Abstract
An enhanced understanding of the temporal dynamics of intraspecific diversity is anticipated to improve the adequacy of conservation priorities, methods and metrics. We report on the comparative genetic composition of ex- and in-situ landrace cultivar populations from a potato diversity hotspot in the Andes. A total of 989 landrace cultivars belonging to contemporary custodian-farmer in situ collections from central Peru were compared with 173 accessions from a spatially analogous, but temporally differential ex situ composite genotype reference (CGR) set using 15 nuclear microsatellite markers. A total of 173 alleles were detected, with 129 alleles (74.6%) being shared between both populations. Both populations contain exclusive allelic diversity with 32 and 12 unique alleles belonging to the ex- and in-situ population, respectively. The mean unbiased expected heterozygosity values of the ex- and in-situ population are very similar, 0.749 versus 0.727, with a slightly wider range and standard deviation encountered for the in situ population. Analysis of Molecular Variance shows that 98.8% of the total variation is found within both populations, while the fixation index (Fst = 0.01236) corroborates that the populations are not well differentiated. Surprisingly, only 41.0% of the ex situ population encounters a similar landrace cultivar in 23.4% of the in situ population at a non-stringent threshold similarity coefficient of 0.80. While the ex- and in-situ population under comparison show similarities and unique features at the allelic level, their landrace cultivar composition is surprisingly distinct. Results affirm that crop evolution is an ongoing phenomenon and that change in fixed geographies is occurring.
1. Introduction
The potato is a highly heterozygous outcrossing crop with ploidy levels ranging from the diploid to the pentaploid level. In the Andes, four cultivated species occur: (i) Solanum tuberosum (diploid, triploid and tetraploid Andigenum group); (ii) S. ajanhuiri, (iii) S. juzepzukii, (iv) S. curtilobum [1,2]. The crop is characterized by high levels of intraspecific diversity with thousands of landrace cultivars occurring from 42° south (Chile) to 5° north (Colombia) of the equator. A landrace cultivar is a farmer-selected variety and a product of the natural and cultural environment in which it has developed [3]. It has not been exposed to formal crop breeding schemes and is clearly distinct, uniform and stable in its characteristics and, when propagated by appropriate means, retains those characteristics [4]. By far the highest landrace cultivar diversity is found within Solanum tuberosum, particularly the tetraploid Andigenum group, which constitutes 77% of the total accessions of potato landrace cultivars maintained ex situ by the International Potato Center (CIP). This collection has been intensively characterized and rationalized to reduce redundancy.
From the late 1920s till the early 1990s, potato collection expeditions by American [5], Dutch [6], English [7], Peruvian [8,9,10] and Russian [11] scientists, among researchers from many other nationalities, have contributed to the establishment of important ex situ collections that are currently maintained in gene banks around the world. The signature of the Convention of Biological Diversity (CBD) in 1992 and the adoption of national regulatory frameworks for access to genetic resources in Andean countries have led to a significant reduction of international potato collection missions. Nevertheless, these ex situ collections continue to provide valuable genetic diversity to potato breeding programs around the world in order to continue to deliver bred cultivars adjusted to a changing world in its climatic, as well as human dimensions. At the same time, interventions designed to support in situ conservation have increased throughout the Andes [12]. In situ conservation of cultivated potato genetic resources in the Andes is a contemporary phenomenon, and diverse landrace cultivar pools continue to be actively managed by farmers [13,14]. It is unlikely that present-day landrace cultivar populations and their inherent genetic diversity are an exact copy of what was found on-farm at the time of the collection missions. However, systematic comparisons of the structure of landrace cultivar populations in fixed geographical spaces along a well-defined timeline are scarce, as it is generally difficult to quantify change. A few cases, however, have been documented. For example, Monteros [14] showed that between 2008–2010, more potato landrace cultivars were found in the provinces of Chimborazo and Loja in Ecuador compared to what was reported by collecting missions in the 1970s and 1980s. Vigouroux et al. [15], comparing sorghum cultivars in the same region of Niger between 1976 and 2003, found no major changes in the main cultivars or in their genetic diversity. However, these authors observed a significant shift in adaptive traits.
Temporal variation in genetic structure and landrace cultivar composition in fixed geographical space is a likely consequence of complex biological or social change processes. Initial gaps in baseline data for timeline comparisons, such as species, landrace cultivar, hotspot or infrastructure biases [16], make comparative analysis difficult. The occurrence of potato landrace cultivars in direct proximity to wild relatives, somatic mutations and farmer selection practices are likely to have maintained ongoing diversification and evolution [17,18,19]. Also, landrace cultivar loss and possible genetic erosion may have occurred as a result of extreme natural and human-induced events. Rapidly changing social and environmental conditions in the Andes, such as migration, increased farmer mobility and involvement in off-farm employment, diffusion of bred and commercial landrace cultivars and changing seed-tuber procurement practices, are also likely to play their part in the dynamics of landrace cultivar populations in fixed geographical spaces. Indeed, it is now well recognized that the loss of crop genetic diversity is a complex and context-dependent process conditioned by local differences in environment, culture and economy [20]. However, potato landrace cultivar loss and genetic erosion are not directly correlated, since the potato is a highly heterozygous crop sharing alleles across geography and ploidy. Such genetic features of the crop make the present study particularly relevant in order to distinguish loss of genotypes from loss of alleles, which are usually confounded for autogamous crops.
Different molecular marker systems have been used to compare the population genetic structure of ex- and in-situ populations of crops and wild plant species [21,22,23]. Simple sequence repeat (SSR) or microsatellite markers have also been used for this purpose, including for maize [24], beans [25] and grasses [26], and, indeed, are a robust tool to research the genetic structure and diversity of crop populations [27,28,29]. SSR markers are ideal for paternity determination, identity verification, comparative genetics and population genetic studies, because of their high specificity, high polymorphism, good reproducibility and unambiguous scorability [30,31]. For potato, they are widely used for research concerning genetic diversity [32,33], phylogeny and gene pool structure [2,34,35], as well as routine identification of duplicates [36]. So far, SSR markers have not been used to compare ex- and in situ populations of potato landrace cultivars along a well-defined timeline. Here, we provide a benchmark comparison for a spatially restricted area within the potato’s center of origin and diversity.
Systematic time-line comparisons of the population genetics and composition of potato landrace cultivars in well-defined hotspots within the potato’s center of origin will enhance our understanding of the temporal dynamics of intraspecific diversity. This, in turn, can help to inform conservation priorities, as well as adequate methods and metrics. Knowing whether the allelic and landrace cultivar in situ diversity has significantly shifted or remained relatively static within a 40-year timeframe may provide lessons for what crop conservation scientists consider to be geographically representative populations and how we conceptualize farmer-driven in situ conservation. Is the long-term conservation of landrace cultivars and inherent allelic diversity static or dynamic? What does temporal (non-)change teach us about spatial dynamics and representativeness of diversity in fixed geographies? When we monitor in situ conservation, should we consider total, new or lost or unique diversity as indicators? Or a combination? These types of research questions are particularly important for the International Potato Center (CIP), as the institution has recently embarked on the establishment of long-term monitoring sites in selected hotspots throughout the potato’s center of origin and diversity with a view to document how agrobiodiversity is evolving in light of socioeconomic and environmental change.
