.; et al. Effects of surface functionalization on the surface phage coverage and the subsequent performance of phage-immobilized magnetoelastic biosensors. Biosens. Bioelectron. 2011, 26, 2361–2367. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Horikawa, S.; Li, S.; Wikle, H.C.; Chin, B.A. A surface-scanning coil detector for real-time, in-situ detection of bacteria on fresh food surfaces. Biosens. Bioelectron. 2013, 50, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Grimes, C.A.; Roy, S.C.; Rani, S.; Cai, Q. Theory, Instrumentation and Applications of Magnetoelastic Resonance Sensors: A Review. Sensors 2011, 11, 2809–2844. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Q.; Li, Y.G.; Chen, H.Q.; Horikawa, S.; Shen, W.; Simonian, A.; Chin, B.A. Direct detection of Salmonella typhimurium on fresh produce using phage-based magnetoelastic biosensors. Biosens. Bioelectron. 2010, 26, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Park, M.K.; Park, J.W.; Wikle, H.C.; Chin, B.A. Evaluation of phage-based magnetoelastic biosensors for direct detection of Salmonella Typhimurium on spinach leaves. Sens. Actuators B: Chem. 2013, 176, 1134–1140. [Google Scholar] [CrossRef]
- Huang, S.; Yang, H.; Lakshmanan, R.S.; Johnson, M.L.; Wan, J.; Chen, I.H.; Wikle, H.C., III; Petrenko, V.A.; Barbaree, J.M.; Chin, B.A. Sequential detection of Salmonella typhimurium and Bacillus anthracis spores using magnetoelastic biosensors. Biosens. Bioelectron. 2009, 24, 1730–1736. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Lakshmanan, R.S.; Mathison, L.C.; Petrenko, V.A.; Chin, B.A. Phage coated magnetoelastic micro-biosensors for real-time detection of Bacillus anthracis spores. Sens. Actuators B: Chem. 2009, 137, 501–506. [Google Scholar] [CrossRef]
- Park, M.K.; Park, J.W.; Wikle, H.C.; Chin, B.A. Comparison of phage-based magnetoelastic biosensors with taqman- based quantitative real-time PCR for the detection of salmonella typhimurium directly grown on tomato surfaces. Biosens. Bioelectron. 2012, 3, 113–118. [Google Scholar] [CrossRef]
- Park, M.K.; Weerakoon, K.A.; Oh, J.H.; Chin, B.A. The analytical comparison of phage-based magnetoelastic biosensor with TaqMan-based quantitative PCR method to detect Salmonella Typhimurium on cantaloupes. Food Control 2013, 33, 330–336. [Google Scholar] [CrossRef]
- Sorokulova, I.B.; Olsen, E.V.; Chen, I.H.; Fiebor, B.; Barbaree, J.M.; Vodyanoy, V.J.; Chin, B.A.; Petrenko, V.A. Landscape phage probes for Salmonella typhimurium. J. Microbiol. Methods 2005, 63, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Olsen, E.V.; Sorokulova, I.B.; Petrenko, V.A.; Chen, I.H.; Barbaree, J.M.; Vodyanoy, V.J. Affinity-selected filamentous bacteriophage as a probe for acoustic wave biodetectors of Salmonella typhimurium. Biosens. Bioelectron. 2006, 21, 1434–1442. [Google Scholar] [CrossRef] [PubMed]
- Andrews, W.H.; Jacobson, A.; Hammack, T. Salmonella. In Bacteriological Analytical Manual-Salmonella; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2014; Chapter 5. [Google Scholar]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Horikawa, S.; Hu, J.; Wikle, H.C., III; Chen, I.-H.; Du, S.; Liu, Y.; Chin, B.A. Detection of Salmonella Typhimurium on Spinach Using Phage-Based Magnetoelastic Biosensors. Sensors 2017, 17, 386. https://doi.org/10.3390/s17020386
Wang F, Horikawa S, Hu J, Wikle HC III, Chen I-H, Du S, Liu Y, Chin BA. Detection of Salmonella Typhimurium on Spinach Using Phage-Based Magnetoelastic Biosensors. Sensors. 2017; 17(2):386. https://doi.org/10.3390/s17020386
Chicago/Turabian StyleWang, Fengen, Shin Horikawa, Jiajia Hu, Howard C. Wikle, III, I-Hsuan Chen, Songtao Du, Yuzhe Liu, and Bryan A. Chin. 2017. "Detection of Salmonella Typhimurium on Spinach Using Phage-Based Magnetoelastic Biosensors" Sensors 17, no. 2: 386. https://doi.org/10.3390/s17020386
APA StyleWang, F., Horikawa, S., Hu, J., Wikle, H. C., III, Chen, I.-H., Du, S., Liu, Y., & Chin, B. A. (2017). Detection of Salmonella Typhimurium on Spinach Using Phage-Based Magnetoelastic Biosensors. Sensors, 17(2), 386. https://doi.org/10.3390/s17020386





