Vision Marker-Based In Situ Examination of Bacterial Growth in Liquid Culture Media
Abstract
:1. Introduction
2. Materials and Methods
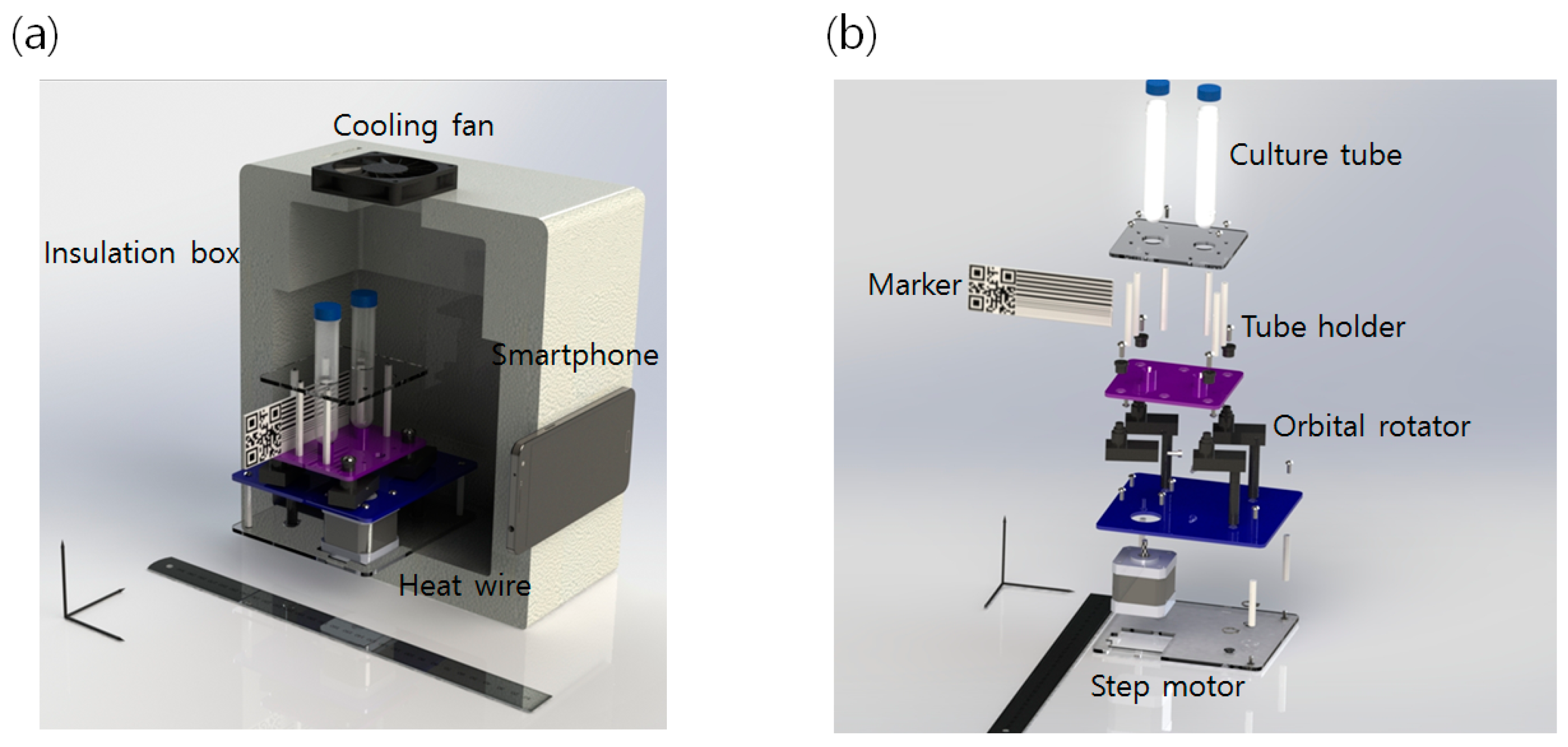
2.1. Platform Overview
2.2. Bacterial Culture and Experiment Settings
2.3. Image Processing Algorithm
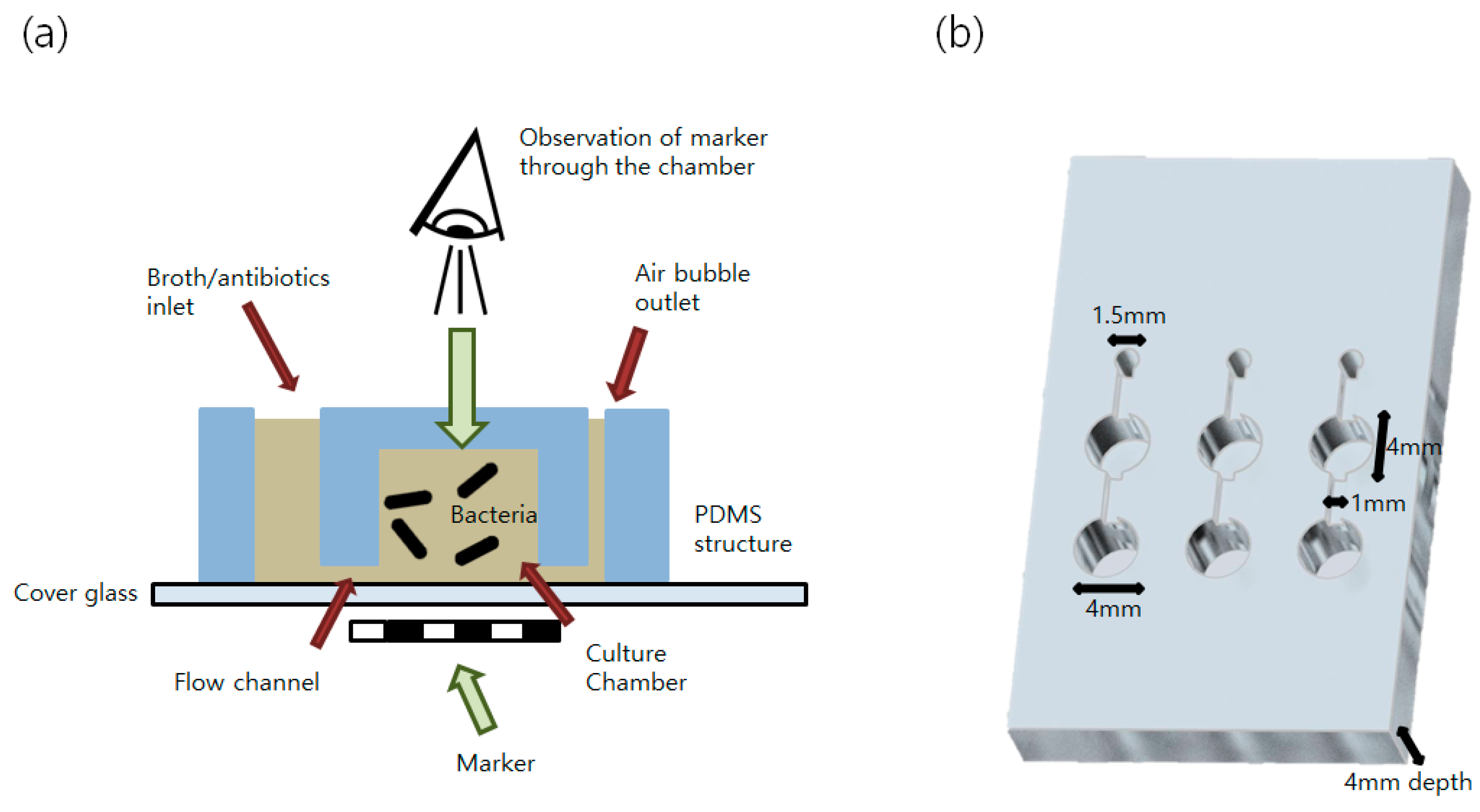
2.4. Preparation of Millifluidic Culture Environment
3. Results and Discussion