2. Experimental Section
2.1. Site Description
Huancavelica is one of Peru’s 24 departments (19 of which produce potato). The department covers 22,131 km2 and 80% of its territory and is located between 3,000 and 4,500 m above sea level. The total annual potato cropping area covers around 12,000 ha and is comprised of more than 86,000 individual farm units [37]. Huancavelica is a well-recognized hotspot of intraspecific diversity of the potato with a total stock of well over 400 distinct landrace cultivars reported to be present in its territory [38]. The highlands of Huancavelica used to be very isolated, but have opened-up as peace returned to the countryside in the late 1990s, and newly paved roads were constructed between 2003 and 2010. The earliest and most voluminous CIP collection from Huancavelica was established between 1972 and 1974 (n = 344; 64.8%), with accessions being added to the ex situ collection in 1985 (n = 7; 1.3%) and 1992–1994 (n = 180; 33.9%). Today, it consists of 143 accessions kept as tubers and 108 accessions conserved as in vitro materials for long-term storage and distribution.
2.2. Materials
DNA was extracted from leaf samples from a total of 989 landrace cultivars representing the total diversity conserved by 8 custodian farmers in their family stocks. These custodian farmers are individuals with large family heritage collections of landrace cultivars. Individual farmers contributed a minimum of 57 and maximum of 175 landrace cultivars. All together, they provided the total landrace cultivar sample defined as the in situ population for the purpose of the research reported here. The in situ population covered 3 of Huancavelica’s 7 provinces and 5 of the department’s 565 farmer communities (comunidades campesinas) along a north-south pseudo-transect. The participating custodian farmers were well recognized by their home communities as having particularly rich and representative landrace cultivar collections. Landrace cultivars were established in on-farm trials, with single replicate plots containing 5 to 10 plants per landrace cultivar. A single DNA sample was taken from one representative plant from each farmer-recognized landrace cultivar. The total in situ population has been molecularly characterized with 18 SSR markers [13]. CIP routinely uses a well-defined composite genotype reference (CGR) set consisting of 742 accessions for comparative purposes. It concerns a representative subpopulation containing 16% of the landrace accessions of the gene bank collection with adequate geographical, ploidy, species and genetic coverage. The CGR set has been molecularly characterized with 50 different SSR markers [2]. We used a spatially restricted subset (the ex situ population) of 173 landrace cultivar accessions from the department of Huancavelica and its 3 neighboring departments (Lima, Junín and Ayacucho) for the purpose of comparing a spatially well-defined and representative ex situ population with a contemporary in situ population. A wider geospatial coverage for the ex situ sample was chosen to compensate for the fact that the original gene bank collection had been rationalized in the early 1990s. Duplicate accessions from Huancavelica that were eliminated from the CIP gene bank collection as part of this exercise are partially captured by using an expanded geographical range for the representative ex situ population. Landrace accessions from neighboring departments were also included, because past seed-tuber exchange is likely to have resulted in considerable overlap between the landrace cultivar portfolios of these departments (Figure 1). Both the ex- and in-situ populations are composed of diploid and polyploid landrace cultivars (2n = 2x = 24 to 2n = 5x = 60) belonging to multiple cultivated species and cultivar groups.
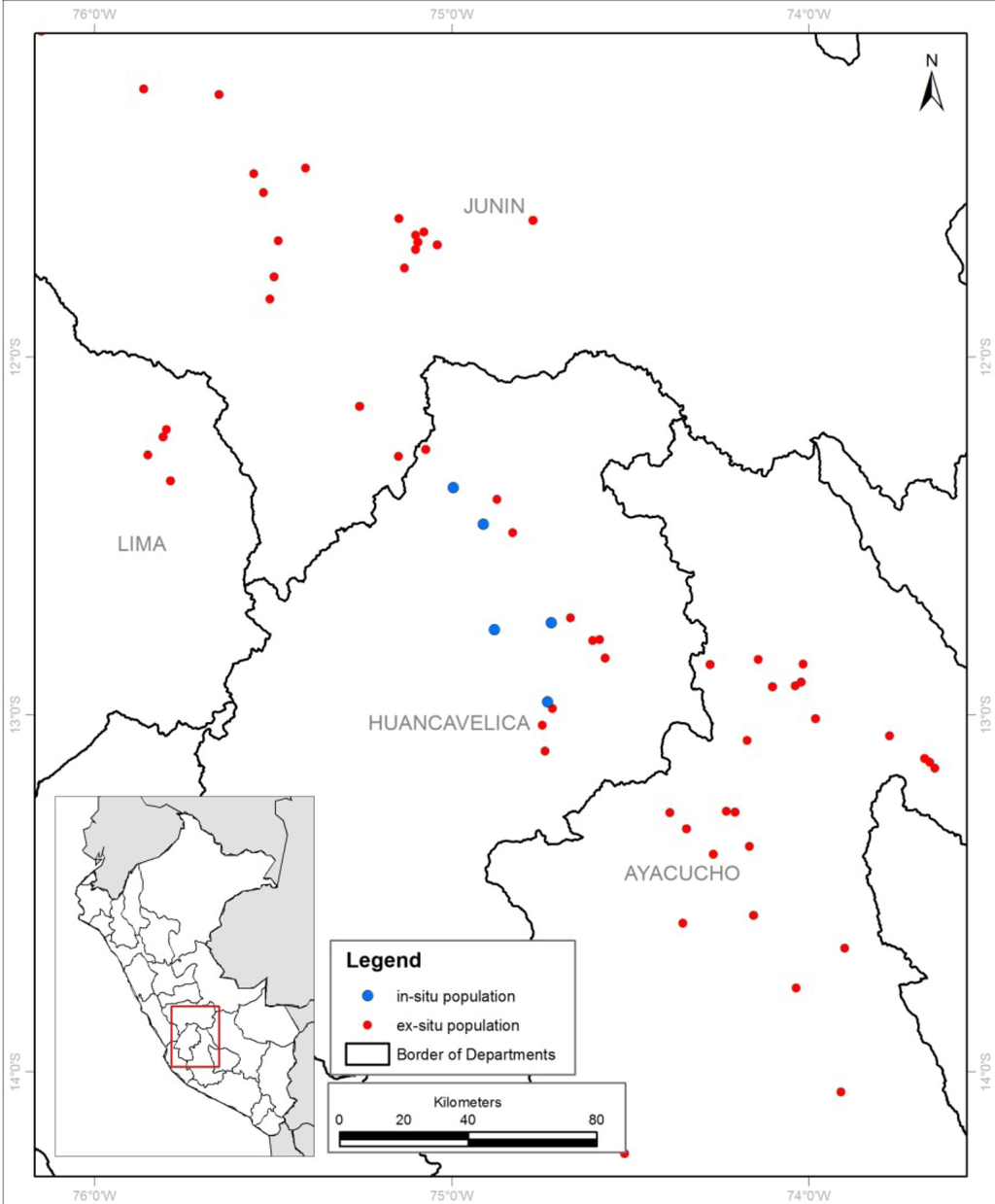
Figure 1.
Geospatial coverage of the samples composing the in- and ex-situ population.
