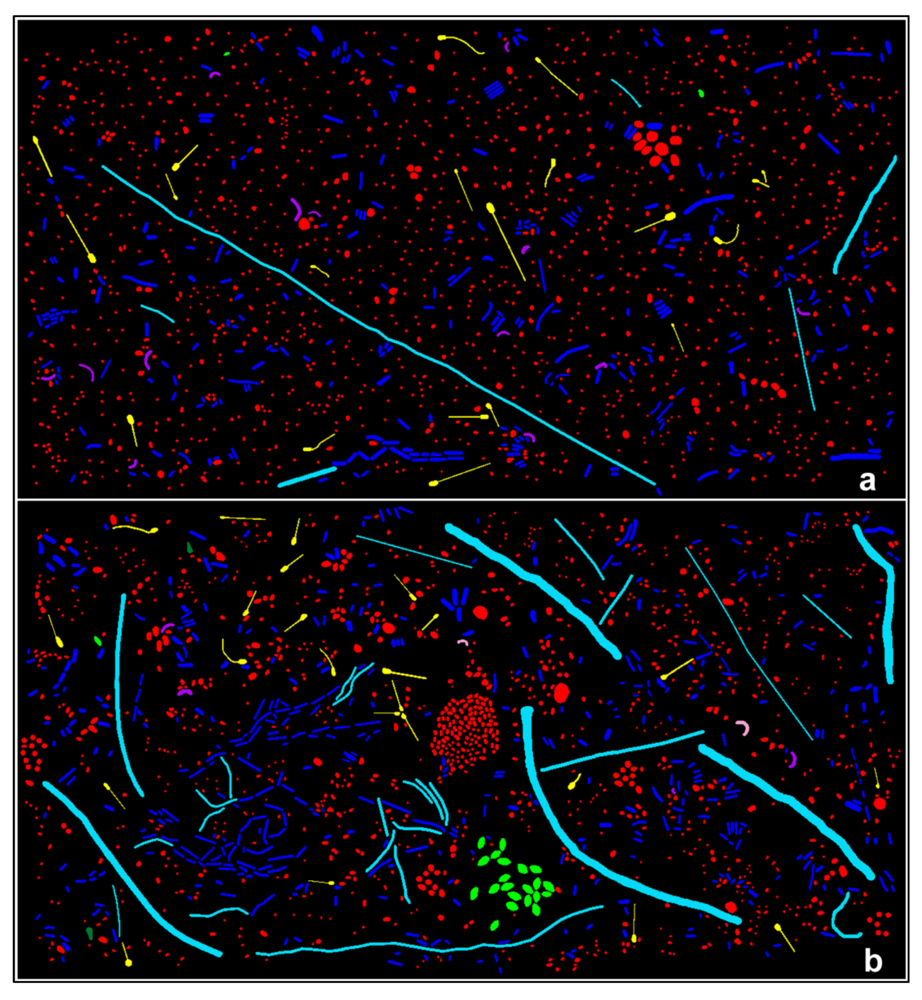
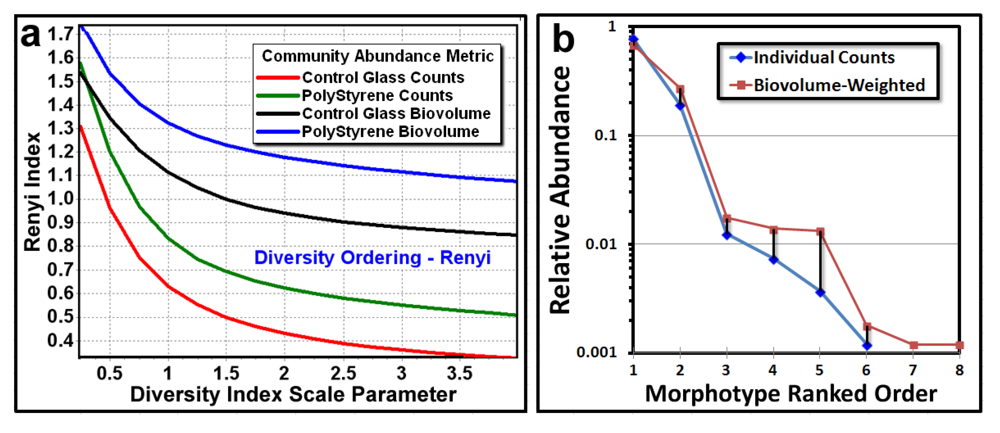
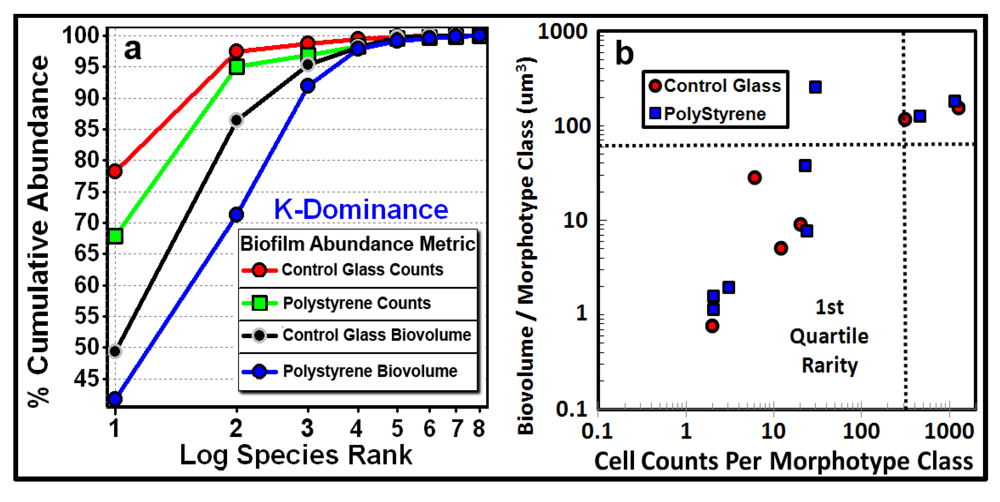
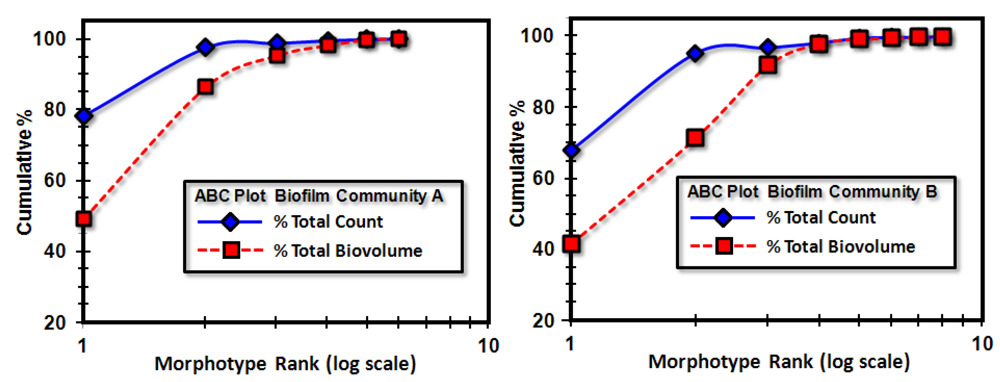
3.3.1. Background
Spatial patterns of distribution among community members influence many processes that are ecologically important [
43]. Because many ecological processes are scale-dependent, their study should include a spatial component measured at the scale in which the process occurs [
43]. A major goal of spatial analysis in ecophysiological studies is to define what a measured characteristic at one location can reveal about that same characteristic at neighboring locations. Analyses of
in situ spatial ecology are designed to scrutinize distribution patterns at a given spatial scale and produce predictive ecological models of colonization behavior that help to reveal the ecophysiological processes occurring in that habitat [
43].
The quantification of spatial heterogeneity is necessary to elucidate relationships between ecological processes and spatial patterns [
43]. The essence of the statistical pattern analysis is to distinguish between spatial distributions of the organisms that can be explained by random chance
versus those that cannot. The benchmark of that assessment tests the null hypothesis that the patterns have
complete spatial randomness represented by a Poisson distribution with means equal to variance. Complete spatial randomness implies that no microbial interactions affect the events resulting in their spatial pattern of colonization. In contrast, statistically significant deviations from complete randomness in spatial patterns can reflect phenotypic ecophysiological adaptations in spatially structured landscapes, indicating that localized and/or regionalized microbial interactions have affected their colonization behavior resulting in the spatial pattern present.
Non-random spatial structures may not only result from ecological interactions; they may also play an essential functional role in organizing the interactions that dictate their ecophysiology and stability [
44]. Spatial patterns of microbial colonization that are
