Imaging and Force Recognition of Single Molecular Behaviors Using Atomic Force Microscopy
Abstract
:1. Introduction
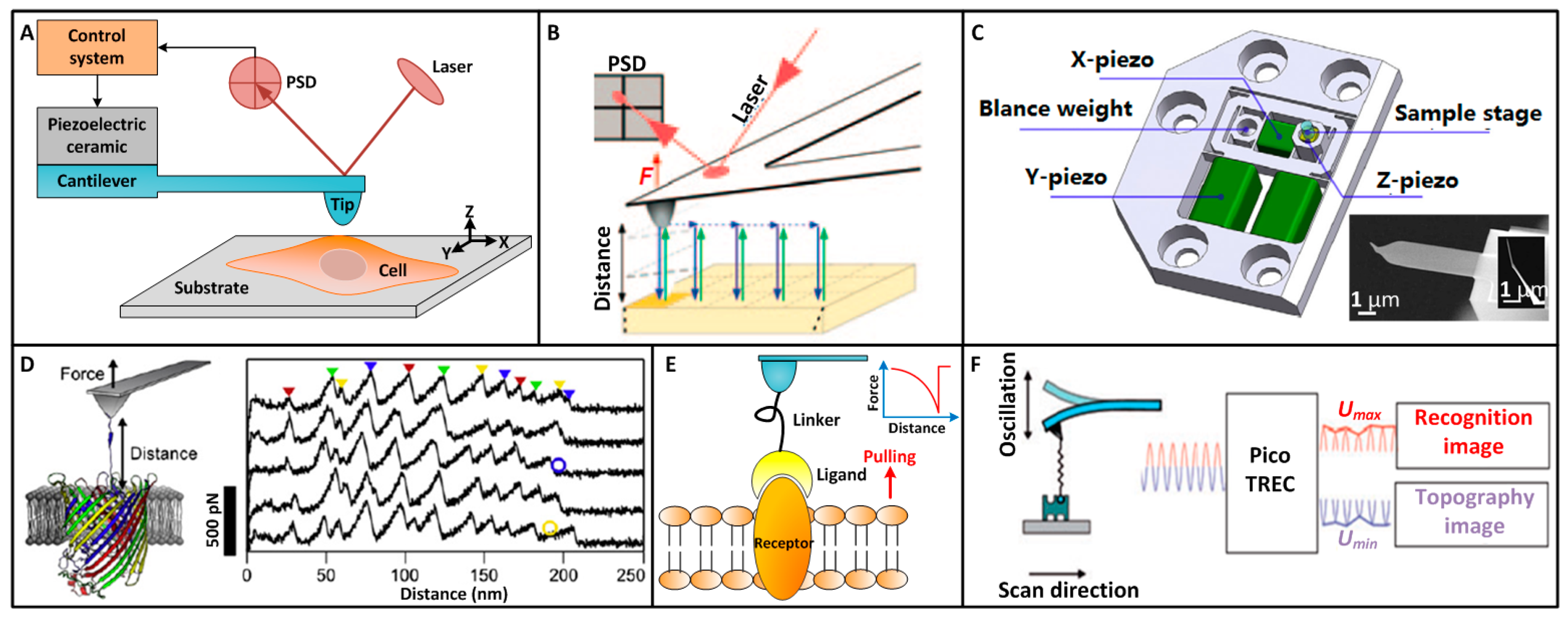
2. AFM Single-Molecule Techniques
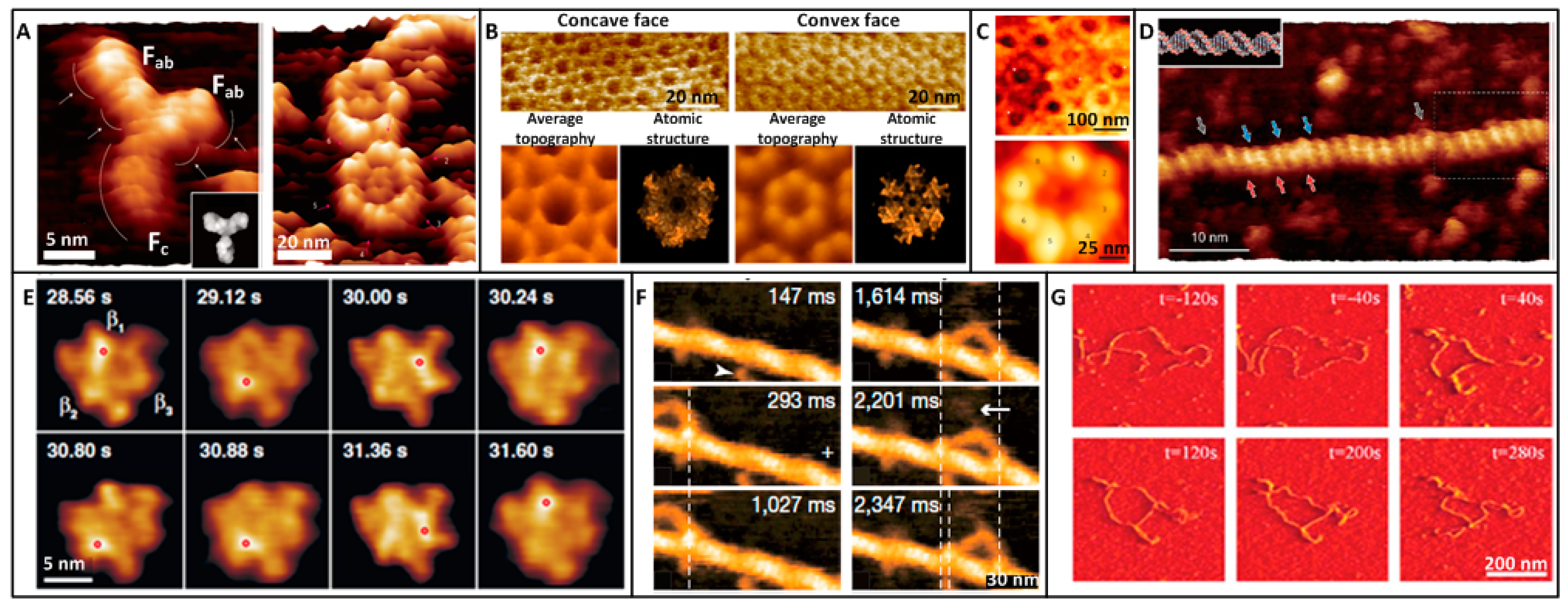
3. Molecular Imaging
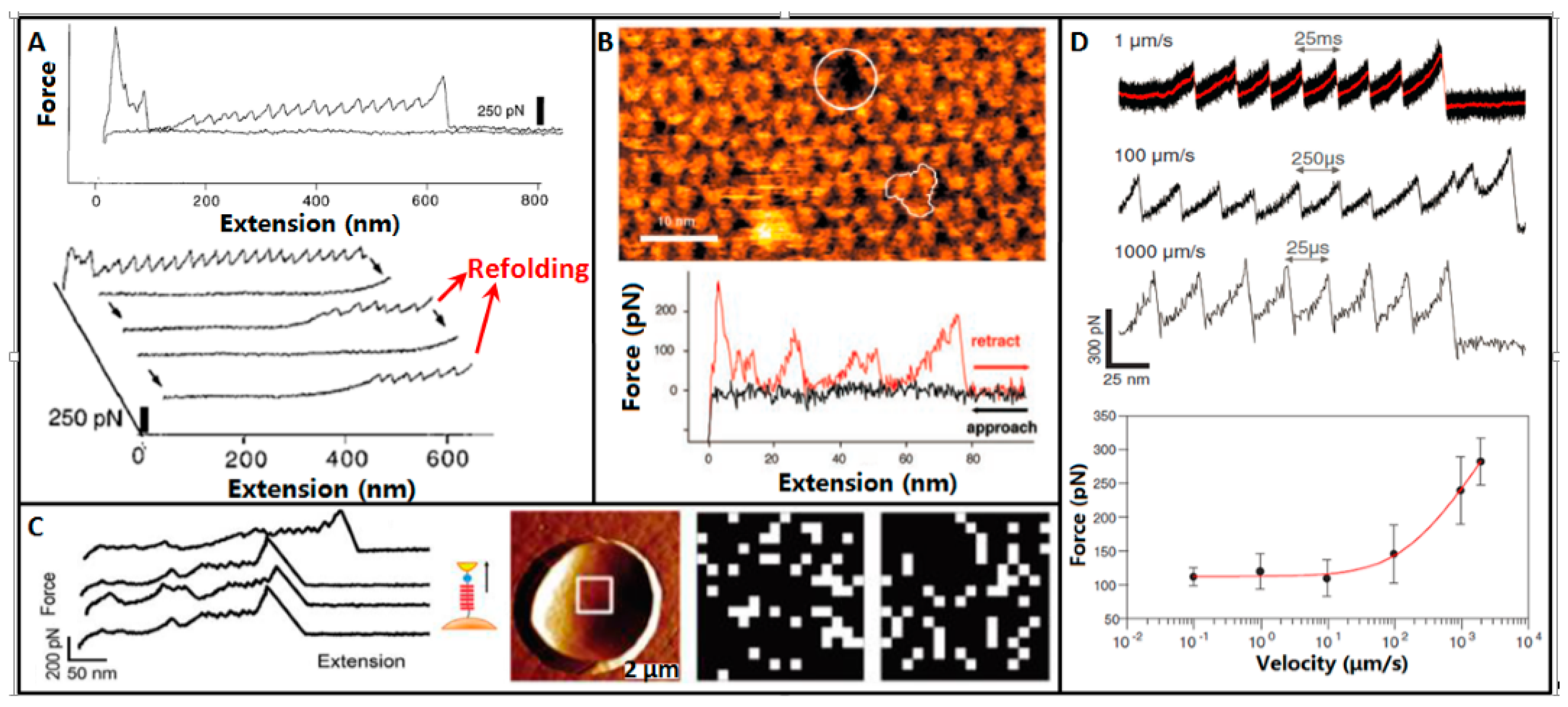
4. Molecular Mechanics
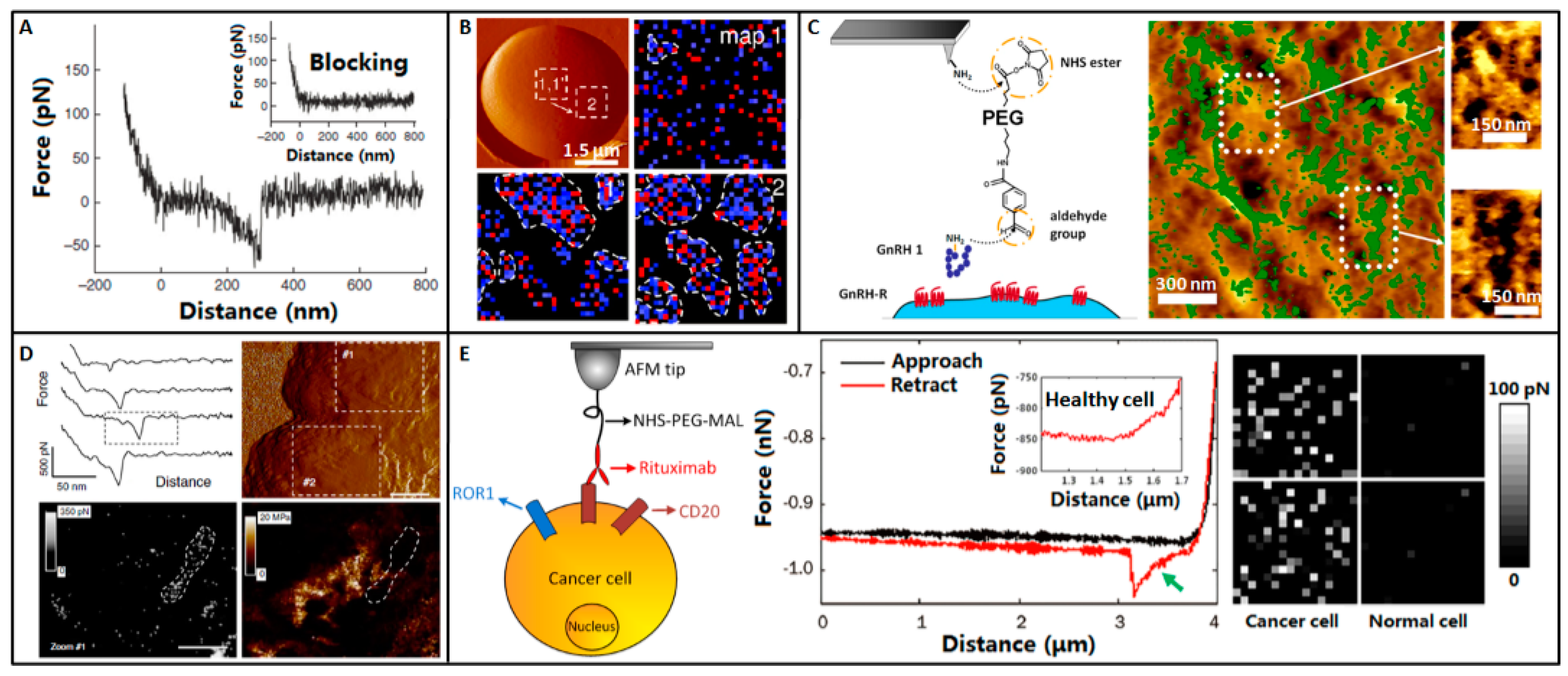
5. Molecular Recognition
6. Molecular Activities on Cell Surface
7. Challenges and Outlook
Acknowledgments
Conflicts of Interest
Appendix A
| Typical Applications | Tip Material | Tip Radius | Cantilever Spring Constant | References |
|---|---|---|---|---|
| Antibody imaging on mica | Silicon | <10 nm | 42 N/m | [58] |
| Membrane protein imaging on mica | Silicon nitride | ≤20 nm | 0.09 N/m | [59] |
| High-speed imaging of molecules | Silicon nitride | ~4 nm | 0.1–0.2 N/m | [12] |
| High-speed imaging of living cell | Silicon nitride | <10 nm | 0.1 N/m | [44] |
| Imaging DNA dynamics in water | Silicon nitride | <10 nm | 0.32 N/m | [63] |
| Unfolding single proteins on cells | Silicon nitride | <10 nm | 0.01–0.025 N/m | [69] |
| High-speed unfolding proteins | Silicon nitride | <10 nm | ~0.1 N/m | [71] |
| SMFS on cell surface | Silicon nitride | ~10 nm | 0.008–0.021 N/m | [90] |
| TREC imaging on cell surface | Silicon | 8 nm | 0.14 N/m | [91] |
| PFT multiparametric imaging | Silicon nitride | ~20 nm | 0.45–0.77 N/m | [92] |
References
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Josan, J.S.; Vagner, J.; Caplan, M.R.; Hruby, V.J.; Mash, E.A.; Lynch, R.M.; Morse, D.L.; Gillies, R.J. Heterobivalent ligands target cell-surface receptor combinations in vivo. Proc. Natl. Acad. Sci. USA 2012, 109, 21295–21300. [Google Scholar] [CrossRef] [PubMed]
- Dupres, V.; Alsteens, D.; Wilk, S.; Hansen, B.; Heinisch, J.J.; Dufrene, Y.F. The yeast Wsc1 cell surface sensor behaves like a nanospring in vivo. Nat. Chem. Biol. 2009, 5, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Hinterdorfer, P.; Garcia-Parajo, M.F.; Dufrene, Y.F. Single-molecule imaging of cell surfaces using near-field nanoscopy. Acc. Chem. Res. 2012, 45, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Young, R.M.; Staudt, L.M. Targeting pathological B cell receptor signaling in lymphoid malignancies. Nat. Rev. Drug Discov. 2013, 12, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, L.; Xi, N.; Wang, Y. Progress in measuring biophysical properties of membrane proteins with AFM single-molecule force spectroscopy. Chin. Sci. Bull. 2014, 59, 2717–2725. [Google Scholar] [CrossRef]
- Schubeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Ashworth, A. The DNA damage response and cancer therapy. Nature 2012, 481, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Rambo, R.P.; Tainer, J.A. Super-resolution in solution x-ray scattering and its applications to structural systems biology. Ann. Rev. Biophys. 2013, 42, 415–441. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Leiro, R.; Scheres, S.H.W. Unravelling biological macromolecules with cryo-electron microscopy. Nature 2016, 537, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef] [PubMed]