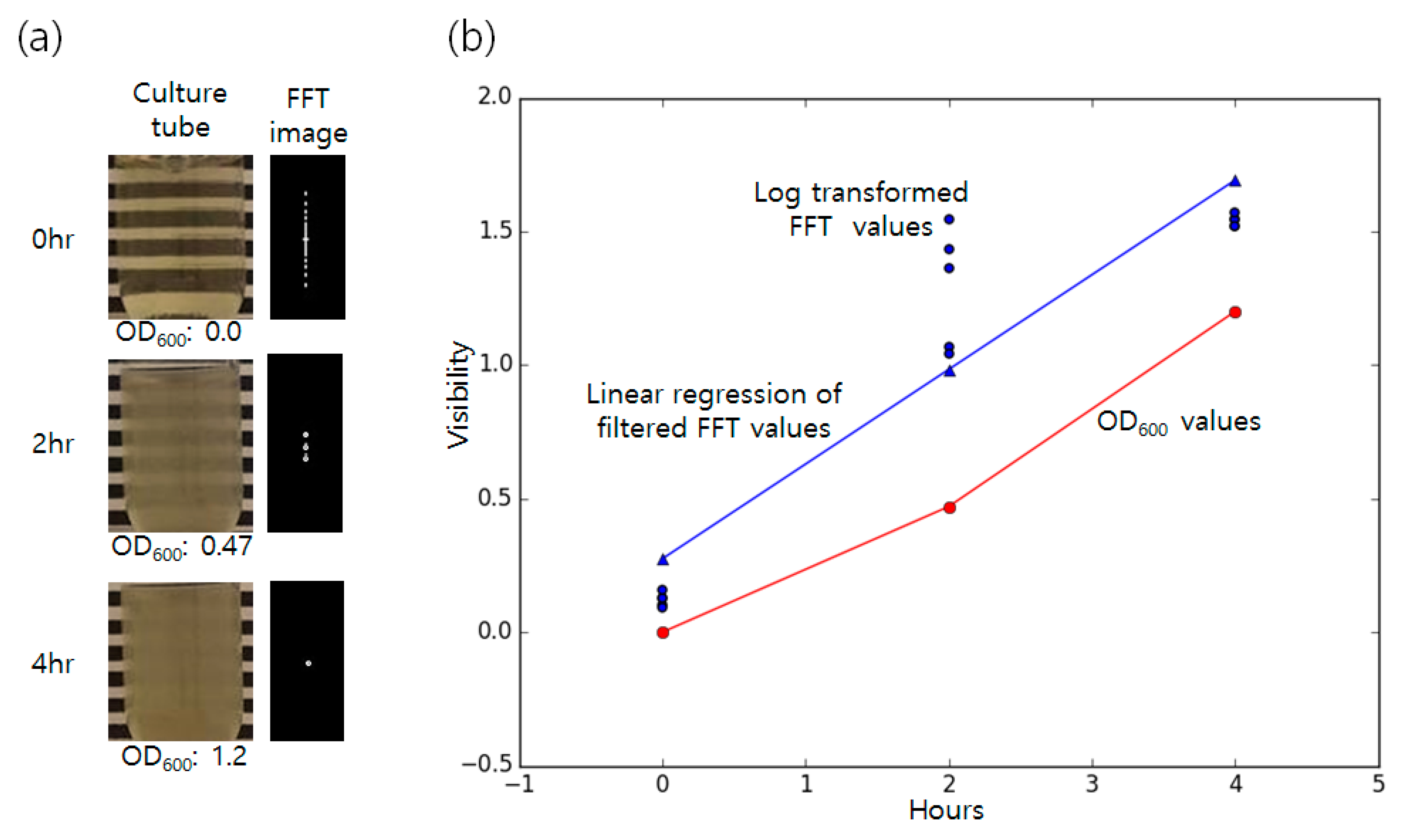
3.1. Detection of Bacterial Growth in Liquid Broth
3.2. Expansion of the Concept to the Lab on a Chip Device
4. Conclusions and Future Works
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Anzalone, G.C.; Glover, A.G.; Pearce, J.M. Open-Source Colorimeter. Sensors 2013, 13, 5338–5346. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, H.; Lim, H.; Myung, H. A Low Cost/Low Power Open Source Sensor System for Automated Tuberculosis Drug Susceptibility Testing. Sensors 2016, 16, 942. [Google Scholar] [CrossRef] [PubMed]
- Toyoura, M.; Aruga, H.; Turk, M. Detecting Markers in Blurred and Defocused Images. In Proceedings of the 2013 International Conference on Cyberworlds (CW 2013), Yokohama, Japan, 21–23 October 2013; pp. 183–190.