aggregated (clustered) imply positive (cooperative and/or mutualistic) interactions among neighboring cells that have promoted each other’s growth ecophysiology. Extreme examples of this relationship occur in interspecies coaggregates of cross-feeding, e.g., syntrophic microbial species residing in methanogenic communities. Aggregated patterns of distribution in landscape structures also result from the scale-dependent heterogeneous fractal variability in limiting resource partitioning, and reflect the high efficiency at which cells actively disperse and cooperatively position themselves spatially and physiologically when faced with the interactive forces of microbial coexistence to optimize their allocation of nutrient resources on a local competitive scale [
26]. In contrast, patterns of spatial
uniformity (regularity) imply negative (inhibitory and/or antagonistic) interactions that have resulted in their maximally separated, over-dispersed, self-avoiding colonization behavior. This information is of significant ecophysiological importance because spatial heterogeneity resulting from both types of nonrandom patterns between individuals (aggregated or uniform) tends to stabilize ecological systems [
44,
45] and can explain much of the species diversity that coexists in a community colonizing a habitat [
26]. Key issues in spatial pattern analysis that typically follow statistical rejection of the null hypothesis of complete spatial randomness include whether the pattern exhibits uniform regularity or coaggregation, the spatial scale at which the pattern of interaction is defined (local
vs. regional), and the statistical strength of that pattern’s departure from randomness [
45].
Does location really matter in microbial ecophysiology? Do the patterns of microbial spatial distribution represent “ecological music” or are they no more than random noise? The common answer is
yes indeed,
location does matter and structured patterns of microbial spatial distribution can be magnificently symphonic. Spatial segregation of morphotypes colonizing the same habitat can provide insights into their feeding behavior, trophic level, food web dynamics and reproductive capacity. This trend has consistently been found in our spatial ecology studies of microbial biofilms that develop in a variety of natural and managed habitats, including plant rhizoplanes and phylloplanes [
46,
47,
48,
49], freshwater streambed pebbles [
50], and microscope slides suspended in various river/lake ecosystems [
2,
51,
52].