- Kodera, N.; Yamamoto, D.; Ishikawa, R.; Ando, T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature 2010, 468, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Heinisch, J.J.; Lipke, P.N.; Beaussart, A.; El KiratChatel, S.; Dupres, V.; Alsteens, D.; Dufrene, Y.F. Atomic force microscopy—looking at mechanosensors on the cell surface. J. Cell Sci. 2012, 125, 4189–4195. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.J.; Dufrene, Y.F. Atomic force microscopy: A nanoscopic window on the cell surface. Trends. Cell Biol. 2011, 21, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Ando, T. High-speed atomic force microscopy coming of age. Nanotechnology 2012, 23, 062001. [Google Scholar] [CrossRef] [PubMed]
- Katan, A.J.; Dekker, C. High-speed AFM reveals the dynamics of single biomolecules at the nanometer scale. Cell 2011, 147, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Woodside, M.T.; Block, S.M. Reconstructing folding energy landscapes by single-molecule force spectroscopy. Ann. Rev. Biophys. 2014, 43, 19–39. [Google Scholar] [CrossRef] [PubMed]
- Dufrene, Y.F.; Martinez-Martin, D.; Medalsy, I.; Alsteens, D.; Muller, D.J. Multiparametric imaging of biological systems by force-distance curve-based AFM. Nat. Methods 2013, 10, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Uchihashi, T.; Scheuring, S. Filming biomolecular processes by high-speed atomic force microscopy. Chem. Rev. 2014, 114, 3120–3188. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Wang, H. The structure and function of cell membranes examined by atomic force microscopy and single-molecule force spectroscopy. Chem. Soc. Rev. 2015, 44, 3617–3638. [Google Scholar] [CrossRef] [PubMed]
- Whited, A.M.; Park, P.S. Atomic force microscopy: A multifaceted tool to study membrane proteins and their interactions with ligands. Biochim. Biophys. Acta 2014, 1838, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Schillers, H.; Medalsy, I.; Hu, S.; Slade, A.L.; Shaw, J.E. Peakforce tapping resolves individual microvilli on living cells. J. Mol. Recognit. 2016, 29, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Zhao, W.; Shang, X.; Jiang, J.; Ji, H.; Tang, Z.; Wang, H. Direct evidence of lipid rafts by in situ atomic force microscopy. Small 2012, 8, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Hecht, E.; Thompson, K.; Frick, M.; Wittekindt, O.H.; Dietl, P.; Mizaikoff, B.; Kranz, C. Combined atomic force microscopy-fluorescence microscopy: Analyzing exocytosis in alveolar type II cells. Anal. Chem. 2012, 84, 5716–5722. [Google Scholar] [CrossRef] [PubMed]
