Re-Evaluating Causal Modeling with Mantel Tests in Landscape Genetics
Abstract
:1. Introduction

2. Experimental Section
2.1. Study Area and Resistance Hypotheses

| Model Acronym | Model Description |
|---|---|
| EH | Minimum resistance at high elevations (1,500 m) |
| EHFH | Minimum resistance in forest (strong) at high elevations |
| EHFL | Minimum resistance in forest (weak) at high elevations |
| EHRH | Minimum resistance at high elevations with high resistance of roads |
| EHRL | Minimum resistance at high elevations with weak resistance of roads |
| EL | Minimum resistance at low elevations (500 m) |
| ELFH | Minimum resistance at in forest (strong) at low elevations |
| ELFL | Minimum resistance in forest (weak) at low elevations |
| ELRH | Minimum resistance at low elevations with high resistance of roads |
| ELRL | Minimum resistance at low elevations with weak resistance of roads |
| EM | Minimum resistance at middle elevations (1,000 m) |
| EMFH | Minimum resistance in forest (strong) at middle elevations |
| EMFL | Minimum resistance in forest (weak) at middle elevations |
| EMRH | Minimum resistance at middle elevations with high resistance of roads |
| EMRL | Minimum resistance at middle elevations with weak resistance of roads |
| FH | Minimum resistance in forest (strong) |
| FHEHRH | Minimum resistance in forest (strong) at high elevations with high resistance of roads |
| FHEHRL | Minimum resistance in forest (strong) at high elevations with weak resistance of roads |
| FHELRH | Minimum resistance in forest (strong) at low elevations with high resistance of roads |