Microbial colonization of local areas low in nutrient availability leads to poor productivity with a tendency to form overdispersed spatial patterns indicative of intense competition, whereas colonization of nutritionally enriched microenvironments results in increased growth that flourishes as aggregated patterns of local microcolony biofilms [
51,
52]. Central to these negative (conflicting) and positive (cooperative) interactions are various molecular cell–cell communication events that regulate the ecophysiology affecting microbial colonization behavior and biofilm architecture [
52]. Also, knowing the location and intensity of clustered behavior for microorganisms can improve the understanding of the underlying processes that generate and sustain the interdependent microbe-environment relationships within biofilm architectures and the spatial scale at which they occur [
51,
52]. Thus, modeling spatial patterns of microbial communities at multiple spatial scales is crucial to understand their ecophysiological functioning fully. Indeed, no study of microbial ecophysiology is complete without an
in situ spatial consideration of their activities within the habitat, simply because everything is not randomly located everywhere. Applying spatial statistics to analyze microbial biofilm architectures can also provide insight on the ecological forces that underlie the basic mechanisms of the colonization behavior that created them.
3.3.2. Point Pattern Spatial Distribution Analysis
CMEIAS is designed to analyze three different categories of microbial spatial distribution. The first category is a plot-less “point pattern,” based on extraction of the in situ micrometer scaled distances between each bacterial cell and its nearest neighbor within the biofilm landscape. Spatial attributes included in these analyses are the X|Y Cartesian coordinates of each cell’s centroid position (relative to the assigned 0, 0 landmark position located at the lower left corner of the image), the μm scaled distance from each object’s centroid to its first and second nearest bacterial neighbors, and the empirical distribution function of first nearest neighbor distances.
Several analyses of spatial point patterns were performed on the two biofilm communities for this study. The first involved a calculation of the Clark and Evan’s Randomness Index intended to test for departure from complete spatial randomness in the overall landscape structure. This spatial statistic is computed from data of the first nearest neighbor distances between cells and the spatial density within the biofilm landscape. The mean neighboring distances were greater for observed than expected values, Z values were greater than the threshold border of 1.96, and R indices were greater than 1.00 (
Table 6). These results detect statistically significant departure of complete spatial randomness for spatial patterns in both biofilm assemblages, including uniform patterns signalling negative over-dispersed conflicts of self-avoiding colonization behavior that are more intense in biofilms of Community A than in Community B (
p values of 3.21 × 10
−64 and 2.78 × 10
−5, respectively).
Table 6.
Clark and Evans spatial point pattern test for complete spatial randomness of cells in biofilm communities A and B. Calculations are based on landscape areas defined by the convex hull of point distributions with Donnelly edge correction [
8].
Table 6.
Clark and Evans spatial point pattern test for complete spatial randomness of cells in biofilm communities A and B. Calculations are based on landscape areas defined by the convex hull of point distributions with Donnelly edge correction [8].
| Point Pattern Statistic | Community A | Community B |
|---|
| Observed Mean Distance | 1.5125 | 1.3191 |
| Expected Mean Distance | 1.2331 | 1.2494 |
| Z test statistic | 16.92 | 4.2168 |
| R Index | 1.266 | 1.056 |
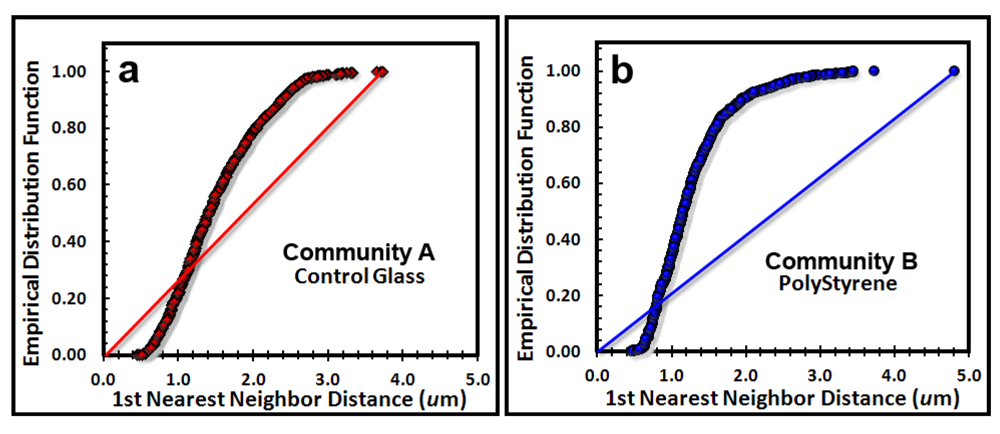
The next point pattern analysis evaluated the
Empirical Distribution Function (EDF) of the cells’ spatial distribution, which produces a plot that compares the cumulative ranking of the first nearest neighbor distances between individual cells in the sample to the theoretical distribution that would result if the pattern were completely random (indicated by a diagonal random trendline that extends from the XY intercept to the maximum nearest neighbor distance found in the analysis). Datapoints indicate a uniform pattern of distribution when they form a tight group with little range of first neighbor distance, an aggregated distribution when the curve ascends to form a distinct extended asymptote at EDF of 1.00 above the random diagonal trendline, and a random distribution when the EDF curve ascend with a shallower slope closer to the diagonal trendline. Differences in intensity of aggregated patterns are indicated by their relative distance above the diagonal trendline of complete spatial randomness. The empirical distribution plot for the biofilm landscapes (
Figure 6a,b) indicate a greater portion of uniformly dispersed pattern for Community A (consistent with Clark and Evans R index,
Table 6), and a steeper curve displaced further above the blue diagonal trendline for Community B, indicating that Community B has a more intense aggregated pattern than does Community A, whose EDF curve is shallower and closer to the red diagonal trendline.