- Oesterhelt, F.; Osterhelt, D.; Pfeiffer, M.; Engel, A.; Gaub, H.E.; Muller, D.J. Unfolding pathways of individual bacteriorhodopsins. Science 2000, 288, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Heu, C.; Berquand, A.; Elie-Caille, C.; Nicod, L. Glyphosate-induced stiffening of HaCaT keratinocytes, a peak force tapping study on living cells. J. Struct. Biol. 2012, 178, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xiao, X.; Liu, L.; Xi, N.; Wang, Y. Rapid recognition and functional analysis of membrane proteins on human cancer cells using atomic force microscopy. J. Immunol. Methods 2016, 436, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Pfeundschuh, M.; Martinez-Martin, D.; Mulvihill, E.; Wegmann, S.; Muller, D.J. Multiparametric high-resolution imaging of native proteins by force-distance curve-based AFM. Nat. Protoc. 2014, 9, 1113–1130. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, L.; Xi, N.; Wang, Y.; Dong, Z.; Tabata, O.; Xiao, X.; Zhang, W. Imaging and measuring the rituximab-induced changes of mechanical properties in B-lymphoma cells using atomic force microscopy. Biochem. Biophys. Res. Commun. 2011, 404, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Rasool, H.I.; Palanisamy, V.; Mathiesen, C.; Schmidt, M.; Wong, D.T.; Gimzewski, J.K. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano 2010, 4, 1921–1926. [Google Scholar] [CrossRef] [PubMed]
- Walczyk, W.; Schon, P.M.; Schonherr, H. The effect of peakforce tapping mode AFM imaging on the apparent shape of surface nanobubbles. J. Phys. Condens. Matter 2013, 25, 184005. [Google Scholar] [CrossRef] [PubMed]
- Hoh, J.H.; Schoenenberger, C.A. Surface morphology and mechanical properties of MDCK monolayers by atomic force microscopy. J. Cell Sci. 1994, 107, 1105–1114. [Google Scholar] [PubMed]
- Rico, F.; Su, C.; Scheuring, S. Mechanical mapping of single membrane proteins at submolecular resolution. Nano Lett. 2011, 11, 3983–3986. [Google Scholar] [CrossRef] [PubMed]
- Medalsy, I.; Hensen, U.; Muller, D.J. Imaging and quantifying chemical and physical properties of native proteins at molecular resolution by force-volume AFM. Angew. Chem. Int. Ed. 2011, 50, 12103–12108. [Google Scholar] [CrossRef] [PubMed]
- Pfreundschuh, M.; Hensen, U.; Muller, D.J. Quantitative imaging of the electrostatic field and potential generated by a transmembrane protein pore at subnanometer resolution. Nano Lett. 2013, 13, 5585–5593. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Uchihashi, T.; Kodera, N.; Yamamoto, D.; Miyagi, A.; Taniguchi, M.; Yamashita, H. High-speed AFM and nano-visualization of biomolecular processes. Eur. J. Physiol. 2008, 456, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Ando, T. High-speed AFM imaging. Curr. Opin. Struct. Biol. 2014, 28, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Uchihashi, T.; Fukuma, T. High-speed atomic force microscopy for nano-visualization of dynamic biomolecular processes. Prog. Surf. Sci. 2008, 83, 337–437. [Google Scholar] [CrossRef]
- Uchihashi, T.; Iino, R.; Ando, T.; Noji, H. High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science 2011, 333, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Preiner, J.; Kodera, N.; Tang, J.; Ebner, A.; Brameshuber, M.; Blaas, D.; Gelbmann, N.; Gruber, H.J.; Ando, T.; Hinterdorfer, P. IgGs are made for walking on bacterial and viral surfaces. Nat. Commun. 2014, 5, 4394. [Google Scholar] [CrossRef] [PubMed]