| FHELRL | Minimum resistance in forest (strong) at low elevations with weak resistance of roads |
| FHEMRH | Minimum resistance in forest (strong) at middle elevations with high resistance of roads |
| FHEMRL | Minimum resistance in forest (strong) at middle elevations with weak resistance of roads |
| FHRH | Minimum resistance in forest (strong) with high resistance of roads |
| FHRL | Minimum resistance in forest (strong) with low resistance of roads |
| FL | Minimum resistance in forest (weak) |
| FLEHRH | Minimum resistance in forest (weak) at high elevations with high resistance of roads |
| FLEHRL | Minimum resistance in forest (weak) at high elevations with weak resistance of roads |
| FLELRH | Minimum resistance in forest (weak) at low elevations with high resistance of roads |
| FLELRL | Minimum resistance in forest (weak) at low elevations with weak resistance of roads |
| FLEMRH | Minimum resistance in forest (weak) at middle elevations with high resistance of roads |
| FLEMRL | Minimum resistance in forest (weak) at middle elevations with weak resistance of roads |
| FLRH | Minimum resistance in forest (weak) with high resistance of roads |
| FLRL | Minimum resistance in forest (weak) with low resistance of roads |
| RH | Strong resistance of roads |
| RL | Weak resistance of roads |
2.2. Landscape Genetic Simulation with CDPOP
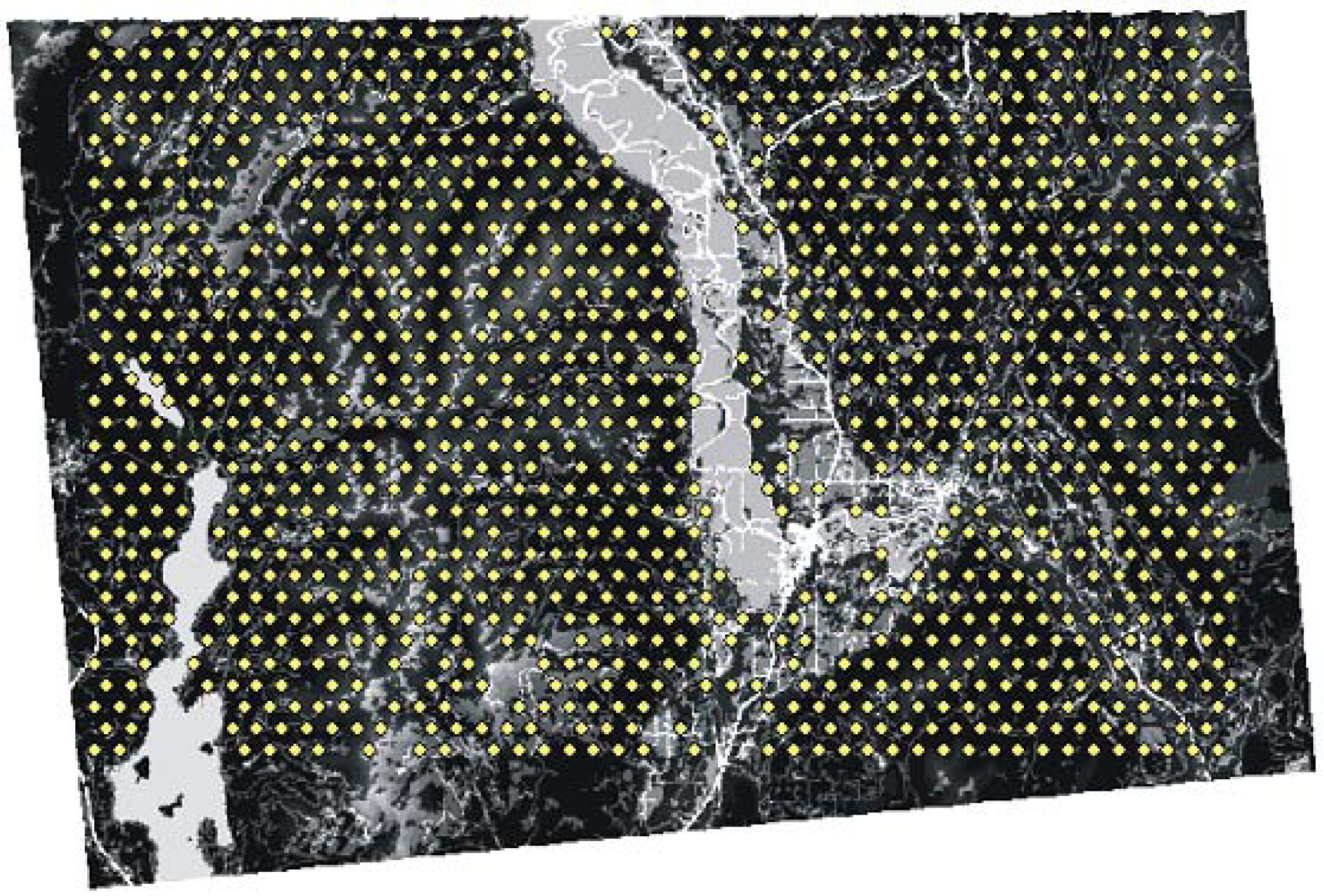
2.3. Evaluating Reliability of the Cushman et al. (2006) Causal Modeling Framework