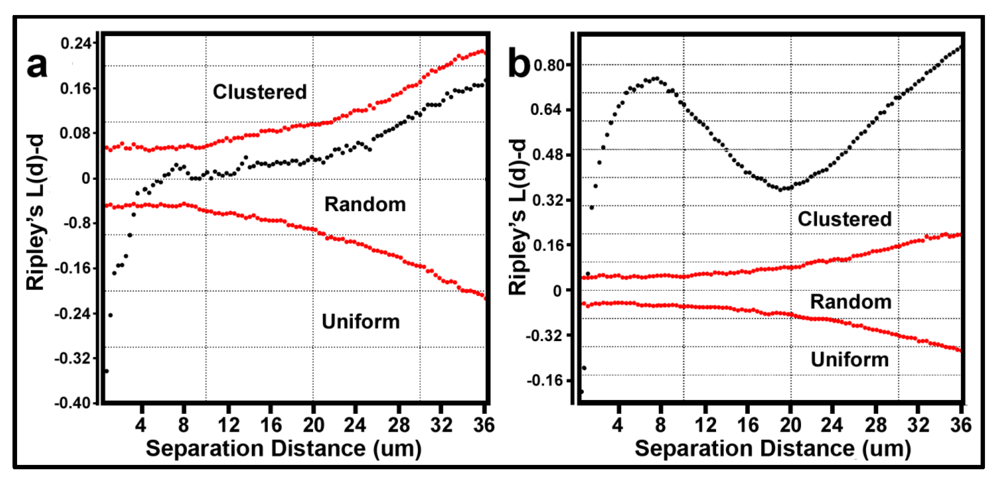
Ripley’s K [
53] multi-distance spatial cluster analysis is a useful sequel to the EDF analysis. This is a second-order, point distribution statistic that evaluates the co-occurrence of separation distances between pairs of object points to determine if the point pattern changes with distance of the spatial scale of analysis. The
K(d) function measures the average count of objects enclosed within circles of radius
d that are centered on every object point in the landscape divided by the mean spatial density of objects in that landscape. A plot of all
K(d) functions
vs. all radial separation distances for all objects in the landscape indicates if the pattern is uniformly dispersed, clustered or enclosed within a Monte Carlo simulation of the confidence envelope representing the 95% critical limits for a test of complete spatial randomness. Observed
K(d) values represent
uniform spatial distributions when located below the confidence envelope of spatial randomness, and represent
clustered distributions when located above the confidence envelope. The intensity of clustering or uniformity is indicated by the relative proportion and location of observed points whose separation distances lie above or below the statistically defined 95% envelope of spatial randomness, respectively.
Figure 6.
Cumulative empirical distribution function of the first nearest neighbor distances between individual bacteria within biofilm assemblages of community A (a) and community B (b). The diagonal trendline of complete spatial randomness is indicated in both plots for comparison.
Figure 6.
Cumulative empirical distribution function of the first nearest neighbor distances between individual bacteria within biofilm assemblages of community A (a) and community B (b). The diagonal trendline of complete spatial randomness is indicated in both plots for comparison.
The Donnelly edge-corrected [
7,
8] and standardized Ripley K (L(d)-d) [
8] values of point pattern characteristics for all bacteria within biofilm community assemblages developing on the plain glass and polystyrene substrata are plotted in
Figure 7a,b respectively. Several features of these two Ripley K plots reveal statistically significant information that dramatically distinguishes the spatial patterns of the microbes at different spatial scales in these two biofilm assemblages. First, some cells in both biofilm landscapes are uniformly equidistant from each other. These occur at eight different separation distances in community A and two different separation distances in community B. Second, the majority of separation distances is represented by cells with random spatial distribution in community A, and by cells that are spatially aggregated in community B (note the larger Y-axis scale in
Figure 7b). Third, the distribution of K functions for spatially aggregated cells in Community B has a discrete mode of local interactions at separation distances of approximately 6–8 μm (up to 18 μm) and another mode of regional interactions at a maximum of 36 μm. These interesting results indicate that the biofilm landscape that developed on the polystyrene substratum is significantly more spatially aggregated than the biofilm developed on plain glass. This distinction of colonization behavior revealed by the Ripley K analysis illustrates how the spatial structure of second-order interactions within microbial biofilms can be significantly influenced by the (physico)chemistry of the substratum upon which it develops.
Figure 7.
Donnelly edge-corrected and standardized Ripley K plots of point pattern characteristics for all bacteria within biofilm community assemblages of communities A and B developing on the (a) plain glass and (b) polystyrene substrata. The results indicate that most cells are randomly distributed in Community A and clustered in Community B.