2.3. Methods
Standard procedures as practiced at CIP were applied, including DNA extraction with DNeasy 96 plant kits, high throughput genotyping with a LI-COR 4300 DNA Analysis System and SSR allele scoring with SAGA Generation 2 software (LI-COR). A genetic identity kit with highly polymorphic SSR markers was used for genetic fingerprinting [36]. A primer combination was chosen to cover the whole genome and for high Polymorphism Index content [39]. The alleles were scored as present (1) or absent (0), while missing data were scored as ‘‘9’’. The total fingerprinted in situ population (n = 989) was compared with the spatially restricted ex situ CGR set (n = 173) from central Peru. The 989 and 173 landrace cultivars from the in- and ex-situ populations had good quality data (<0.3% missing data). Data from 15 SSR primer combinations were shared between both populations and used for comparative analysis (Table 1). The ploidy level of most landrace cultivars was determined applying microscopy and/or flow cytometry with a Partec® ploidy analyzer. The complete dataset is available as supplementary data and can be accessed from the website [40]

Table 1.
Coverage of the simple sequence repeat (SSR) markers applied.
| SSR Name | Source of markers (*) | Repeat motif | Map location (chromosome ) |
|---|---|---|---|
| STM5127 | SCRI | (TCT)n | I |
| STG0006 | TIGR | (AC)n | II |
| STI0036 | IDAHO | (AC)n (TC)imp | II |
| STG0010 | TIGR | (TG)n | III |
| STI0032 | IDAHO | (GGA)n | V |
| STM0031 | SCRI | (AC)n (AC)n GCAC (AC)n (GCAC)n | VII |
| STI0003 | IDAHO | (ACC)n | VIII |
| STI0022 | IDAHO | (ACCCG)n | VIII |
| STGBSS | SCRI | (TCT)n | VIII |
| STM1052 | SCRI | (AT)n GT (AT)n (GT)n | IX |
| STI0014 | IDAHO | (TGG)n (AGG)n | IX |
| STI0023 | IDAHO | (CAG)n | X |
| STM1106 | SCRI | (ATT)n | X |
| STM0037 | SCRI | (TC)n (AC)n AA (AC)n (AT)n | XI |
| STI0030 | IDAHO | (ATT)n | XII |
* SCRI (Scottish Crop Research Institute (currently the James Hutton Institute); Dundee, Scotland, UK); TIGR (The Institute of Genomic Research; Rockville, USA); IDAHO (University of Idaho; Moscow, USA).
2.4. Statistical Analysis UPGMA
Dissimilarity analysis for the populations was conducted using the Jaccard coefficient and UPGMA clustering method applying NTSYS-pc 2.2 software [41]. Principal Component Analysis (PCA) was conducted, and dissimilarity trees (dendrograms) were constructed using an unweighted neighbor joining (NJ) clustering method for a dissimilarity matrix calculated with the Jaccard’s coefficient using DARwin 5.0 software [42]. Unbiased expected heterozygosity (Heu) values were calculated for both populations and all SSR markers. Additionally, relative allele frequencies for the in- and ex-situ populations were calculated. The population genetic structure of the spatially restricted ex situ and contemporary in situ populations were compared applying Analysis of Molecular Variance (AMOVA) with Arlequin 3.5.1.3 software [43].
3. Results
A total of 173 alleles were detected within the two populations under comparison with a total of 161 and 141 alleles encountered in the ex- and in-situ populations, respectively. A total of 129 alleles occurred in both the ex- and in-situ populations, while 32 and 12 alleles belonged uniquely to the ex- and in-situ populations, representing 19.9% and 8.5%, respectively, of the total number of alleles accounted for within each population. In all cases, the largest proportion of alleles detected with individual SSR markers were shared between populations (Figure 2). Most markers detected unique alleles for both the ex- and in-situ populations. Five individual markers detected unique alleles restricted to one of the populations: STI0030, STI0032, STM1052, STG0006 and STI0022.
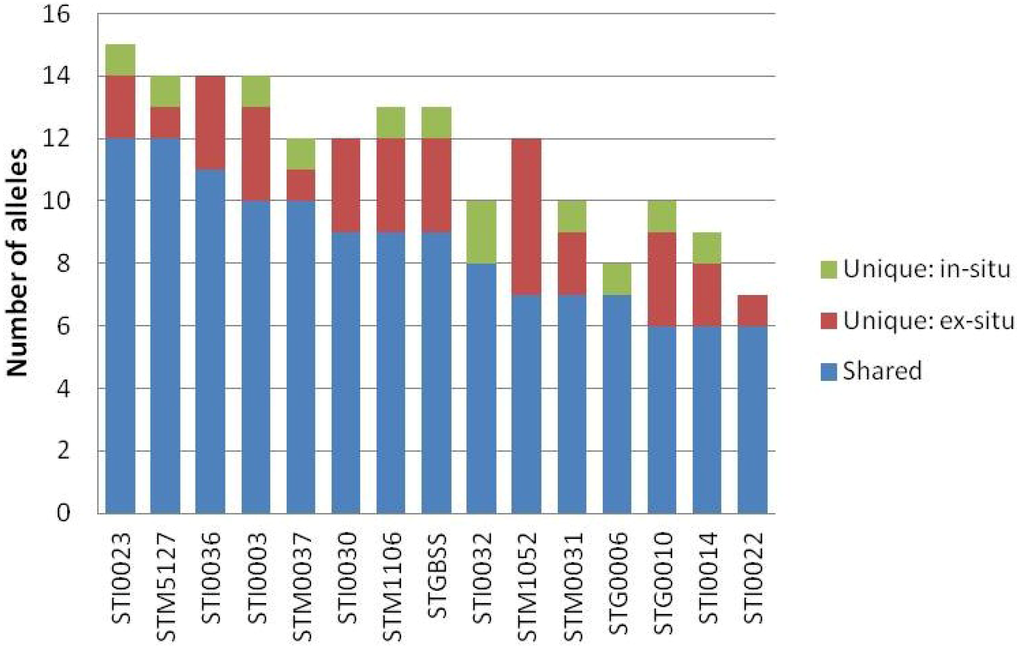
Figure 2.
Absolute number of alleles shared between the ex- and in-situ populations and absolute number of alleles restricted to each population detected with each of 15 SSR.
Most of the alleles detected in the ex- and in-situ population were frequent (Table 2). The in situ population is characterized by a high presence of rare alleles indicating uneven distribution patterns in farmer’s landrace cultivar stocks. All of the in situ population’s unique alleles were rare, with an average frequency of 0.3% (minimum 0.1%, maximum 0.7%). A high proportion of scarce rather than rare alleles within the ex situ population alludes to a comparatively more even allele distribution among different landrace cultivars. Most of the ex situ population’s unique alleles were scarce, with an average frequency of 1.3% (minimum 0.6%, maximum 5.2%).

Table 2.
Summary of relative allele frequencies (f) by population (%).
| Population | Landrace cultivars (n) | Frequent (f > 10%) | Moderate (f < 10%) | Scarce (f < 5%) | Rare (f < 1%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | ||
| Ex situ | 173 | 72 | 44.7 | 13 | 8.1 | 52 | 32.3 | 24 | 14.9 |
| In situ | 989 | 68 | 48.2 | 16 | 11.3 | 24 | 17.0 | 33 | 23.4 |
Allelic diversity was slightly higher for the ex situ as compared to the in situ population, with average unbiased expected heterozygosity (Heu) values of 0.749 and 0.727, respectively. The difference of the allelic diversity for these two populations was non-significant (Figure 3). Heu values for 13 out of 15 SSR markers were higher for the ex situ as compared to the in situ population, with differences ranging from a minimum of 0.011 for marker STM5127 to a maximum of 0.056 for marker STI0023. Heu values for two SSR markers were higher for the in situ population, with a difference of 0.001 and 0.023 for markers STGBSS and STM0031, respectively.