| Test Number | Dependent Variable | Independent Variable | Covariate | Expected Outcome |
|---|---|---|---|---|
| 1 | Genetic Distance | Landscape Resistance Model Cost Distance | Isolation by Distance | Significant |
| 2 | Genetic Distance | Landscape Resistance Model Cost Distance | Isolation by Barrier | Significant |
| 3 | Genetic Distance | Isolation by Distance | Landscape.Resistance Model Cost Distance | Not Significant |
| 4 | Genetic Distance | Isolation by Barrier | Landscape.Resistance.Model Cost Distance | Not Significant |
2.4. Evaluating Reliability of the Wasserman et al. (2010) Causal Modeling Framework
3. Results and Discussion
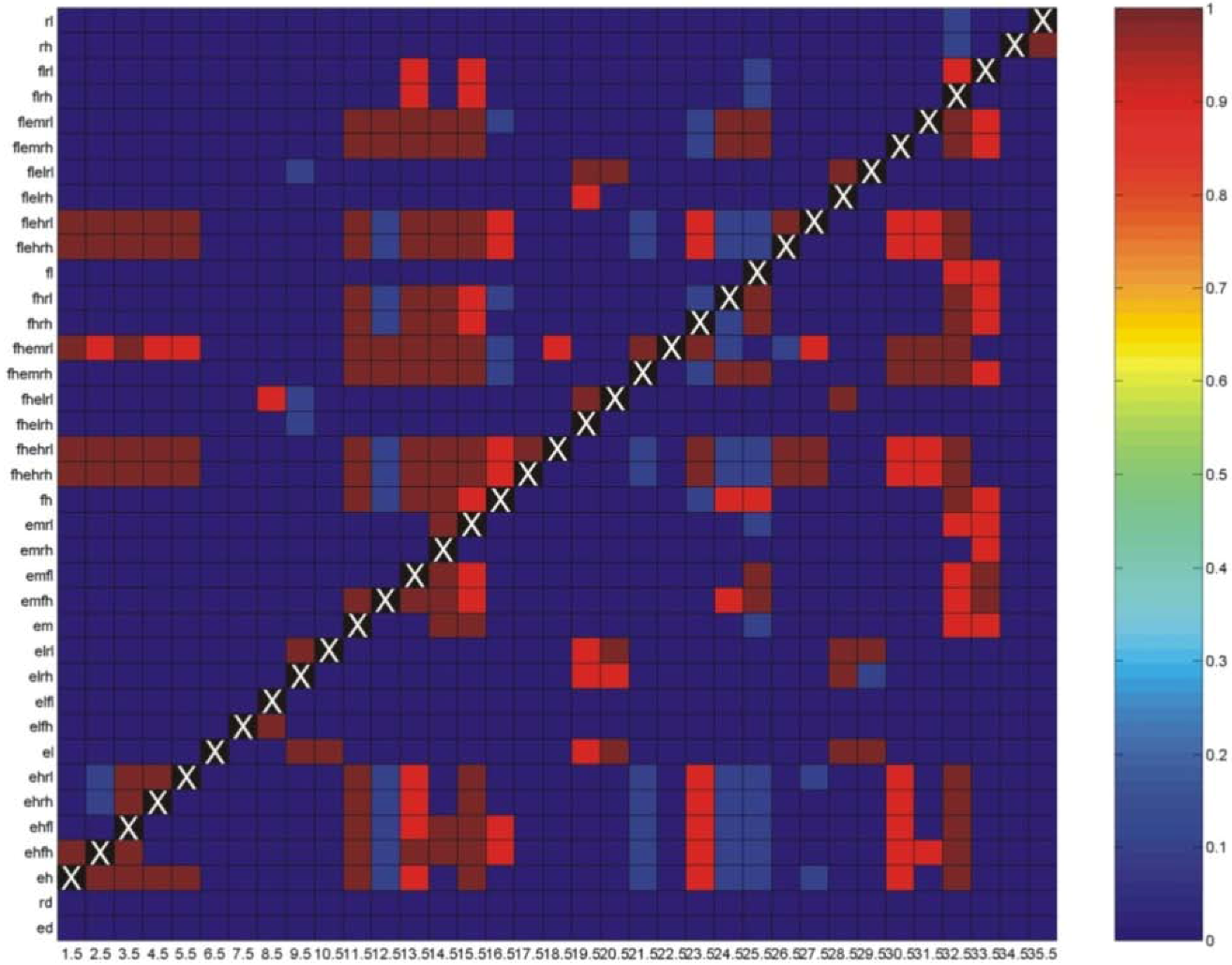
3.1. Correlations among Resistance Hypotheses
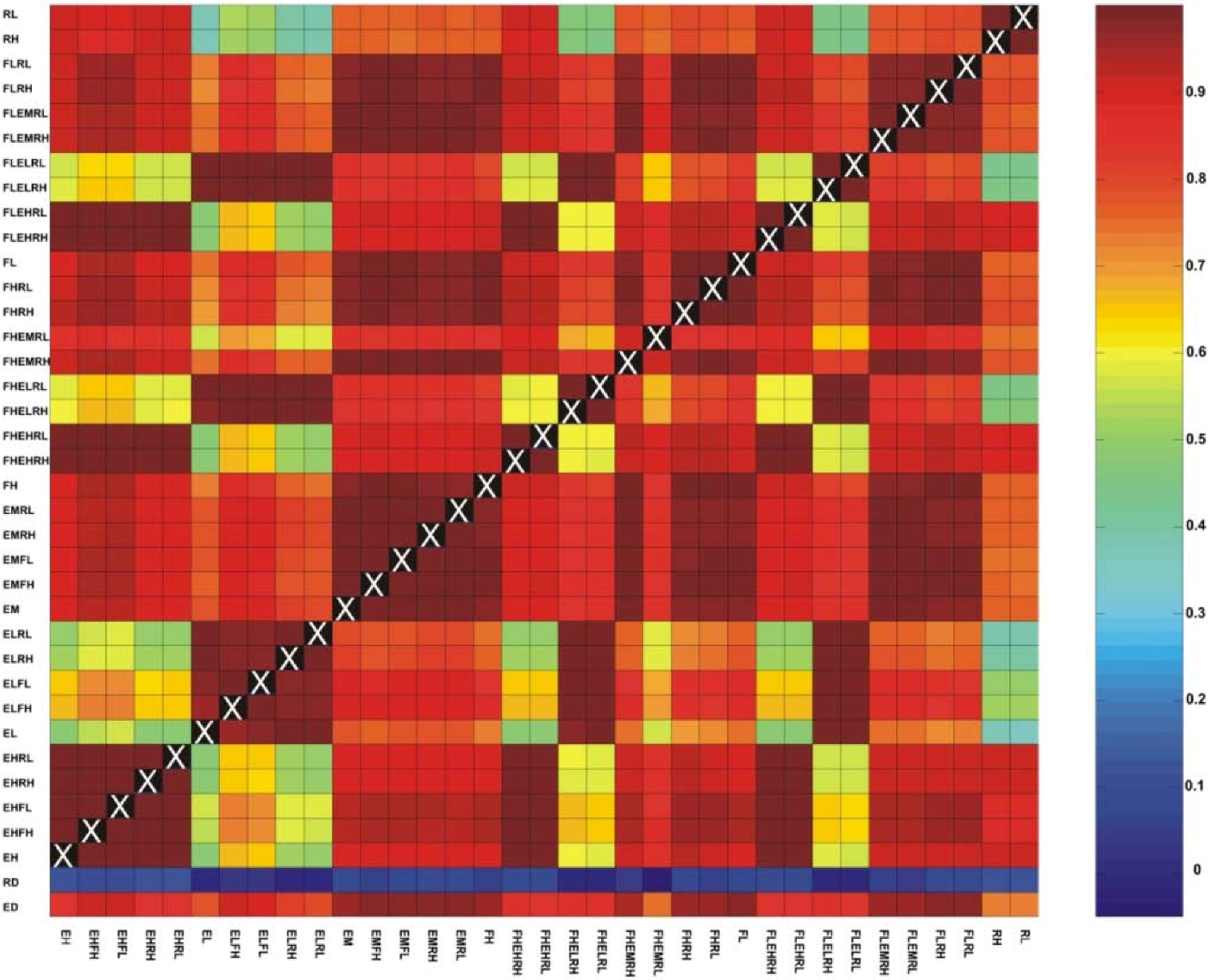
3.2. Evaluating Reliability of Causal Modeling
| Test Number | Logistic Regression Model | Estimate | Std. Error | Z value | Pr(>|z|) | |
|---|---|---|---|---|---|---|
| 1 | IBD | Model | Intercept | 10.207 | 6.276 | 1.626 | 0.1039 |
| DD | −17.451 | 7.855 | −2.222 | 0.0263 | ||
| 2 | IBB | Model | Intercept | −2.8887 | 0.3562 | −8.111 | 5.03 x 10-16 |
| DD | 22.7708 | 3.9155 | 5.816 | 6.04 x 10-9 | ||
| 3 | True | Alternative | Intercept | −36.6 | 1.187 | −30.82 | <2 × 10−16 |
| DD | 36.472 | 1.211 | 30.11 | <2 × 10−16 | ||
| 4 | Alternative | True | Intercept | −3.7234 | 0.1289 | −28.88 | <2 × 10−16 |
| DD | 3.916 | 0.1471 | 26.62 | <2 × 10−16 |
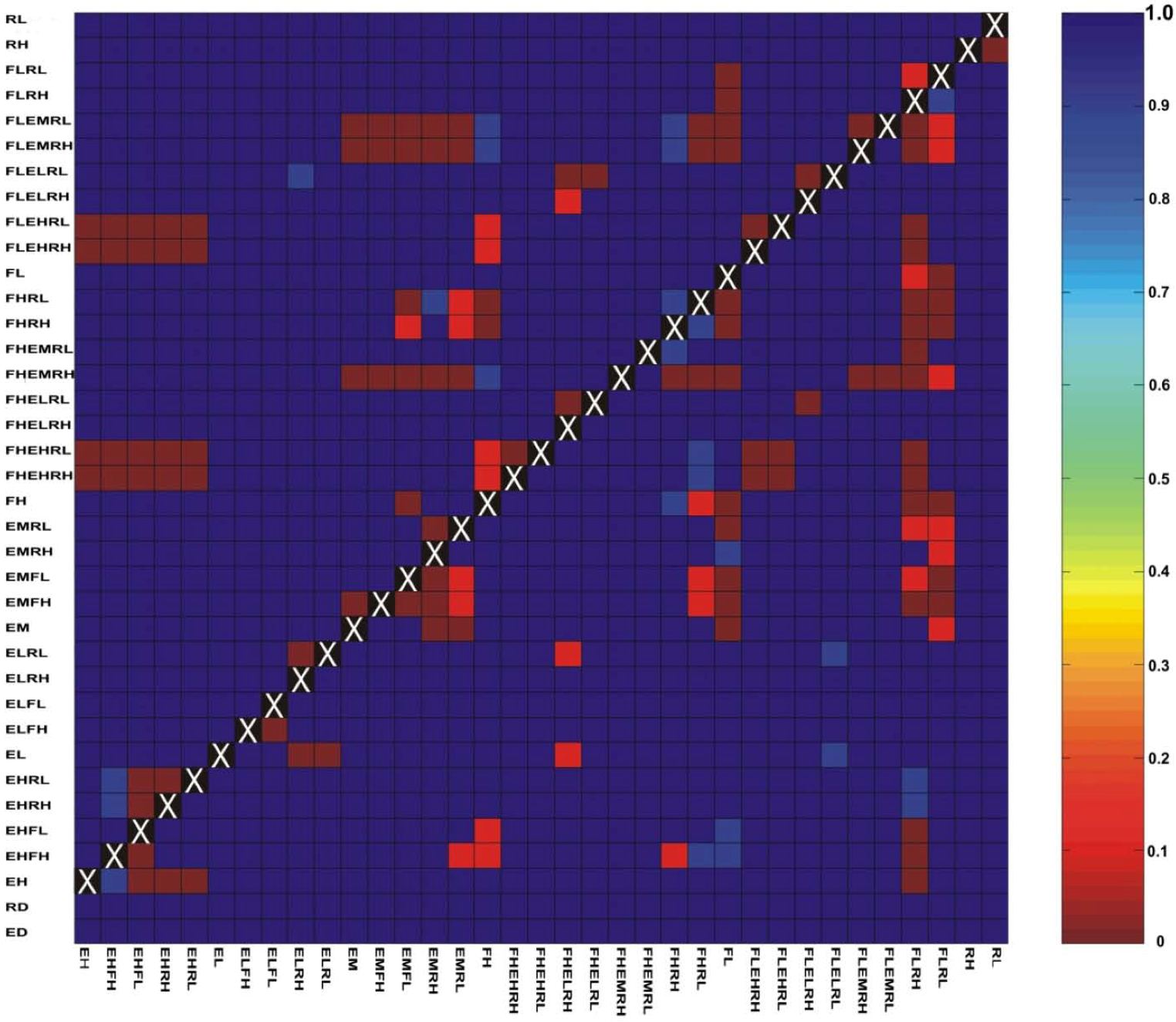
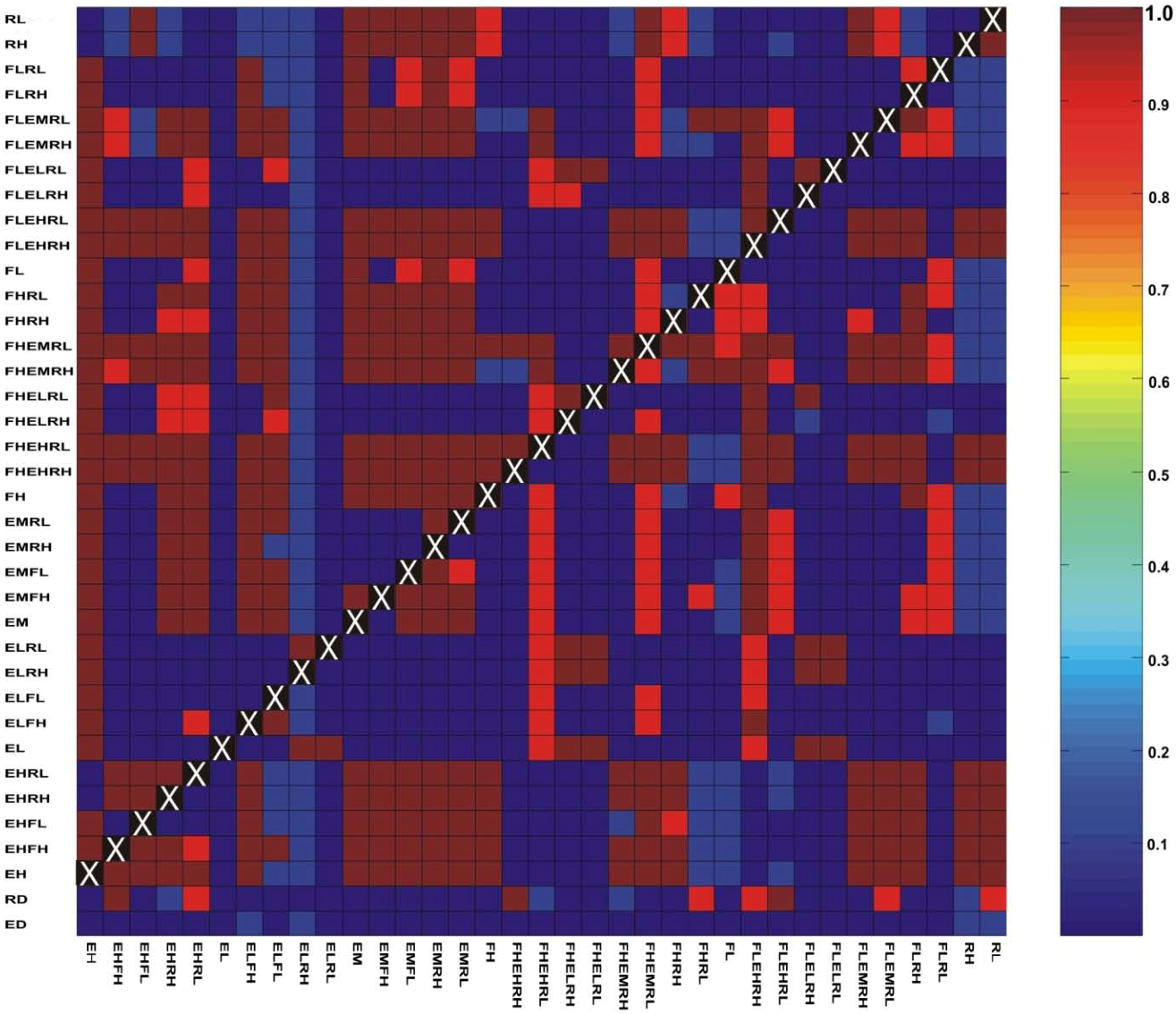
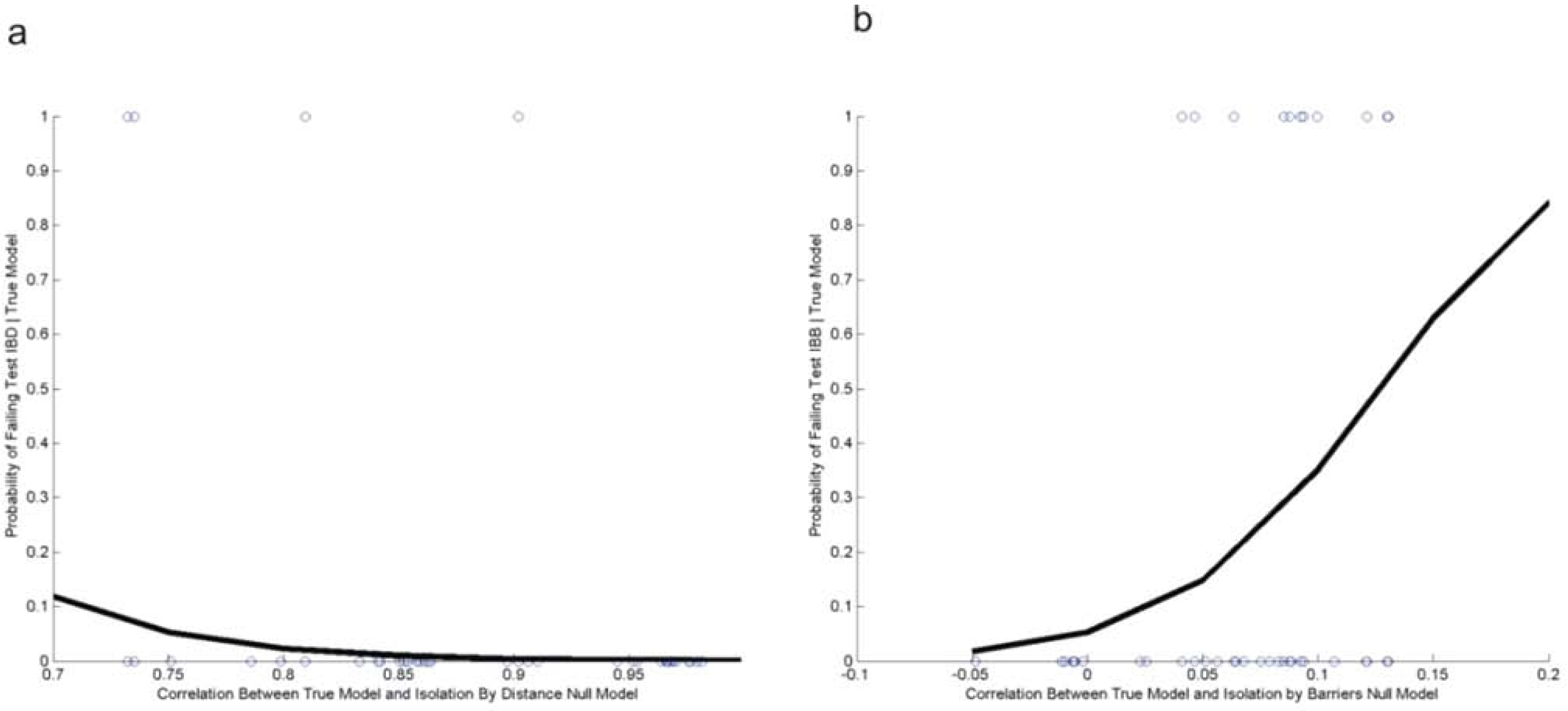
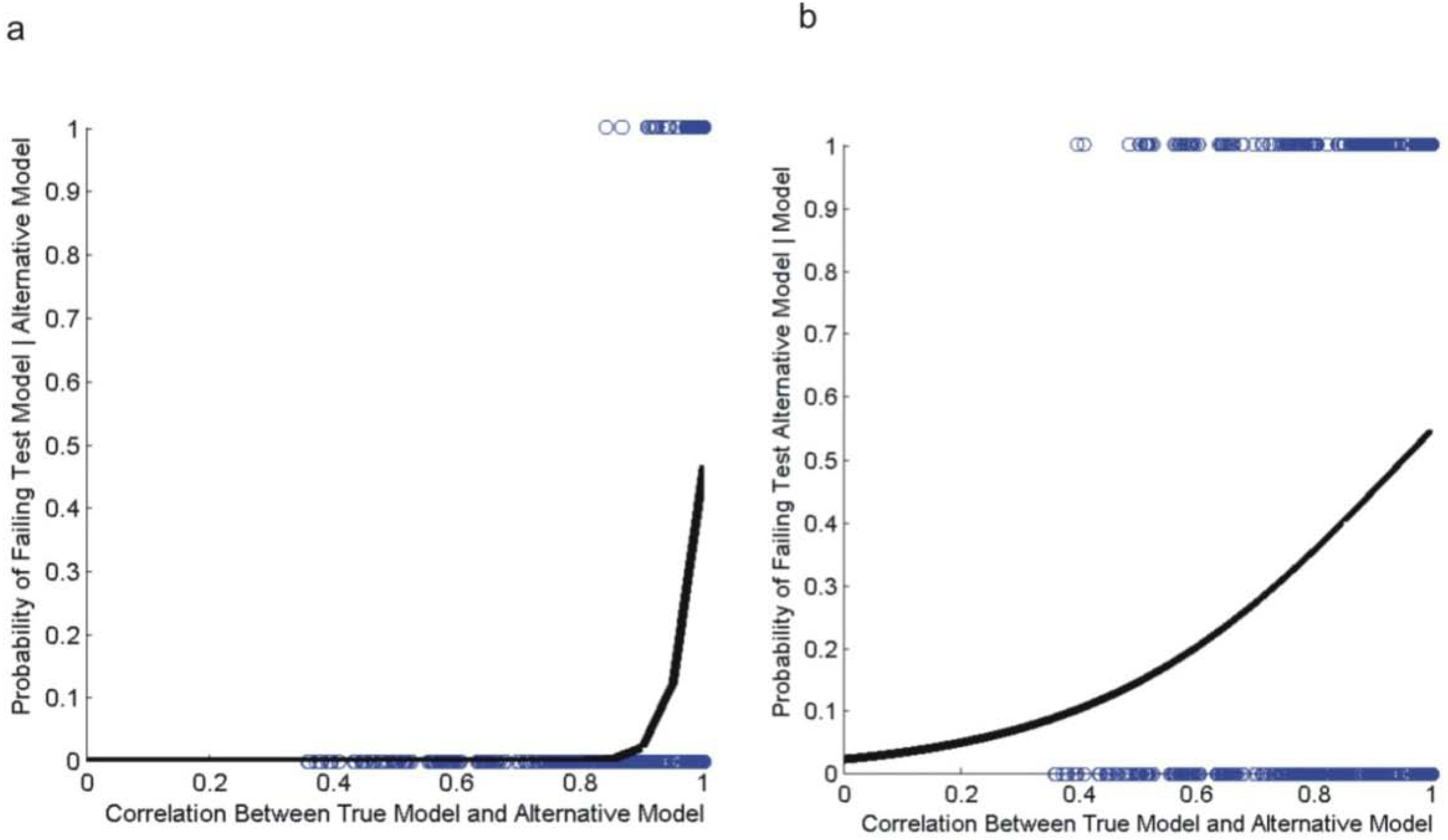