- Casuso, I.; Khao, J.; Chami, M.; Paul-Gilloteaux, P.; Husain, M.; Duneau, J.P.; Stahlberg, H.; Sturgis, J.N.; Scheuring, S. Characterization of the motion of membrane proteins using high-speed atomic force microscopy. Nat. Nanotechnol. 2012, 7, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Colom, A.; Casuso, I.; Rico, F.; Scheuring, S. A hybrid high-speed atomic force-optical microscope for visualizing single membrane proteins on eukaryotic cells. Nat. Commun. 2013, 4, 2155. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Sakai, N.; Yoshida, A.; Uekusa, Y.; Yagi, A.; Imaoka, Y.; Ito, S.; Karaki, K.; Takeyasu, K. High-speed atomic force microscopy combined with inverted optical microscopy for studying cellular events. Sci. Rep. 2013, 3, 213. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Sakai, N.; Uekusa, Y.; Deguchi, K.; Gilmore, J.L.; Kumeta, M.; Ito, S.; Takeyasu, K. Probing in vivo dynamics of mitochondria and cortical actin networks using high-speed atomic force/fluorescence microscopy. Genes Cells 2015, 20, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Uchihashi, T.; Ando, T.; Yasuda, R. Long-tip high-speed atomic force microscopy for nanometer-scale imaging in live cells. Sci. Rep. 2015, 5, 8724. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Uchihashi, T.; Kobashi, T.; Shibata, M.; Nishiyama, J.; Yasuda, R.; Ando, T. Wide-area scanner for high-speed atomic force microscopy. Rev. Sci. Instrum. 2013, 84, 053702. [Google Scholar] [CrossRef] [PubMed]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.A.; Martins, I.C.; Santos, N.C. Atomic force microscopy and force spectroscopy on the assessment of protein folding and functionality. Arch. Biochem. Biophys. 2013, 531, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Thoma, J.; Bosshart, P.; Pfreundschuh, M.; Muller, D.J. Out but not in: The large transmembrane β-barrel protein fhua unfolds but cannot refold via β-hairpins. Structure 2012, 20, 2185–2190. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.J.; Engel, A. Atomic force microscopy and spectroscopy of native membrane proteins. Nat. Protoc. 2007, 2, 2191–2197. [Google Scholar] [CrossRef] [PubMed]
- Senapati, S.; Lindsay, S. Recent progress in molecular recognition imaging using atomic force microscopy. Acc. Chem. Res. 2016, 49, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, L.; Xi, N.; Wang, Y. Nanoscale monitoring of drug actions on cell membrane using atomic force microscopy. Acta Pharmacol. Sin. 2015, 36, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Hinterdorfer, P.; Dufrene, Y.F. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods 2006, 3, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhang, X.; Xia, T.; Fang, X. Living cell study at the single-molecule and single-cell levels by atomic force microscopy. Nanomedicine 2012, 7, 1625–1637. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Wang, Y.M.; Huang, L.S.; Lin, S. Atomic force microscopy: Determination of unbinding force, off rate and energy barrier for protein-ligand interaction. Micron 2007, 38, 446–461. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, L.; Xi, N.; Wang, Y.; Dong, Z.; Xiao, X.; Zhang, W. Mapping CD20 molecules on the lymphoma cell surface using atomic force microscopy. Chin. Sci. Bull. 2013, 58, 1516–1519. [Google Scholar] [CrossRef]