AMOVA provides additional insights into the molecular variation of the ex- and in-situ populations. The Fst value of 0.01236 indicates that the populations are not very well differentiated. In other words, the two populations share most of the allelic diversity. Most variation is encountered within rather than among populations, reaffirming that most alleles are shared between populations with comparable allele frequencies. Unique alleles of each population were either rare or scarce, partially explaining the modest percentage of variation of 1.24% found among the two populations (Table 3).
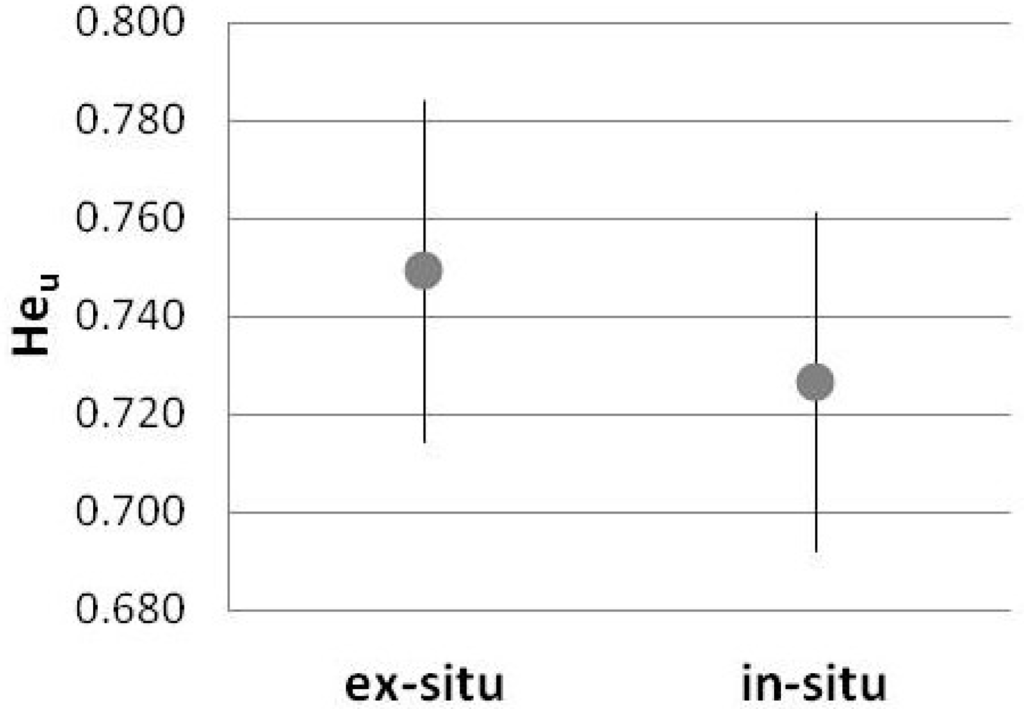
Figure 3.
Unbiased expected heterozygosity mean (Heu; CI 95%) considering all microsatellite loci for the ex- and in-situ population.

Table 3.
Comparison of the ex and in-situ populations based on Analysis of Molecular Variance (AMOVA). Fst, fixation index.
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation |
|---|---|---|---|---|
| Among the in situ and ex situ populations | 1 | 64.3 | 0.17184 Va | 1.24 |
| Within the in situ and ex situ populations | 1,160 | 15,922.2 | 13.72606 Vb | 98.76 |
| Total | 1,161 | 15,986.6 | 13.90 | 100 |
| Fixation Index: | Significance test (1023 permutations): | |||
| Fst: 0.01236 | Va and Fst, p is <0.001 | |||
Beyond allelic structure and diversity, a comparative analysis was conducted for the number and percentage of landrace cultivars with equivalents shared between both populations at different threshold levels as defined by the Jaccard similarity coefficient (Figure 4). Considering a stringent threshold level equivalent to a similarity coefficient of 1.00, a total of 16 accessions from the ex situ population (n = 173) match with 20 putatively identical landrace cultivars from the in situ population (n = 989). Only very few landrace cultivars are shared between the two populations, with 157 out of 173 and 969 out of 989 cultivars relative to the total sample size belonging uniquely to the ex- and in-situ population. At a non-stringent threshold level, considering a similarity coefficient of 0.80, a total of 71 accessions from the ex situ population match with 231 shared landrace cultivars from the in situ population. In other words, at this level, 41.0% of the ex situ population encounters a relatively similar landrace cultivar in 23.4% of the in situ populations. Even at this non-stringent level, 102 out of 173 and 758 out of 989 cultivars, respectively, belong uniquely to the ex- and in-situ population. Allelic diversity, which is slightly higher within the ex situ population, is, thus, not automatically matched by a higher percentage and number of unique landrace cultivars when compared with the in situ populations.
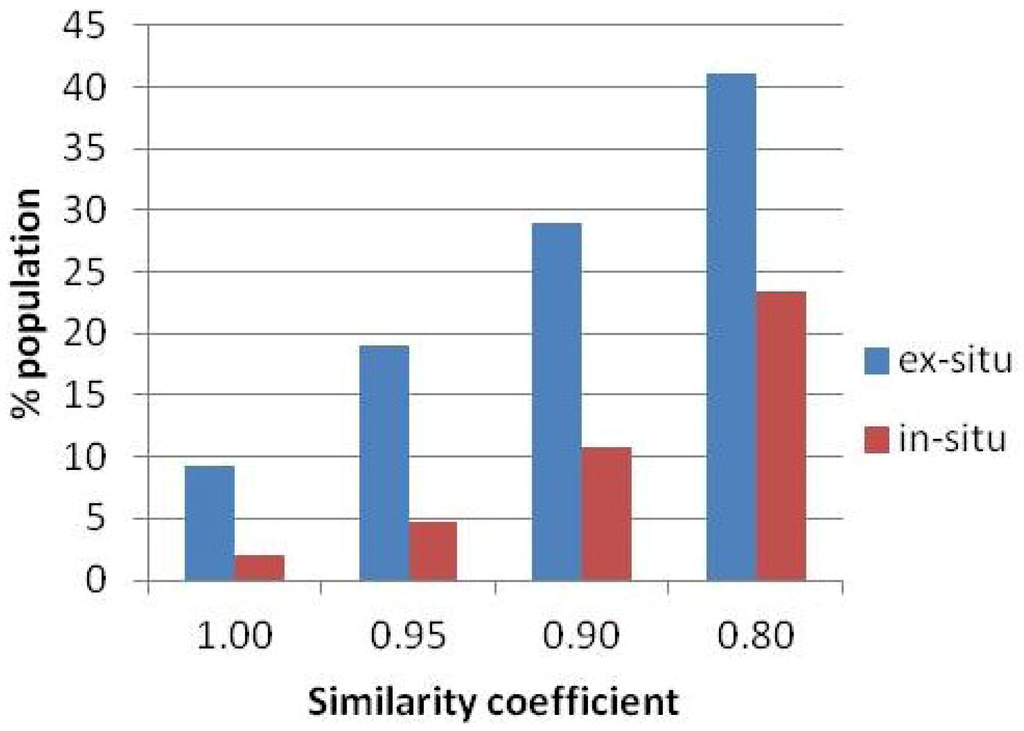
Figure 4.