3.3. Evaluation of Hypotheses
3.4. Implications for Landscape Genetic Analyses
4. Conclusions
Acknowledgments
References
- Manel, S.; Schwartz, M.K.; Luikart, G.; Taberlet, P. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 2003, 18, 189–197. [Google Scholar] [CrossRef]
- Holderegger, R.; Wagner, H.H. Landscape genetics. Bioscience 2008, 58, 199–207. [Google Scholar] [CrossRef]
- Balkenhol, N.; Gugerli, F.; Cushman, S.A.; Waits, L.P.; Coulon, A.; Arntzen, J.W.; Holderegger, R.; Wagner, H.H.; Arens, P.; Campagne, P.; et al. Identifying future research needs in landscape genetics: where to from here? Landscape Ecol. 2009, 24, 455–463. [Google Scholar] [CrossRef]
- Segelbacher, G.; Cushman, S.A.; Epperson, B.K.; Fortin, M.-J.; Francois, O.; Hardy, O.J.; Holderegger, R.; Manel, S. Applications of landscape genetics in conservation biology: concepts and challenges. Conserv. Genet. 2010, 11, 375–385. [Google Scholar] [CrossRef]
- Spear, S.F.; Peterson, C.R.; Matocq, M.D.; Storfer, A. Landscape genetics of the blotched tiger salamander (Ambystoma tigrinum melanostictum). Mol. Ecol. 2005, 14, 2553–2564. [Google Scholar] [CrossRef]
- Cushman, S.A.; McKelvey, K.S.; Hayden, J.; Schwartz, M.K. Gene-flow in complex landscapes: testing multiple models with causal modeling. Am. Nat. 2006, 168, 486–499. [Google Scholar] [CrossRef]
- McRae, B.H.; Beier, P. Circuit theory predicts gene flow in plant and animal populations. Proc. Natl. Acad. Sci. USA 2007, 104, 19885–19890. [Google Scholar] [CrossRef]