Figure 7.
Donnelly edge-corrected and standardized Ripley K plots of point pattern characteristics for all bacteria within biofilm community assemblages of communities A and B developing on the (a) plain glass and (b) polystyrene substrata. The results indicate that most cells are randomly distributed in Community A and clustered in Community B.
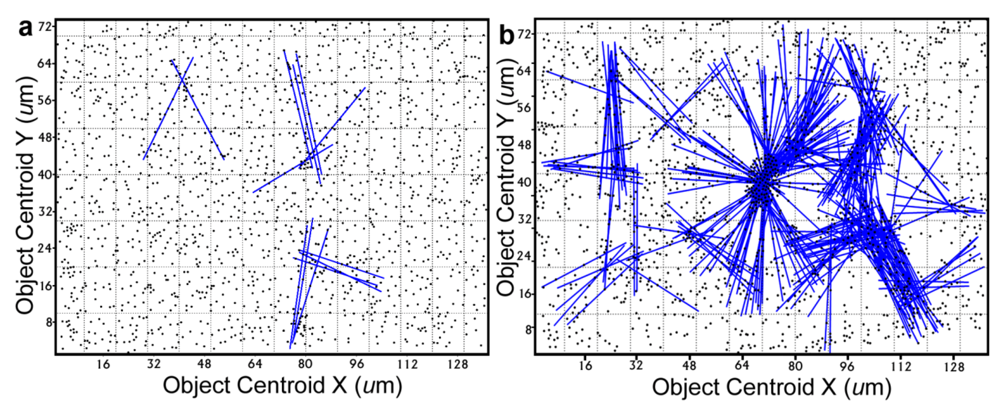
Further definition of the location, intensity and radial dimension of bacterial interactions emanating from discrete spatial aggregates within biofilms can be predicted from 2-dimensional plots of their
line segment patterns. These plots indicate the statistically significant
linear point alignments (
Figure 8a,b) whose angular orientations dramatically illustrate the directionality of multiple intersecting “hot spot” epicenters of interpoint interactions [
8,
54] that are strongest within clusters of closely neighboring bacteria in the biofilm communities. As anticipated, the numbers of linear point alignments and clustered “hot-spot” epicenters of positive interpoint interactions between closely aggregated bacteria are significantly greater in the biofilm community B, and they point in all compass directions (
Table 7).
Figure 8.
Line segment pattern plots of statistically significant linear point alignments whose intersecting angular orientations locate numerous “hot-spot” epicenters of clustered bacterial interactions within the biofilms communities A (a) and B (b).
Figure 8.
Line segment pattern plots of statistically significant linear point alignments whose intersecting angular orientations locate numerous “hot-spot” epicenters of clustered bacterial interactions within the biofilms communities A (a) and B (b).
Table 7.
Intensity of the line segment patterns indicating clustered bacterial interactions in
Figure 8a,b.
Table 7.
Intensity of the line segment patterns indicating clustered bacterial interactions in Figure 8a,b.
| Biofilm Landscape | Point Alignment Lines | Point Alignment Epicenters |
|---|
| Community A | 13 | 4 |
| Community B | 242 | 31 |
3.3.4. Geostatistical Spatial Distribution Analysis
The third category of spatial pattern analysis performed on spatial data is called a geostatistical analysis. This robust method of analysis measures the degree of dependency among observations in a geographic space to evaluate the continuity or continuous variation of spatial patterns over that entire spatial domain [
55]. Geostatistics tests whether a user-defined, continuously distributed variable is
spatial autocorrelated,
i.e., exhibits spatial structure. The result of the analysis quantifies the spatial uncertainty about the high irregularity of regionalized variables and the scale of their spatial correlation. It indicates whether the events among pairs of cells at one location influence that same measured variable associated with their cell neighbors at more distant locations. Patterns displaying spatial autocorrelation indicate that operations of colonization behavior involve a spatially explicit process rather than occur randomly and independent of their location. It quantifies the resemblance of the measured parameter between neighbors as a function of their separation distance. Geospatial data are autocorrelated when close neighbors are more similar (as in aggregated distributions) than are neighbors locate further apart [
56]. When found, the autocorrelation can be accurately modeled using its weighted average from neighboring sampled locations to connect various spatially dependent relationships derived from regionalized variable theory, plus make optimal, statistically rigorous interpolation (kriging) maps of the parameter at unmeasured locations within that spatial domain.
Geostatistical analysis and krig mapping require that the relevant parameter being analyzed (the so-called “Z variate”) is a quantitative (non-binary) metric that is continuously distributed over the spatial domain, and that the sampling sites are
georeferenced at known
X, Y Cartesian coordinates relative to a landmark position in the same landscape (commonly set to 0, 0 coordinates at the lower left corner of the image) [
10,
55]. We developed the
CMEIAS cluster index as the major Z-variate to conduct geostatistical analyses of bacterial surface colonization behavior at single-cell resolution [
57]. CMEIAS computes this cluster index as the inverse of the separation distance between the object centroid of each bacterial cell and its first nearest cell neighbor. This index is an example of a local indicator of spatial association whose magnitude reflects the intensity of each cell’s clustered distribution in relation to other cells in its local environment. Used as such, it performs well as a sensitive sensor of positive, spatially autocorrelated cell-to-cell interactions that cooperatively promote bacterial colonization behavior
in situ [
51,
52,
57]. Cells with a high cluster index are typically arranged in aggregated patterns that facilitate cell communications, resulting in positive metabolic cooperations that promote their localized growth into populations of microcolony biofilms [
47,
51,
52,
57,
58].