Comparison of the percentage (%) of shared equivalent landrace cultivars between the ex- and in-situ populations based on different threshold values of the Jaccard similarity coefficient.
Although most alleles are shared between the two populations, the same is not true at the level of landrace cultivars. Most landrace cultivars are actually unique to either the ex- or in-situ populations. In terms of absolute numbers, the in situ population holds most unique landrace cultivars, even though, as expected, dissimilarity analysis within each population indicates that the percentage of similar landrace cultivars within the in situ population is much higher compared to the ex situ population: 7.5 versus 8.2%, 22.5 versus 35.9% and 31.2% versus 64.2% at a similarity coefficient of 1.00, 0.90 and 0.80 of the ex- and in-situ population, respectively. In other words, at a similarity coefficient of 1.00, 0.90 and 0.80, the ex- and in-situ populations, respectively, contain 160 and 908, 134 and 634, 119 and 354 unique landrace cultivars.
Figure 5 shows a comparison of the ex- and in-situ populations based on a Principal Component Analysis (PCA) and dissimilarity analysis with DARwin software. Both graphs visually reaffirm that the in situ population consists of numerous unique landrace cultivars that do not overlap with landrace accessions from the ex situ population. A comparison of each sub-branch of the dissimilarity tree at similarity coefficients of 0.4, 0.5 and 0.6, respectively, evidences that 21 (2.1%), 335 (33.9%) and 570 (57.6%) out of the 989 landrace cultivars from the in situ population are located in isolated subgroups with no accession from the ex situ population present in the sub-branch. This analysis also shows that five out of 39 (12.8%), 55 out of 136 (40.4%) and 126 out of 239 (52.6%) subgroups existing at similarity coefficients of 0.4, 0.5 and 0.6, respectively, are unrepresented by an accession from the CGR set or ex situ population. In other words, selected areas of the PCA graph and branches of the dendrogram are exclusively composed of sub-populations of the overall in situ population, pointing to the presence of unique landrace cultivars that do not have a near-similar match in the ex situ population.
Ploidy levels were determined for 825 out of the 989 landrace cultivars from the in situ population (83.4%). For this subsample (n = 825), the ploidy distribution was as follows: 29.0% diploids (2n = 2x = 34), 20.0% triploids (2n = 3x = 36), 50.9% tetraploids (2n = 4x = 48) and 0.1% pentaploids (2n = 5x = 60). The ploidy distribution of the ex situ population consisted of 32.4%, 34.7%, 30.6% and 2.3% of di-, tri-, tetra- and penta-ploid accessions, respectively. The distribution pattern of the in situ population is closer to what one would expect based on previous reports, with tetraploids generally being dominant [8], followed by diploids, which are known to be particularly common in the central Andes of Peru [44], triploids and rather low levels of pentaploids belonging to the bitter species, Solanum curtilobum [45].
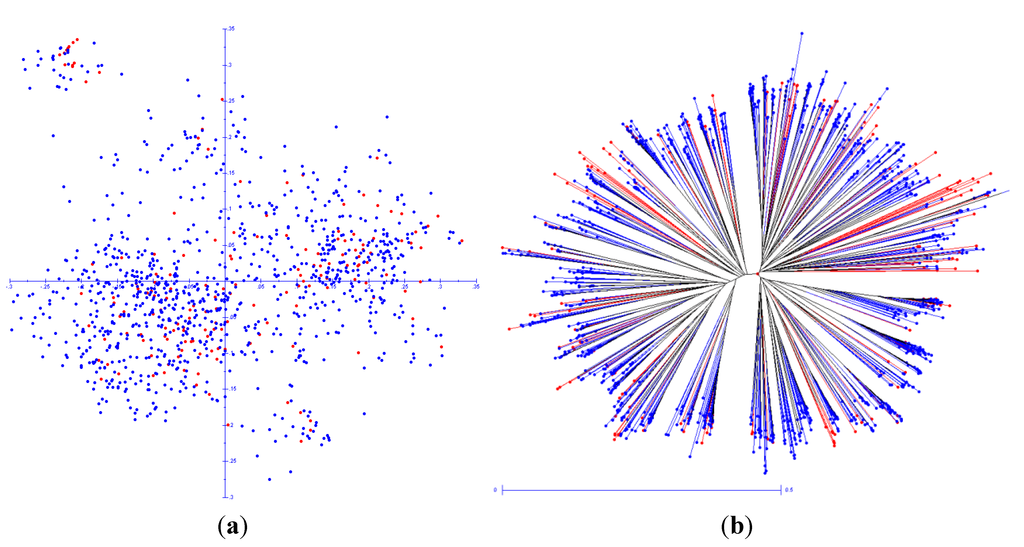
Figure 5.
Principal Component Analysis (PCA) and unweighted neighbor joining (NJ) dissimilarity tree comparing the contemporary in situ population (n = 989; Huancavelica department) with an International Potato Center (CIP)’s spatially restricted ex situ composite genotype reference (CGR) population (n = 173; Huancavelica, Ayacucho Junín and Lima departments). (a) Principal Component Analysis; (b) radial dissimilarity tree.
4. Discussion
4.1. SSR-Based Population Genetics
A temporal comparison of the populations separated over a near 40-year time period shows relative stability in the sense that three quarters of the alleles are shared between both populations, and 98.8% of variation based on AMOVA is encountered within rather than among populations. The outcome of this time-line comparison is similar to findings from localized spatial analysis in the Andes, which have shown that the majority of allelic variation is contained within geographies and landrace populations [46]. It reaffirms that potato landrace cultivars do share a large proportion of alleles across species, ploidy, space and time. This can be attributed to common parentage, interspecific hybridization and seed-tuber exchange [2,47,48,49].
The fact that most unique alleles are either scarce (ex situ) or rare (in situ) suggests modest genetic change at the margins of the allele frequency distribution pattern. This, in turn, can also be interpreted as evolution with an influx of “new” and efflux of “old” alleles. A total of 32 alleles, or 19.9% of the total number of alleles detected in the ex- situ population, were apparently not detected in situ, indicating that genetic erosion sensu stricto within a geographical confined space might be occurring at the allele level. Strictly speaking, ex situ conservation is not completely static. Seed-tuber and in vitro conservation needs regular regenerations. Therefore, management practices and somaclonal variations could potentially be additional sources of genetic change of the ex situ conserved population. Another factor that may explain some of these differences is the possible mixture of some landrace cultivar accessions during long-term ex-situ conservation. Clearly, the detection of exclusive alleles ex-situ reaffirms the importance of conservation in gene banks. The fact that the comparison of the ex- and in-situ populations resulted in a non-significant difference of the allelic diversity and representative allelic diversity coverage as shown by the low fixation index (Fst value) suggests that the spatially restricted CGR set is a genetically representative subset of the overall gene bank collection.