- Pérez-Espona, S.; Pérez-Barbería, F.J.; McLeod, J.E.; Jiggins, C.D.; Gordon, I.J.; Pemberton, J.M. Landscape features affect gene flow of Scottish Highland red deer (Cervus elaphus). Mol. Ecol. 2008, 17, 981–996. [Google Scholar] [CrossRef]
- Lee-Yaw, J.A.; Davidson, A.; McRae, B.H.; Green, D.M. Do landscape processes predict phylogeographic patterns in the wood frog? Mol. Ecol. 2009, 18, 1863–1874. [Google Scholar] [CrossRef]
- Shirk, A.; Wallin, D.O.; Cushman, S.A.; Rice, R.C.; Warheit, C. Inferring landscape effects on gene flow: a new multi-scale model selection framework. Mol. Ecol. 2010, 19, 3603–1619. [Google Scholar] [CrossRef]
- Wasserman, T.N.; Cushman, S.A.; Schwartz, M.K.; Wallin, D.O. Spatial scaling and multi-model inference in landscape genetics: Martes americana in northern Idaho. Landscape Ecol. 2010, 25, 1601–1612. [Google Scholar] [CrossRef]
- Murphy, M.A.; Evans, J.S.; Storfer, A. Quantifying Bufo boreas connectivity in Yellowstone National Park with landscape genetics. Ecology 2010, 91, 252–261. [Google Scholar] [CrossRef]
- Krist, F.J.; Brown, D.G. GIS modeling of paleo-indian period caribou migrations and viewsheds in northeastern lower Michigan. Photogramm. Eng. Rem. S. 1994, 60, 1129–1137. [Google Scholar]
- Walker, R.; Craighead, L. Analyzing wildlife movement corridors in Montana using GIS. Proceedings of the Esri User Conference '97. Available online: http://gis.Esri.com/library/userconf/proc97/proc97/to150/pap116/p116.htm (accessed on 15 February 2013). [Green Version]