- Li, M.; Xiao, X.; Zhang, W.; Liu, L.; Xi, N.; Wang, Y. AFM analysis of the multiple types of molecular interactions involved in rituximab lymphoma therapy on patient tumor cells and NK cells. Cell. Immunol. 2014, 290, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Ido, S.; Kimiya, H.; Kobayashi, K.; Kominami, H.; Matsushige, K.; Yamada, H. Immunoactive two-dimensional self-assembly of monoclonal antibodies in aqueous solution revealed by atomic force microscopy. Nat. Mater. 2014, 13, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Sutter, M.; Faulkner, M.; Aussignargues, C.; Paasch, B.C.; Barrett, S.; Kerfeld, C.A.; Liu, L.N. Visualization of bacterial microcompartment facet assembly using high-speed atomic force microscopy. Nano Lett. 2016, 16, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Sakiyama, Y.; Mazur, A.; Kapinos, L.E.; Lim, R.Y.H. Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high-speed atomic force microscopy. Nat. Nanotechnol. 2016, 11, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Ido, S.; Kimura, K.; Oyabu, N.; Kobayashi, K.; Tsukada, M.; Matsushige, K.; Yamada, H. Beyond the helix pitch: Direct visualization of native DNA in aqueous solution. ACS Nano 2013, 7, 1817–1822. [Google Scholar] [CrossRef] [PubMed]
- Lyubchenko, Y.L.; Shlyakhtenko, L.S.; Ando, T. Imaging of nucleic acids with atomic force microscopy. Methods 2011, 54, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Sarduy, L.; Longo, G.; Dietler, G.; Kasas, S. Time-lapse AFM imaging of DNA conformational changes induced by daunorubicin. Nano Lett. 2013, 13, 5679–5684. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Sugiyama, H. Single-molecule imaging of dynamic motions of biomolecules in DNA origami nanostructures using high-speed atomic force microscopy. Acc. Chem. Res. 2014, 47, 1645–1653. [Google Scholar] [CrossRef] [PubMed]
- Bujalowski, P.J.; Oberhauser, A.F. Tracking unfolding and refolding reactions of single proteins using atomic force microscopy methods. Methods 2013, 60, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Rief, M.; Gautel, M.; Oesterhelt, F.; Fernandez, J.M.; Gaub, H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 1997, 276, 1109–1112. [Google Scholar] [CrossRef] [PubMed]
- Fisher, T.E.; Marszalek, P.E.; Fernandez, J.M. Stretching single molecules into novel conformations using the atomic force microscope. Nat. Struct. Biol. 2000, 7, 719–724. [Google Scholar] [PubMed]
- Li, M.; Liu, L.; Xi, N.; Wang, Y.; Dong, Z.; Xiao, X.; Zhang, W. Progress of AFM single-cell and single-molecule morphology imaging. Chin. Sci. Bull. 2013, 58, 3177–3182. [Google Scholar] [CrossRef]