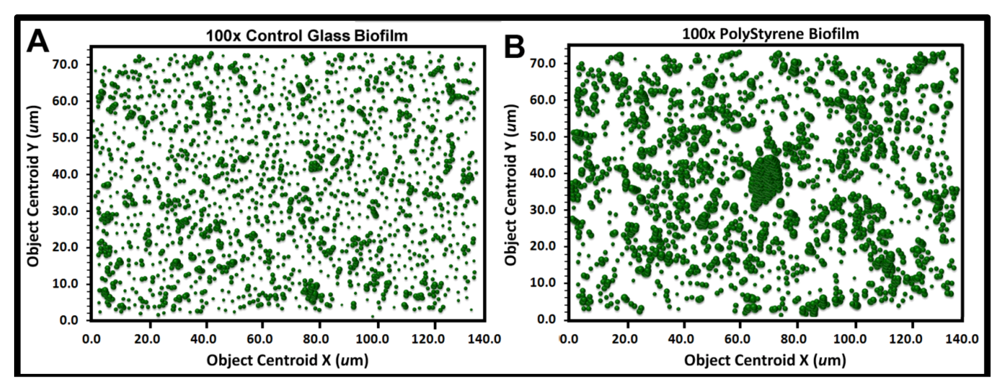
The heterogeneity in cluster index assigned to cells within the landscape domain of the biofilm can be evaluated several ways. One method is to produce a 3-dimensional
georeferenced bubble scatterplot where each cell in the biofilm landscape is represented by a bubble whose diameter (width) is weighted by its individual CMEIAS cluster index. The information provided by this spatial aggregation analysis reveals the significant variable of spatial proximity among individual cells in the biofilm over the entire landscape domain, which is more important than a quorum of high population density
per se in governing the success of cell–cell communication within biofilms [
12]. First, an object analysis is performed on the spatially calibrated image to extract the XYZ data from every cell, indicating its X,Y Cartesian posting locations relative to the landmark origin and the Z coordinate of its cluster index. These georeferenced data are then used to produce the 3-dimensional bubble scatterplot, indicating the X, Y spatial position of every cell and the magnitude of its associated cluster index. This analysis shows the proximity and intensity of spatial coaggregation where positive cell–cell interactions affecting colonization behavior are likely to occur.
Figure 9a,b show the bubble plots for all cells in the biofilm assemblages of communities A and B, respectively. The significantly larger and more numerous clustered bubbles in the biofilm landscape of community B clearly indicate higher spatial aggregation of cells, with implication that its spatial heterogeneity would define the intensity of diffusion gradients of chemical signals and other bioactive metabolites that influence their biofilm ecophysiological activities, including colonization behavior.
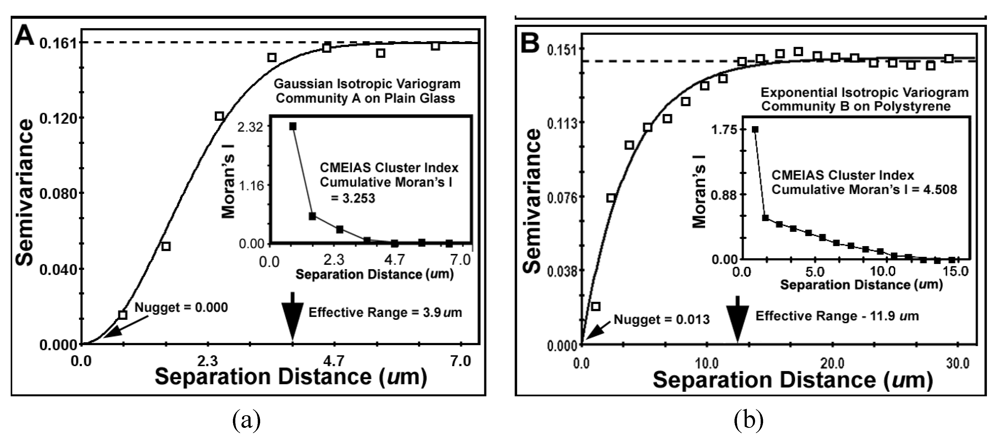
The second type of geostatistical analysis is designed to produce the variogram plot, which defines the extent that the measured Z-variate for each cell exhibits spatial dependence between pairs of sample locations. This plot relates the uncertainty of regionalized variables with the distances that they autocorrelate. It statistically describes how autocorrelated values at two points begin to become different as the separation distance between them increases. That information provides the profoundly important and statistically defendable estimate of the real micrometer range of spatial scale of separation distances at which individual neighboring cells can influence each other’s Z-variate.