- McRae, B.H. Isolation by resistance. Evolution 2006, 60, 1551–1561. [Google Scholar]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Smouse, P.E.; Long, J.C.; Sokal, R.R. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 1986, 35, 627–632. [Google Scholar] [CrossRef]
- Raufaste, N.; Rousset, F. Are partial Mantel tests adequate? Evolution 2001, 55, 1703–1705. [Google Scholar]
- Castellano, S.; Balletto, E. Is the partial Mantel test inadequate? Evolution 2002, 56, 1871–1873. [Google Scholar]
- Legendre, P.; Fortin, M.-J. Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Mol. Ecol. Res. 2010, 10, 831–844. [Google Scholar] [CrossRef]
- Guillot, G.; Rousset, F. On the use of simple and partial Mantel tests in the presence of spatial auto-correlation. 2011, arXi, 1112.0651v1. [Google Scholar]
- Meirmans, P.G. The trouble with isolation by distance. Mol. Ecol. 2012, 21, 2839–2846. [Google Scholar] [CrossRef]
- Amos, J.; Bennet, A.F.; Mac Nally, R.; Newell, G.; Radford, J.Q.; Pavlova, A.; Thompson, J.; White, M.; Sunnucks, P. Predicting landscape genetic consequences of habitat loss, fragmentation and mobility for species of woodland birds. Plos One 2012, 7, e30888. [Google Scholar]
- Legendre, P.; Troussellier, M. Aquatic heterotrophic bacteria: modeling in the presence of spatial autocorrelation. Limnol. Oceanogr. 1988, 33, 1055–1067. [Google Scholar] [CrossRef]
- Legendre, P. Spatial autocorrelation: trouble or new paradigm? Ecology 1993, 74, 1659–1673. [Google Scholar] [CrossRef]
- Cushman, S.A.; Landguth, E.L. Spurious correlations and inference in landscape genetics. Mol. Ecol. 2010, 19, 3592–3602. [Google Scholar] [CrossRef]