- Alsteens, D.; Dupres, V.; Klotz, S.A.; Gaur, N.K.; Lipke, P.N.; Dufrene, Y.F. Unfolding individual als5p adhesion proteins on live cells. ACS Nano 2009, 3, 1677–1682. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Hu, C.; Hu, X.; Hu, X.; Xiao, A.; Perkins, T.T.; Li, H. Direct observation of the reversible two-state unfolding and refolding of an α/β protein by single-molecule atomic force microscopy. Angew. Chem. 2015, 127, 10059–10063. [Google Scholar] [CrossRef]
- Rico, F.; Gonzalez, L.; Casuso, I.; Puig-Vidal, M.; Scheuring, S. High-speed force spectroscopy unfolds titin at the velocity of molecular dynamics simulations. Science 2013, 342, 741–743. [Google Scholar] [CrossRef] [PubMed]
- Florin, E.L.; Moy, V.T.; Gaub, H.E. Adhesion forces between individual ligand-receptor pairs. Science 1994, 264, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Hinterdorfer, P.; Baumgartner, W.; Gruber, H.J.; Schilcher, K.; Schindler, H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA 1996, 93, 3477–3481. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.A.; Santos, N.C. Atomic force microscopy-based force spectroscopy-biological and biomedical applications. IUBMB Life 2012, 64, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, A.; Ros, R. Single-molecule force spectroscopy: A method for quantitative analysis of ligand-receptor interactions. Nanomedicine 2010, 5, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Ebner, A.; Wildling, L.; Kamruzzahan, A.S.M.; Rankl, C.; Wruss, J.; Hahn, C.D.; Holzl, M.; Zhu, R.; Kienberger, F.; Blaas, D.; Hinterdorfer, P.; Gruber, H.J. A new, simple method for linking of antibodies to atomic force microscopy tips. Bioconjug. Chem. 2007, 18, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Stroh, C.; Wang, H.; Bash, R.; Ashcroft, B.; Nelson, J.; Gruber, H.; Lohr, D.; Lindsay, S.M.; Hinterdorfer, P. Single-molecule recognition imaging microscopy. Proc. Natl. Acad. Sci. USA 2004, 101, 12503–12507. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Rupprecht, A.; Ebner, A.; Haselgrubler, T.; Gruber, H.J.; Hinterdorfer, P.; Pohl, E.E. Mapping the nucleotide binding site of uncoupling protein 1 using atomic force microscopy. J. Am. Chem. Soc. 2013, 135, 3640–3646. [Google Scholar] [CrossRef] [PubMed]
- Oddershede, L.B. Force probing of individual molecules inside the living cell is now a reality. Nat. Chem. Biol. 2012, 8, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Taatjes, D.J.; Quinn, A.S.; Rand, J.H.; Jena, B.P. Atomic force microscopy: High resolution dynamic imaging of cellular and molecular structure in health and disease. J. Cell. Physiol. 2013, 228, 1949–1955. [Google Scholar] [CrossRef] [PubMed]
- Pfister, G.; Stroh, C.M.; Perschinka, H.; Kind, M.; Knoflach, M.; Hinterdorfer, P.; Wick, G. Detection of HSP60 on the membrane surface of stressed human endothelial cells by atomic force and confocal microscopy. J. Cell Sci. 2005, 118, 1587–1594. [Google Scholar] [CrossRef]