The results of key metrics derived from the geostatistical autocorrelation analysis are presented in
Table 9 and
Figure 10a,b. The computed mathematical models of the geostatistical data for both communities made a statistically highly significant fit (r
2) indicating that the cluster index Z-variate is
spatial autocorrelated. These best-fit models are isotropic, indicating that variations in the cluster index occur in all compass directions (not anisotropic), consistent with the vectors of linear point alignments indicated in the line segment pattern plots in
Figure 8a,b. This lack of anisotropy at the scale measured predictably reflects the freedom of rotation of the biofilm substratum (microscope slides) and their dangling orientations while connected to the fishing pole line in the flowing river ecosystem. The very low
nugget values for both communities indicate minimal discontinuity in the data,
i.e., that the amount of measured geospatial microstructure of the cluster index was adequately represented by the number of sampling points and that they were sampled at the proper spatial scale.
Figure 9.
Georeferenced bubble plots of the biofilm landscapes containing community A (a) and B (b). The distribution of each individual bacterial cell is represented as a 3-dimensional bubble whose width is weighted by its Cluster index. Note the larger aggregated bubbles for the biofilm assemblage of community B.
Figure 9.
Georeferenced bubble plots of the biofilm landscapes containing community A (a) and B (b). The distribution of each individual bacterial cell is represented as a 3-dimensional bubble whose width is weighted by its Cluster index. Note the larger aggregated bubbles for the biofilm assemblage of community B.
The
effective ranges of autocorrelated separation distance between sampling points indicate the real spatial scale of the first order maximal radial distance at which individual cells influence their neighbor’s spatial aggregation. In this example, the effective range represents the maximal radial distance at which each individual cell can still influence its neighbor’s clustered colonization behavior, enabling it to grow into an aggregated microcolony biofilm
in situ. This corresponds to the first (local) scale of aggregated influence in community B found by the Ripley K point-pattern analysis (
Figure 7b). Interestingly, the Ripley’s K point-pattern also indicated a second peak of regional aggregated influence occurring between cells at a separation distance of approximately 36 μm in the biofilm assemblage of community B (
Figure 7b).
Table 9.
Parameters of the best-fit geostatistical models derived from the isotropic variogram of the CMEIAS cluster index for microbial cells within biofilm assemblages of communities A and B.
Table 9.
Parameters of the best-fit geostatistical models derived from the isotropic variogram of the CMEIAS cluster index for microbial cells within biofilm assemblages of communities A and B.
| Parameter | Biofilm Community A | Biofilm Community B |
|---|
| Best Fit Variogram Model | Gaussian | Exponential |
| r2 of Correlation Coefficient | 0.992 | 0.971 |
| Nugget (variance of Y at X = 0 distance) | 0.0001 | 0.0128 |
| Effective Range (Radius of Autocorrelation, μm) | 3.9 | 11.9 |
| Cumulative Global Moran’s Index | +3.253 | +4.508 |
Figure 10.
Variogram plots of the spatially autocorrelated Z-variate of Cluster Index for all bacteria in the biofilm landscapes of communities A (a) and B (b). Shown are differences in (1) sample variance (dashed lines), (2) the mathematical model best fitted to the data (solid lines and its associated statistics), (3) the effective ranges of separation distance (large arrows at the X axis), (4) the nuggets (small arrows at the Y axis) and (5) the Moran’s Index insert plot of positively autocorrelated Z-variate intensity.
Figure 10.
Variogram plots of the spatially autocorrelated Z-variate of Cluster Index for all bacteria in the biofilm landscapes of communities A (a) and B (b). Shown are differences in (1) sample variance (dashed lines), (2) the mathematical model best fitted to the data (solid lines and its associated statistics), (3) the effective ranges of separation distance (large arrows at the X axis), (4) the nuggets (small arrows at the Y axis) and (5) the Moran’s Index insert plot of positively autocorrelated Z-variate intensity.
The global
Moran’s Index represents the intensity of spatial dependence in the autocorrelated Z-variate of aggregated pattern relationship between paired observations in the neighborhood. The positive autocorrelated values of the Moran’s Index (
Table 9) for communities A and B indicate that the cell clustering behavior is significantly more spatially dependent than would be expected if the underlying spatial processes of colonization were random across the geographic landscape structure. From the microbial ecology perspective, spatial patterns of distribution with positive Moran’s autocorrelated indices imply an active cooperation in aggregated colonization behavior involving cell-to-cell interactions that positively affect their spatial distribution over a defined radial scale within the landscape domain. The radial distance of that influence is ~3-fold larger for community B than community A (
Table 9), and encapsulates 100% of the cells based on their first nearest neighbor distance. Ecophysiological processes resulting in this type of positive autocorrelation would indicate that the measured Z variate between neighboring cells is a function of their spatial location within the domain, and predictably could include,
inter alia, nutritional cross-feeding, elaboration of signal molecules that activate genes positively affecting cell growth into microcolonies, localized detoxification/degradation of extracellular metabolic wastes, increased sequestration of limiting nutrients, and a biofilm matrix of protective extracellular polysaccharides providing them with a defensive refuge from predatory activity and restricted diffusion of antimicrobials. Negative Moran indices would imply strong competitive and/or inhibitory cell-to-cell interactions.