- Cushman, S.A.; Lewis, J. Movement behavior explains genetic differentiation in American black bear. Landscape Ecol. 2010, 25, 1613–1625. [Google Scholar] [CrossRef]
- Wasserman, T.N.; Cushman, S.A.; Shirk, A.S.; Landguth, E.L.; Littell, J.S. Simulating the effects of climate change on population connectivity of American marten (Mates americana) in the northern Rocky Mountains, USA. Landscape Ecol. 2012, 27, 211–225. [Google Scholar] [CrossRef]
- Wasserman, T.N.; Cushman, S.A.; Littell, J.S.; Landguth, E.L. Population connectivity and genetic diversity of American marten (Martes Americana) in the United States northern Rocky Mountains in a climate change context. Conserv. Genet. 2012. [Google Scholar] [CrossRef]
- Landguth, E.L.; Cushman, S.A. CDPOP: An individual-based, cost-distance spatial population genetics model. Mol. Ecol. Resour. 2010, 10, 156–161. [Google Scholar] [CrossRef]
- Short Bull, R.A.; Cushman, S.A.; Mace, R.; Chilton, T.; Kendall, K.C.; Landguth, E.L.; Schwartz, M.K.; McKelvey, K.S.; Allendorf, F.W.; Luikart, G. Why replication is important in landscape genetics: American black bear in the Rocky Mountains. Mol. Ecol. 2011, 20, 1092–1107. [Google Scholar]
- Cushman, S.A.; Landguth, E.L. Multi-species connectivity in the northern Rocky Mountains. Ecol Model. 2012, 231, 101–112. [Google Scholar] [CrossRef]
- Cushman, S.A.; Landguth, E.L. Ecological associations, dispersal ability and landscape connectivity in the northern Rocky Mountains. Available online: http://www.fs.fed.us/rm/pubs/rmrs_rp090.pdf (accessed on 15 February 2013).