- Puntheeranurak, T.; Wildling, L.; Gruber, H.J.; Kinne, R.K.H.; Hinterdorfer, P. Ligands on the string: Single-molecule AFM studies on the interaction of antibodies and substrates with the Na+-glucose co-transporter SGLT1 in living cells. J. Cell Sci. 2006, 119, 2960–2967. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, Q.; Shi, X.; Ma, X.; Yang, H.; Chen, Y.G.; Fang, X. Single-molecule force spectroscopy study of interaction between transforming growth factor β1 and its receptor in living cells. J. Phys. Chem. B 2007, 111, 13619–13625. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, B.H.; Louban, I.; Djandji, D.; Wabnitz, G.H.; Deeg, J.; Bulbuc, N.; Samstag, Y.; Gunzer, M.; Spatz, J.P.; Hammerling, G.J. Immune synapse formation determines interaction forces between T cells and antigen-presenting cells measured by atomic force microscopy. Proc. Natl. Acad. Sci. USA 2009, 106, 17852–17857. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.A.; Connell, S.; Miltenberger-Miltenyi, G.; Pereira, S.V.; Tavares, A.; Ariens, R.A.S.; Santos, N.C. Atomic force microscopy-based molecular recognition of a fibrinogen receptor on human erythrocytes. ACS Nano 2010, 4, 4609–4620. [Google Scholar] [CrossRef] [PubMed]
- Beaussart, A.; Alsteens, D.; El-Kirat-Chatel, S.; Lipke, P.N.; Kucharikova, S.; Dijck, P.V.; Dufrene, Y.F. Single-molecule imaging and functional analysis of alsadhesins and mannans during candida albicans morphogenesis. ACS Nano 2012, 6, 10950–10964. [Google Scholar] [PubMed]
- Li, M.; Liu, L.; Xi, N.; Wang, Y.; Xiao, X.; Zhang, W. Imaging and measuring the biophysical properties of Fc gamma receptors on single macrophages using atomic force microscopy. Biochem. Biophys. Res. Commun. 2013, 438, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, J.; Schubert, R.; Chiquet-Ehrismann, R.; Muller, D.J. Deciphering teneurin domains that facilitate cellular recognition, cell-cell adhesion, and neurite outgrowth using atomic force microscopy-based single-cell force spectroscopy. Nano Lett. 2013, 13, 2937–2946. [Google Scholar] [CrossRef] [PubMed]
- Puntheeranurak, T.; Neundlinger, I.; Kinne, R.K.H.; Hinterdorfer, P. Single-molecule recognition force spectroscopy of transmembrane transporters on living cells. Nat. Protoc. 2011, 6, 1443–1452. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Garcia, M.C.; Lipke, P.N.; Dufrene, Y.F. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc. Natl. Acad. Sci. USA 2010, 107, 20744–20749. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chtcheglova, L.A.; Zhu, R.; Hinterdorfer, P.; Zhang, B.; Tang, J. Nanoscale organization of human GnRH-R on human bladder cancer cells. Anal. Chem. 2014, 86, 2458–2464. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Trabelsi, H.; Soumillion, P.; Dufrene, Y.F. Multiparametric atomic force microscopy imaging of single bacteriophages extruding from living bacteria. Nat. Commun. 2013, 4, 2926. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xiao, X.; Liu, L.; Xi, N.; Wang, Y.; Dong, Z.; Zhang, W. Nanoscale mapping and organization analysis of target proteins on cancer cells from B-cell lymphoma patients. Exp. Cell Res. 2013, 319, 2812–2821. [Google Scholar] [CrossRef] [PubMed]