Most of the unique alleles that are now restricted to the ex situ population are scarce, and if taken as a mirror of the original base-line situation for the in situ population in the early 1970s, would consequently have been more prone to loss compared to moderate or frequent alleles. Causes of localized allele loss are likely to involve landrace cultivar loss, which in turn, may be a consequence of multiple underlying causes, ranging from pest and disease pressures to socioeconomic change [50]. Interestingly, 12 unique alleles, representing 8.5% of the total number of alleles detected in the in situ population, were encountered exclusively within the contemporary in situ population, evidencing the origination of “new” allelic diversity within the pre-defined geographical space of Huancavelica as compared to the base-line situation reflected by the spatially restricted CGR set with landrace cultivars from the exact same region and direct neighboring departments. This finding reaffirms the importance of in situ conservation to allow for seed-tuber flows and ongoing evolution. The dynamic nature of in situ conservation can be expected to have resulted in significant changes of the landrace cultivar composition during several decades of gene flow, mutations and anthropogenic selection [51,52,53,54]. This might also have involved the displacement of landrace by bred cultivars, as these have been grown in Peru since the early 1950s. Enhanced integration, road building and temporal migration will surely also have facilitated seed-tuber exchange between formerly isolated regions and, consequently, can also offer a partial explanation for the “new” allelic diversity encountered [55,56].
Conservation measures to avoid genetic erosion should build on an enhanced understanding of socioeconomic and environmental change and its impact on loss or enrichment [57]. It may include such diverse areas as youth engagement, seed system innovations, market linkages and farmer cuisine [58]. Links with gene banks through regular checks of ex- against in-situ populations (and vice versa) may also help prioritization of appropriate conservation interventions, such as the incorporation of rare landrace cultivars into gene banks or the reintroduction of “lost” landrace cultivars when requested by farmer communities.
4.2. SSR-Based Overlap of Landraces
The unexpected finding that most landrace cultivars do not have shared equivalents between the contemporary in situ and spatially restricted CGR set or ex situ population may have several explanations. It may be a direct consequence of the population dynamics of landrace cultivars, which is known to involve such diverse processes as sporadic sexual reproduction, adaptive farmer management, chimerism, rural change and technology options promoted by development interventions [54,59,60]. Farmer seed-tuber exchange of landrace cultivars in Huancavelica is typically very limited during normal cropping seasons. However, during periods of seed shortages following extreme weather events, such as hail, frost and drought, seed flows tend to be more voluminous and offer a partial explanation for cultivar change [61]. Peru’s central highlands were severely affected by political instability and rural violence between 1981 and 1994 [62]. This conflict disrupted agricultural production and is likely to have had a significant impact on the dynamics of cultivar portfolios.
The CGR set from Huancavelica and its direct neighboring departments is likely to have underrepresented the total coverage of landrace cultivars, because by definition, a composite genotype collection is a reduced subset of the complete ex situ gene bank collection. So-called cosmopolitan landrace cultivars that have a wide geographical distribution range generally only have a small number of representatives in the gene bank and may, therefore, have been unrepresented. The rationalization of CIP’s gene bank was conducted without bias for provenance, such that landrace cultivars collected in Huancavelica were eliminated from the collection if they were duplicates with samples from another department. Furthermore, collection gaps may represent an additional cause of the limited overlap of shared landrace cultivars encountered between the ex- and in-situ populations. This is partially a consequence of the tendency that unique genotypes are microgeographically concentrated [46]. Recent in-depth studies at the sub-regional level also tend to sample more extensively compared to earlier potato collectors [13,14].
It is clear that significant allelic coverage between populations is not equivalent to adequate landrace cultivar coverage. The bulk of landrace cultivars are restricted to either the ex- or in-situ population: 102 out of 173 and 758 out of 989, respectively, at a non-stringent similarity coefficient of 0.80. This has interesting implications for the conservation ecology of the vegetatively propagated potato in its center of origin. The finding that only 41.0% of the ex situ population encounters a similar landrace cultivar in 23.4% of the in situ population at a non-stringent threshold similarity coefficient of 0.80 clearly suggests that the population dynamics of potato landrace cultivars in confined mountain geographies may be much more dynamic than previously thought. How much of this dynamism over a near 40-year timeline can be assigned to a “natural” Darwinistic type of selection versus anthropogenic change processes remains unresolved. Yet, it is well-known that the population dynamics of landrace cultivars as compared to wild species is to a much larger extent explained by human factors [63]. A high level of dynamism for landrace cultivars as compared to their inherent allelic diversity also has implications about what we consider as representative for a region based on georeferenced passport data from gene banks. Ex situ landrace cultivar populations from a spatially well-defined hotspot may only represent a partial snapshot of what a situation was at a predefined moment in time.
5. Conclusions
The SSR marker comparison of a large spatially restricted ex- and in-situ population along a well-defined timeline shows that 74.6% of the detected alleles are shared between both populations, and only 1.2% of the molecular variation is encountered among rather than within populations. Overall, the population genetic structure in terms of allele presence versus absence and allele frequency variation can be considered modestly stable. However, out of the total number of alleles detected, both the ex- and in-situ populations also contain 19.9% and 8.5% of unique alleles. These unique alleles are either scarce or rare and seem to represent the tail ends of an evolutionary continuum with a simultaneous influx and efflux of alleles.
The “loss” and origination of “new” alleles within a defined timeframe and confined geographical space do occur in parallel and, thus, provide evidence for the occurrence of ongoing crop evolution within in situ conservation hotspots. The relatively stable but at the same time open nature of the population genetics of the potato in its natural habitat, resulting in both loss and enrichment, reinforces the complementary nature of ex- and in-situ conservation strategies. Ex situ conservation will ensure the availability and use of unique genetic diversity that may at present be non-adapted in its original collection site, while continued in situ management will result in responsive evolution favoring the presence and frequency of alleles to facilitate adaptation to changing environments.
Low levels of overlap of equivalent landrace cultivars shared between the ex- and in-situ populations at stringent (1.00, 0.95) to non-stringent (0.90, 0.80) threshold levels of the Jaccard similarity coefficient suggests that the population dynamics of potato landrace cultivars over a near 40-year time period is much more dynamic than anticipated, particularly more so than changes observed in the population genetics at the allelic level. Both the ex- and in-situ populations predominantly contain unique landrace cultivars, but particularly the contemporary in situ population is characterized by the presence of numerous unique landrace cultivars. A model for more systematic, rigorous and regular monitoring of the landrace cultivar population dynamics in well-defined in situ hotspots is needed to better understand the drivers of change and evolution in different social and environmental settings. Such long-term monitoring would ideally involve interdisciplinary teams of biophysical and social scientists collaborating toward an enhanced understanding of the links between socioeconomic, environmental and crop evolutionary change processes.
Acknowledgments
The authors are grateful for the participation of custodian farmers from Huancavelica in this research and financial support from the government of Spain.
Conflict of Interest
The authors declare no conflict of interest.