- Cushman, S.A.; Landguth, E.L.; Shirk, A.J. Separating the effects of habitat area, fragmentation and matrix resistance on genetic differentiation in complex landscapes. Landscape Ecol. 2012, 27, 369–380. [Google Scholar] [CrossRef]
- Balloux, F. EASYPOP (Version 1.7): A computer program for population genetics simulations. J. Hered. 2001, 92, 301–302. [Google Scholar] [CrossRef]
- Manel, S.; Berthoud, F.; Bellemain, E.; Gaudeul, M.; Luikart, G.; Swenson, J.E.; Waits, L.P.; Taberlet, P.; Consortium, I. A new individual-based spatial approach for identifying genetic discontinuities in natural populations. Mol. Ecol. Resour. 2007, 16, 2031–2043. [Google Scholar] [CrossRef]
- Landguth, E.L.; Cushman, S.A.; Luikart, G.; Murphy, M.A.; Schwartz, M.K.; McKelvey, K.S. Quantifying the lag time to detect barriers in landscape genetics. Mol. Ecol. 2010, 19, 4179–4191. [Google Scholar] [CrossRef]
- Jaquiéry, J.; Broquet, T.; Hirzel, A.H.; Yearsley, J.; Perrin, N. Inferring landscape effects on dispersal from genetic distances: How far can we go? Mol. Ecol. 2011, 20, 692–705. [Google Scholar] [CrossRef]
- Bowcock, A.M.; Ruiz-Linares, A.; Tomfohrde, J.; Minch, E.; Kidd, J.R.; Cavalli-Sforza, L.L. High resolution of human evolutionary trees with polymorphic micorsatellites. Nature 1994, 368, 455–457. [Google Scholar] [CrossRef]
- ESRI (Environmental Systems Research Incorporated). ArcGIS. ESRI, Redlands, CA, USA, 2003.
- Mantel, N.A. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Goslee, S.C.; Urban, D.L. The ecodist package for dissimilarity-based analysis of ecological data. J. Statist. Softw. 2007, 22, 1–19. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2012. Available online: http://www.R-project.org (accessed on 15 February 2013). ISBN 3-900051-07-0.
- Balkenhol, N.; Waits, L.P.; Dezzani, R.J. Statistical approaches in landscape genetics: an evaluation of methods for linking landscape and genetic data. Ecography 2009, 32, 818–830. [Google Scholar] [CrossRef]
- Landguth, E.L.; Fedy, B.C.; Oyler-McCance, S.J.; Garey, A.L.; Emel, S.L.; Mumma, M.; Wagner, H.H.; Fortin, M.-J.; Cushman, S.A. Effects of sample size, number of markers, and allelic richness on the detection of spatial genetic patterns. Mol. Ecol. Resour. 2012, 12, 276–284. [Google Scholar] [CrossRef]
Supplementary
| Model | | | Model | |||||
|---|---|---|---|---|---|---|
| Alpha | Proportion Model | Null Sig (one-step) | Proportion Model | Alt. Models Sig. (two-step) | Proportion Model | Alt. Models Sig. (all-pairs) | Proportion Null | Model Sig (one-step) | Proportion Alt. Models | Model Sig. (two-step) | Proportion Alt. Models | Model Sig. (all-pairs) |
| 0.05 | 1 | 0.828571 | 0.101587 | 0.428571 | 0 | 0.506349 |
| 0.04 | 1 | 0.828571 | 0.111905 | 0.4 | 0.028571 | 0.512698 |
| 0.03 | 1 | 0.828571 | 0.114286 | 0.371429 | 0.028571 | 0.52381 |
| 0.02 | 1 | 0.828571 | 0.119841 | 0.342857 | 0.028571 | 0.536508 |
| 0.01 | 1 | 0.828571 | 0.13254 | 0.342857 | 0.028571 | 0.554762 |
| 0.009 | 1 | 0.828571 | 0.13254 | 0.342857 | 0.028571 | 0.554762 |
| 0.008 | 1 | 0.828571 | 0.13254 | 0.342857 | 0.028571 | 0.554762 |
| 0.007 | 1 | 0.828571 | 0.13254 | 0.342857 | 0.028571 | 0.554762 |
| 0.006 | 1 | 0.828571 | 0.13254 | 0.342857 | 0.028571 | 0.554762 |
| 0.005 | 1 | 0.828571 | 0.13254 | 0.342857 | 0.028571 | 0.554762 |
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Cushman, S.A.; Wasserman, T.N.; Landguth, E.L.; Shirk, A.J. Re-Evaluating Causal Modeling with Mantel Tests in Landscape Genetics. Diversity 2013, 5, 51-72. https://doi.org/10.3390/d5010051
Cushman SA, Wasserman TN, Landguth EL, Shirk AJ. Re-Evaluating Causal Modeling with Mantel Tests in Landscape Genetics. Diversity. 2013; 5(1):51-72. https://doi.org/10.3390/d5010051
Chicago/Turabian StyleCushman, Samuel A., Tzeidle N. Wasserman, Erin L. Landguth, and Andrew J. Shirk. 2013. "Re-Evaluating Causal Modeling with Mantel Tests in Landscape Genetics" Diversity 5, no. 1: 51-72. https://doi.org/10.3390/d5010051
APA StyleCushman, S. A., Wasserman, T. N., Landguth, E. L., & Shirk, A. J. (2013). Re-Evaluating Causal Modeling with Mantel Tests in Landscape Genetics. Diversity, 5(1), 51-72. https://doi.org/10.3390/d5010051