- Grünberger, A.; van Ooyen, J.; Paczia, N.; Rohe, P.; Schiendzielorz, G.; Eggeling, L.; Wiechert, W.; Kohlheyer, D.; Noack, S. Beyond growth rate 0.6: Corynebacterium glutamicum cultivated in highly diluted environments. Biotechnol. Bioeng. 2013, 110, 220–228. [Google Scholar] [PubMed]
- Korea Society for Medical Microbiology. Medical Microbiology, 3rd ed.; Hyunmoon: Seoul, Korea, 2004. (In Korean) [Google Scholar]
- Choi, J.; Yoo, J.; Lee, M.; Kim, E.; Lee, J.; Lee, S.; Joo, S.; Song, S.; Kim, E.; Lee, J.; et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014, 6, 267ra174. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Han, S.; Jeon, J.S.; Yamamoto, K.; Zervantonakis, I.K.; Sudo, R.; Kamm, R.D.; Chung, S. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat. Protoc. 2012, 7, 1247–1259. [Google Scholar] [CrossRef] [PubMed]
- Garnacho-Monteno, J.; Albado-Pallas, T.; Garnacho-Monteno, C.; Cayuela, A.; Jimenez, R.; Barroso, S.; Ortiz-Leyba, C. Timing of adequate antibiotic therapy is greater determinant of outcome than are TNF and IL-10 polymorphism in patients with sepsis. Crit. Care 2008, 10, R111. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Bae, H. Comparison of Marine Luminescence Bacteria and Genetically Modified Luminescence E. coli, for Acute Toxicity of Heavy Metals. J. Korea Soc. Environ. Eng. 2005, 27, 900–906. [Google Scholar]
- Cho, K.; Koo, S.; Kim, J.; Ryu, H. Quantification of Inhibitory Impact of Heavy Metals on the Growth of Escherichia coli. Korean J. Microbiol. Biotechnol. 2004, 32, 341–346. [Google Scholar]
- Kim, K.; Myung, H. Sensor Node for Remote Monitoring of Waterborne Disease-Causing Bacteria. Sensors 2015, 15, 10569–10579. [Google Scholar] [CrossRef] [PubMed]


| OD | FFT | |
|---|---|---|
| Cost | High | Low |
| Maintenance | Hard | Easy |
| Setup | Precise | Rough |
| Automation | Hard | Easy |
| Resolution | Standard | Lower |
| Accuracy | Standard | Bit Lower |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Choi, D.; Lim, H.; Kim, H.; Jeon, J.S. Vision Marker-Based In Situ Examination of Bacterial Growth in Liquid Culture Media. Sensors 2016, 16, 2179. https://doi.org/10.3390/s16122179
Kim K, Choi D, Lim H, Kim H, Jeon JS. Vision Marker-Based In Situ Examination of Bacterial Growth in Liquid Culture Media. Sensors. 2016; 16(12):2179. https://doi.org/10.3390/s16122179
Chicago/Turabian StyleKim, Kyukwang, Duckyu Choi, Hwijoon Lim, Hyeongkeun Kim, and Jessie S. Jeon. 2016. "Vision Marker-Based In Situ Examination of Bacterial Growth in Liquid Culture Media" Sensors 16, no. 12: 2179. https://doi.org/10.3390/s16122179
APA StyleKim, K., Choi, D., Lim, H., Kim, H., & Jeon, J. S. (2016). Vision Marker-Based In Situ Examination of Bacterial Growth in Liquid Culture Media. Sensors, 16(12), 2179. https://doi.org/10.3390/s16122179