References
- Ovchinnikova, A.; Krylova, E.; Gavrilenko, T.; Smekalova, T.; Zhuk, M.; Knapp, S.; Spooner, D.M. Taxonomy of cultivated potatoes (Solanum section Petota: Solanaceae). Bot. J. Linn. Soc. 2011, 165, 107–155. [Google Scholar] [CrossRef]
- Spooner, D.M.; Núñez, J.; Trujillo, G.; Del Rosario Herrera, M.; Guzmán, F; Ghislain, M. Extensive simple sequence repeat genotyping of potato landraces support a major reevaluation of their gene pool structure and classification. Proc. Natl. Acad. Sci. USA 2007, 104, 19398–19403. [Google Scholar]
- Zeven, A.C. Landraces: A review of definitions and classifications. Euphytica 1998, 104, 127–139. [Google Scholar] [CrossRef]
- Brickell, C.D.; Baum, B.R.; Hetterscheid, W.L.A.; Leslie, A.C.; McNeill, J.; Trehane, P.; Vrugtman, F.; Wiersema, J.H. International Code of Nomenclature for Cultivated Plants; International Society for Horticultural Science: Leuven, Belgium, 2004. [Google Scholar]
- Correl, D.S. The Potato and its Wild Relatives; Texas Research Foundation: Renner, TX, USA, 1962. [Google Scholar]
- Hermsen, J.H. Schatgraven in het genencentrum van de aardappel (Solanum tuberosum L.). Vakbl. voor Biol. 1974, 14, 204–208. [Google Scholar]
- Hawkes, J.G. Hunting the Wild Potatoes in the South American Andes: Memories of the British Empire Potato Collecting Expedition to South America 1938–1939; Nijmegen University: Nijmegen, The Netherlands, 2003. [Google Scholar]
- Ochoa, C.M. Las Papas del Perú: base de datos 1947–1997 (In Spanish); UNALM, COSUDE, CIP: Lima, Peru, 2003. [Google Scholar]
- Vargas, C. Las Papas Sudperuanas, Parte I; Universidad Nacional del Cusco: Cusco, Peru, 1949. [Google Scholar]
- Vargas, C. Las Papas Sudperuanas, Parte II; Universidad Nacional del Cusco: Cusco, Peru, 1956. [Google Scholar]
- Loskutov, I.G. Vavilov and His Institute: A History of the World Collection of Plant Genetic Resources in Russia; IPGRI: Rome, Italy, 1999. [Google Scholar]
- Scott, G.J. Plants, people, and the conservation of biodiversity of potatoes in Peru. Nat. Conserv. 2011, 9, 1–18. [Google Scholar] [CrossRef]
- De Haan, S.; Núñez, J.; Bonierbale, M.; Ghislain, M. Multilevel agrobiodiversity and conservation of Andean potatoes in central Peru. Mt. Res. Dev. 2010, 30, 222–231. [Google Scholar] [CrossRef]
- Monteros, A.R. Potato landraces: Description and dynamics in three areas in Ecuador. Ph.D. thesis, Wageningen Agricultural University, Wageningen, The Netherlands, 2011. [Google Scholar]
- Vigouroux, Y.; Mariac, C.; De Mita, S.; Pham, J.L.; Gérard, B.; Kapran, I.; Sagnard, F.; Deu, M.; Chantereau, J.; Ali, A.; et al. Selection for earlier flowering crop associated with climatic variations in the Sahel. PLoS One 2011, 6, e19563. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Garrett, K.A.; Huamán, Z.; Zhang, D.P.; Schreuder, M.; Bonierbale, M. Assessing the geographical representativeness of genebank collections: The case of Bolivian wild potatoes. Conserv. Biol. 2000, 14, 1755–1765. [Google Scholar] [CrossRef]
- Johns, T.; Keen, S.L. Ongoing evolution of the potato on the Altiplano of Western Bolivia. Econ. Bot. 1986, 40, 409–424. [Google Scholar] [CrossRef]
- Rabinowitz, D.; Linder, C.R.; Douches, D.S.; Quiros, C.F. Determining natural hybridization between S. sparsipilum and S. stenotomum in the Andes. Am. Potato J. 1988, 65, 496. [Google Scholar]
- Scarcelli, N.; Tostain, S.; Vigouroux, Y.; Luong, V.; Baco, M.N.; Agbangla, C.; Daïnou, O.; Pham, J.L. Genetic structure of farmer-managed varieties in clonally-propagated crops. Genetica 2011, 139, 1055–1064. [Google Scholar] [CrossRef]
- Brush, S.B. Farmers’ Bounty: Locating Crop Diversity in the Contemporary World; Yale University Press: New Haven, CT, USA, 2004. [Google Scholar]
- Etisham-Ul-Haq, M.; Allnutt, T.R.; Smith-Ramirez, C.; Gardner, M.F.; Armesto, J.J.; Newton, A.C. Patterns of genetic variation in in and ex situ populations of the threatened Chilean vine Berberidopsis corallina, detected using RAPD markers. Ann. Bot. 2001, 87, 813–821. [Google Scholar] [CrossRef]
- Lautherbach, D.; Burkart, M.; Gemeinholzer, B. Rapid genetic differentiation between ex situ and their in situ source populations: An example of the endangered Silene otites (Caryophyllaceae). Bot. J. Linn. Soc. 2012, 168, 64–75. [Google Scholar]
- Malice, M.; Martin, N.; Pissard, A.; Rojas-Beltran, J.A.; Gandarillas, A.; Bertin, P.; Baudoin, J.P. A preliminary study of the genetic diversity of Bolivian oca (Oxalis tuberosa Mol.) varieties maintained in situ and ex situ through the utilization of ISSR molecular markers. Genet. Resour. Crop. Evol. 2007, 54, 685–690. [Google Scholar] [CrossRef]
- Rice, E.B.; Smith, M.E.; Mitchell, S.E.; Kresovich, S. Conservation and change: A comparison of in situ and ex situ conservation of Jala maize germplasm. Crop Sci. 2006, 46, 428–436. [Google Scholar] [CrossRef]
- Gomez, O.J.; Blair, M.W.; Frankow-Lindberg, B.E.; Gullberg, U. Comparative study of common bean (Phaseolus vulgaris L.) landraces conserved ex situ in genebanks and in situ by farmers. Genet. Resour. Crop Evol. 2005, 52, 371–380. [Google Scholar] [CrossRef]
- Che, Y.H.; Yang, Y.P.; Yang, X.M.; Li, X.Q.; Li, L.H. Genetic diversity between ex situ and in situ samples of Agropyron cristatum (L.) Gaertn based on simple sequence repeat molecular markers. Crop Pasture Sci. 2011, 62, 639–644. [Google Scholar] [CrossRef]
- Hamblin, M.T.; Warburton, M.L.; Buckler, E.S. Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PLoS One 2007, 2, e1367. [Google Scholar] [CrossRef]
- Van Inghelandt, D.; Melchinger, A.E.; Lebreton, C.; Stich, B. Population structure and genetic diversity in a commercial maize breeding program assessed with SSR and SNP markers. Theor. Appl. Genet. 2010, 120, 1289–1299. [Google Scholar] [CrossRef]
- Yang, X.; Xu, Y.; Shah, T.; Li, H.; Han, Z.; Li, J.; Yan, J. Comparison of SSRs and SNPs in assessment of genetic relatedness in maize. Genetica 2011, 139, 1045–1054. [Google Scholar] [CrossRef]
- Fregene, M.A.; Suarez, M.; Mkumbira, J.; Kulembeka, H.; Ndedya, E.; Kulaya, A.; Mitchel, S.; Gullberg, U.; Rosling, H.; Dixon, A.G.O.; et al. Simple sequence repeat marker diversity in cassava landraces: Genetic diversity and differentiation in a asexually propagated crop. Theor. Appl. Genet. 2003, 107, 1083–1093. [Google Scholar] [CrossRef]
- Tenzer, I.; Degli Ivanissevich, S.; Morgante, M.; Gessler, C. Identification of microsatellite markers and their application to population genetics of Venturia inaequalis. Phytopathology 1999, 89, 748–753. [Google Scholar] [CrossRef]
- Carputo, D.; Alioto, D.; Aversano, R.; Garramone, R.; Miraglia, V.; Villano, C.; Frusciante, L. Genetic diversity among potato species as revealed by phenotypic resistances and SSR markers. Plant Genet. Resour. 2013. [Google Scholar] [CrossRef]
- Ruiz de Galarreta, J.I.; Barandalla, L.; Lorenzo, R.; Gonzalez, J.; Rios, D.J.; Ritter, E. Microsatellite variation in potato landraces from the island of La Palma. Span. J. Agric. Res. 2007, 5, 186–192. [Google Scholar]
- Ashkenazi, V.; Chani, E.; Lavi, U.; Levy, D.; Hillel, J.; Veilleux, R.E. Development of microsatellites markers in potato and their use in phylogenetic and fingerprinting analysis. Genome 2001, 44, 50–62. [Google Scholar] [CrossRef]
- Ghislain, M.; Andrade, D.; Rodriguez, F.; Hijmans, R.J.; Spooner, D.M. Genetic analysis of the cultivated potato Solanum tuberosum L. Phureja Group using RAPDs and nuclear SSRs. Theor. Appl. Genet. 2006, 113, 1515–1527. [Google Scholar] [CrossRef]
- Ghislain, M.; Nunez, J.; Herrera, M.R.; Pignataro, J.; Guzman, F.; Bonierbale, M.; Spooner, D.M. Robust and highly informative microsatellite-based genetic identity kit for potato. Mol. Breed. 2009, 23, 377–388. [Google Scholar] [CrossRef]
- Rubina, A.; Barreda, J. Atlas del Departamento de Huancavelica (In Spanish); Centro de Estudios y Promoción del Desarrollo (DESCO), Buena Ventura: Lima, Peru, 2000. [Google Scholar]
- CIP. In Catálogo de Variedades de Papa Nativa de Huancavelica–Perú (In Spanish); CIP, FEDECCH: Lima, Peru, 2006.