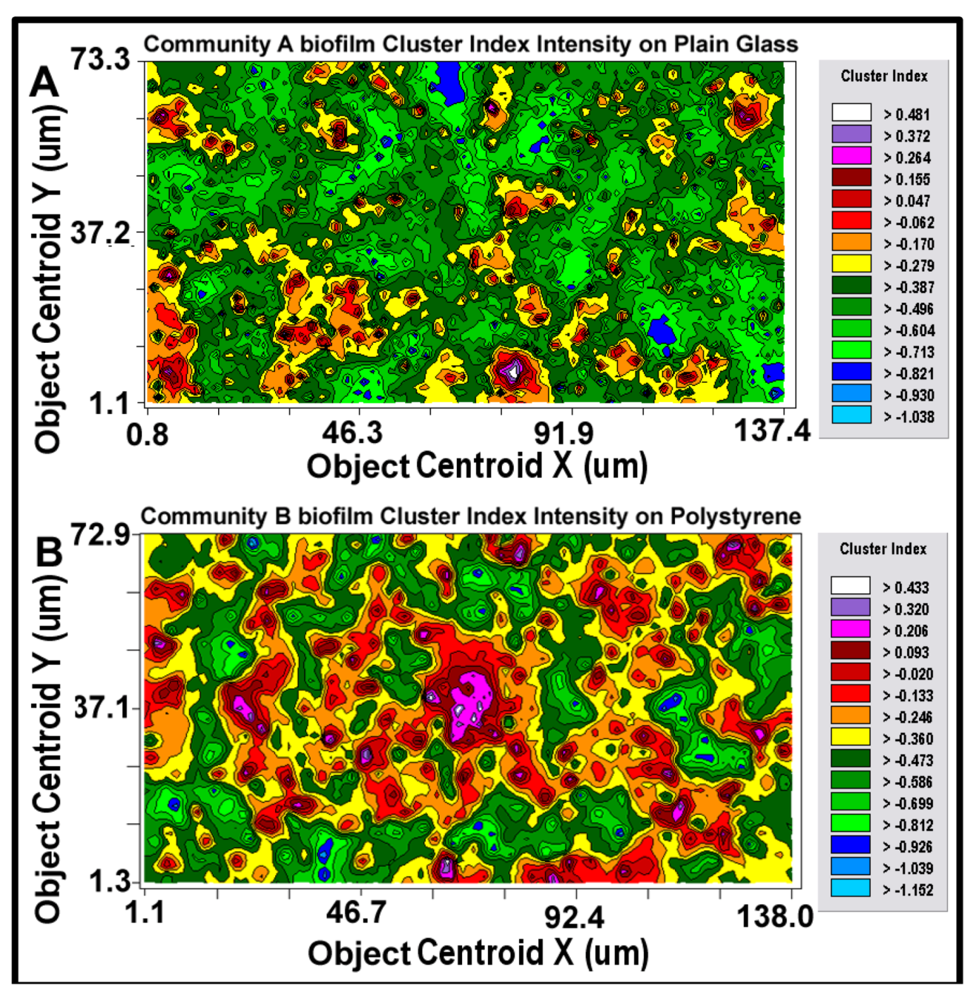
After the best-fit autocorrelation model is computed, the corresponding kriging map can be built (
Figure 11a,b). This kriging method uses the best-fit statistical model to estimate values of the quantitative, regionalized Z-variate parameter and provides a vivid, geostatistically defendable continuous interpolation map of the spatial variability of that parameter, even at places where it has not been measured within the entire spatial domain [
10,
55]. Included in the kriging map are
isopleth lines whose curvature connects points of equal value. The configuration of these contours and the pseudocolored scale associated with them reveal the relative gradient of the Z-variate’s intensity of cell–cell interactions within spatially defined clusters, and estimates that parameter at every spatial location within the landscape domain, like weather maps that interpolate the spatial gradients of temperature and cloud cover over large-scale landmasses.
Figure 11.
2-dimensional kriging maps of the spatial heterogeneity in autocorrelated intensity of positive aggregated colonization behavior of the microbial assemblages in the two biofilm landscapes representing community A (a) and B (b). The kriging map legends show the stepped scale of pseudocolored bins that cover the full range of the Z-variate intensity (CMEIAS cluster index) in each landscape.
Figure 11.
2-dimensional kriging maps of the spatial heterogeneity in autocorrelated intensity of positive aggregated colonization behavior of the microbial assemblages in the two biofilm landscapes representing community A (a) and B (b). The kriging map legends show the stepped scale of pseudocolored bins that cover the full range of the Z-variate intensity (CMEIAS cluster index) in each landscape.
Amazing to find the many significant differences in ecophysiology between the two biofilm communities despite their development only a few inches apart on the fishing line submerged in the flowing Red Cedar River.
Kriging indicates the statistically defendable, intensity-scaled aggregated colonization behavior over a continuum of radial distances in the biofilm landscapes. The pseudocolored scaling and associated isopleth lines in the modeled kriging map (
Figure 11a,b) deliver clearly delineated evidence of autocorrelated centers of highly intensive, local cell–cell aggregation behavior with raised probability of intercellular interactions that are significantly stronger and spatially abundant in the biofilm community B that developed on the polystyrene substratum. These awesome foci of strongly clustered cells colocalize with epicenters of “hot spots” revealed by the point pattern method of linear alignments (
Figure 8a,b), and with the georeferenced posting plots of cells represented by bubbles whose widths are weighted by each individual cell’s cluster index (
Figure 9a,b). Kriging analysis also predicts the
in situ strength of diffusion gradients of extracellular signal molecules that promote/positively regulate these cooperative cell-to-cell interactions within the biofilm [
12,
49,
52].