- Baskar, S.; Kwong, K.Y.; Hofer, T.; Levy, J.M.; Kennedy, M.G.; Lee, E.; Staudt, L.M.; Wilson, W.H.; Wiestner, A.; Rader, C. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B-cell chronic lymphocytic leukamia. Clin. Cancer Res. 2008, 14, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Bicocca, V.T.; Chang, B.H.; Masouleh, B.K.; Muschen, M.; Loriaux, M.M.; Druker, B.J.; Tyner, J.W. Crosstalk between ROR1 and the pre-B cell receptor promotes survival of t(1;19) acute lymphoblastic leukemia. Cancer Cell 2012, 22, 656–667. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xiao, X.; Zhang, W.; Liu, L.; Xi, N.; Wang, Y. Nanoscale distribution of CD20 on B-cell lymphoma tumour cells and its potential role in the clinical efficacy of rituximab. J. Microsc. 2014, 254, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Dufrene, Y.F.; Evans, E.; Engel, A.; Helenius, J.; Gaub, H.E.; Muller, D.J. Five challenges to bringing single-molecule force spectroscopy into living cells. Nat. Methods 2011, 8, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zou, Q. High-speed adaptive contact-mode atomic force microscopy imaging with near-minimum-force. Rev. Sci. Instrum. 2014, 85, 073706. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yadavalli, V.K. Investigating biomolecular recognition at the cell surface using atomic force microscopy. Micron 2014, 60, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Leduc, C.; Si, S.; Gautier, J.; Soto-Ribeiro, M.; Wehrle-Haller, B.; Gautreau, A.; Giannone, G.; Gognet, L.; Lounis, B. A highly specific gold nanoprobe for live-cell single-molecule imaging. Nano Lett. 2013, 13, 1489–1494. [Google Scholar] [CrossRef] [PubMed]
- Lekka, M. Discrimination between normal and cancerous cells using AFM. Bionanoscience 2016, 6, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, D.; Uchihashi, T.; Kodera, N.; Yamashita, H.; Nishikori, S.; Ogura, T.; Shibata, M.; Ando, T. High-speed atomic force microscopy techniques for observing dynamic biomolecular processes. Methods Enzymol. 2010, 475, 541–563. [Google Scholar] [PubMed]

© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Dang, D.; Liu, L.; Xi, N.; Wang, Y. Imaging and Force Recognition of Single Molecular Behaviors Using Atomic Force Microscopy. Sensors 2017, 17, 200. https://doi.org/10.3390/s17010200
Li M, Dang D, Liu L, Xi N, Wang Y. Imaging and Force Recognition of Single Molecular Behaviors Using Atomic Force Microscopy. Sensors. 2017; 17(1):200. https://doi.org/10.3390/s17010200
Chicago/Turabian StyleLi, Mi, Dan Dang, Lianqing Liu, Ning Xi, and Yuechao Wang. 2017. "Imaging and Force Recognition of Single Molecular Behaviors Using Atomic Force Microscopy" Sensors 17, no. 1: 200. https://doi.org/10.3390/s17010200
APA StyleLi, M., Dang, D., Liu, L., Xi, N., & Wang, Y. (2017). Imaging and Force Recognition of Single Molecular Behaviors Using Atomic Force Microscopy. Sensors, 17(1), 200. https://doi.org/10.3390/s17010200