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- CIP. Supplementary dataset. Available online: Supplementary dataset. Available online: https://research.cip.cgiar.org/confluence/x/_aogAw (accessed on 24 April 2013).
- Rohlf, F.J. NTSYSpc: Numerical Taxonomy System, version 2.20; Exeter Publishing: Setauket, NY, USA, 2008. [Google Scholar]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software, version 5.0.158; CIRAD: Montpellier, France, 2006. [Google Scholar]
- Excoffier, L.; Lischer, H.E. Arlequin suite version 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Hawkes, J.G. The Potato: Evolution, Biodiversity & Genetic Resources; Smithsonian Institution Press: Washington, DC, USA, 1990. [Google Scholar]
- Ochoa, C.M. Las Papas de Sudamerica: Peru (In Spanish); Centro Internacional de la Papa: Lima, Peru, 1999. [Google Scholar]
- Zimmerer, K.S.; Douches, D.S. Geographical approaches to crop conservation: The partitioning of genetic diversity in Andean potatoes. Econ. Bot. 1991, 45, 176–189. [Google Scholar] [CrossRef]
- Rodriguez, F.; Ghislain, M.; Clausen, A.M.; Jansky, S.H.; Spooner, D.M. Hybrid origins of cultivated potatoes. Theor. Appl. Genet. 2010, 121, 1187–1198. [Google Scholar] [CrossRef]
- Spooner, D.M.; McLean, K.; Ramsay, G.; Waugh, R.; Bryan, G.J. A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proc. Natl. Acad. Sci. USA 2005, 102, 14694–14699. [Google Scholar] [CrossRef]
- Thiele, G. Informal potato seed systems in the Andes: Why are they important and what should we do with them? World Dev. 1999, 27, 83–99. [Google Scholar] [CrossRef]
- Brush, S.B. Ethnoecology, biodiversity, and modernization in Andean potato agriculture. J. Ethnobiol. 1992, 12, 161–185. [Google Scholar]
- Johns, T.; Huaman, Z.; Ochoa, C.; Schmiediche, P.E. Relationships among wild, weed, and cultivated potatoes in the Solanum ajanhuiri complex. Syst. Bot. 1987, 12, 541–552. [Google Scholar] [CrossRef]
- Quiros, C.F.; Ortega, R.; Van Raamsdonk, L.; Herrera-Montoya, M.; Cisneros, P.; Schmidt, E.; Brush, S.B. Increase of potato genetic resources in their center of diversity: The role of natural outcrossing and selection by Andean farmers. Genet. Resour. Crop Evol. 1992, 39, 107–113. [Google Scholar]
- Rabinowitz, D.; Linder, C.R.; Ortega, R.; Begazo, D.; Murguia, H.; Douches, D.S.; Quiros, C.F. High levels of interspecific hybridization between Solanum sparsipilum and S. stenotomum in experimental plots in the Andes. Am. Potato J. 1990, 67, 73–82. [Google Scholar] [CrossRef]
- Scurrah, M.; Celis-Gamboa, C.; Chumbiauca, S.; Salas, A.; Visser, R.G.F. Hybridization between wild and cultivated potato species in the Peruvian Andes and biosafety implications for the deployment of GM potatoes. Euphytica 2008, 164, 881–892. [Google Scholar]
- Wilson, F. Towards a political economy of roads: Experiences from Peru. Dev. Change 2004, 35, 525–546. [Google Scholar]
- Zimmerer, K.S. Geographies of seed networks for food plants (potato, ulluco) and approaches to agrobiodiversity conservation in the Andes. Soc. Nat. Resour. 2003, 16, 583–601. [Google Scholar] [CrossRef]
- Bellon, M.R. Conceptualizing interventions to support on-farm genetic resource conservation. World Dev. 2004, 32, 159–172. [Google Scholar] [CrossRef]
- Community Biodiversity Management: Promoting Resilience and the Conservation of Plant Genetic Resources; De Boef, W.; De Boef, W.; Subedi, A.; Peroni, N.; Thijssen, M.; O’Keeffe, E. (Eds.) Routledge: Oxford, UK, 2013.
- Brush, S.B.; Taylor, J.E.; Bellon, M.R. Technology adoption and biological diversity in Andean potato agriculture. J. Dev. Econ. 1992, 39, 365–387. [Google Scholar] [CrossRef]
- Zimmerer, K.S. The loss and maintenance of native crops in mountain agriculture. GeoJournal 1992, 27, 61–72. [Google Scholar] [CrossRef]
- De Haan, S.; Almekinders, C.; Thiele, G.; Scurrah, M. Farmer seed systems and infraspecific diversity of potato in Peru’s central Highlands. In Potato Diversity at Height: Multiple Dimensions of Farmer-driven In-situ Conservation in the Andes; Wageningen Agricultural University: Wageningen, The Netherlands, 2009; pp. 133–160. [Google Scholar]
- Pedersen, S.; Tremblay, J.; Errázuriz, C.; Gamarra, J. The sequelae of political violence: Assessing trauma, suffering and dislocation in the Peruvian highlands. Soc. Sci. Med. 2008, 67, 205–217. [Google Scholar] [CrossRef]
- Zimmerer, K.S. Labor shortages and crop diversity in the southern Peruvian Sierra. Geogr. Rev. 1991, 82, 414–432. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).