High Genetic Diversity in Geographically Remote Populations of Endemic and Widespread Coral Reef Angelfishes (genus: Centropyge)
Abstract
:1. Introduction
2. Experimental Section
2.1. Field Collections
2.2. Laboratory Procedures
2.3. Data Analyses
3. Results
3.1. Endemic C. joculator
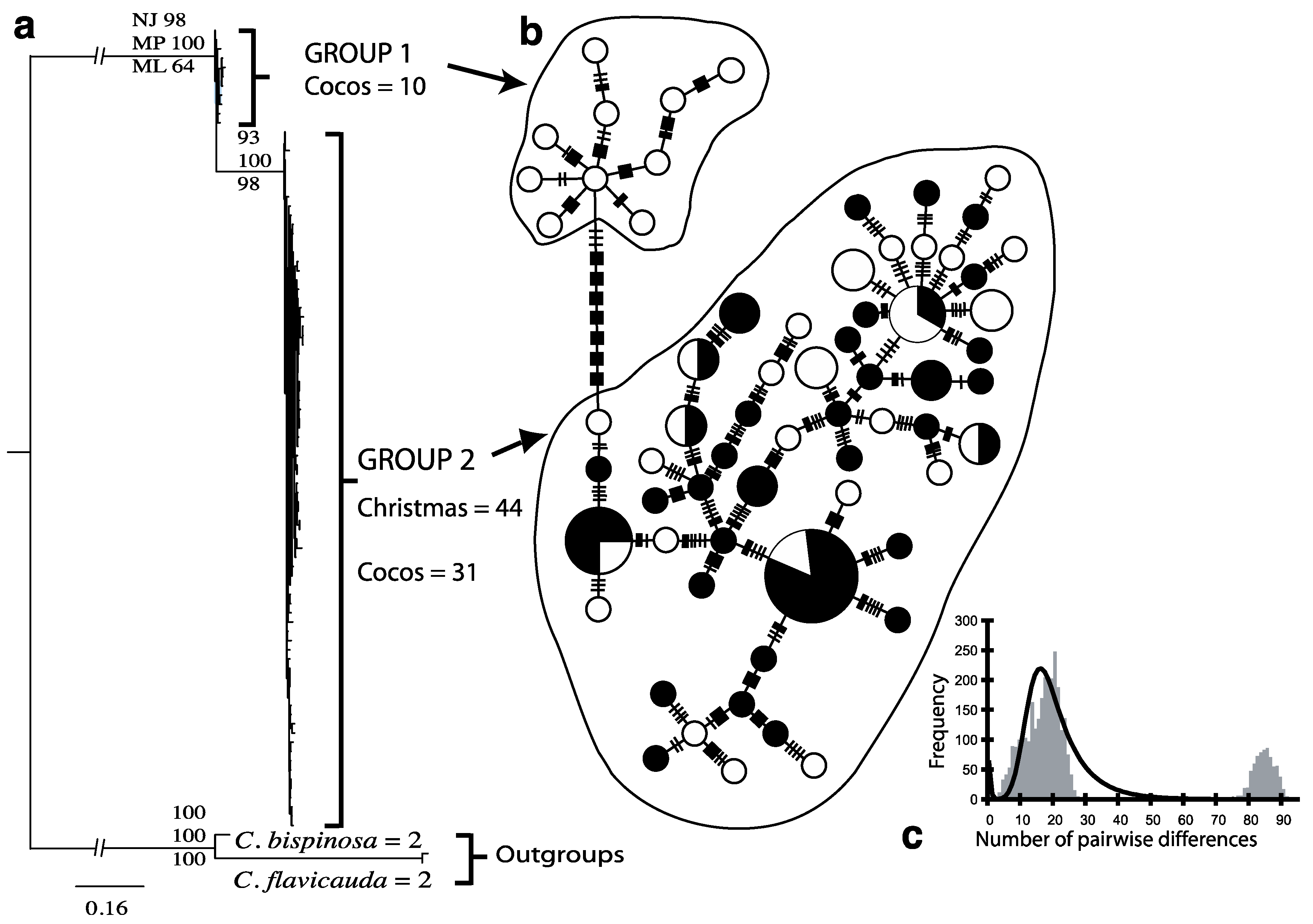
| Species | Location | n | nh | h (±SD) | π%(±SD) | Mean pairwise differences (±SD) |
|---|---|---|---|---|---|---|
| C. flavicauda | Christmas Island | 28 | 28 | 1 ± 0.010 | 2.81 ± 0.39 | 12.08 ± 5.63 |
| C. bispinosa | Christmas Island | 24 | 23 | 0.989 ± 0.012 | 5.78 ± 0.73 | 24.04 ± 10.95 |
| C. joculator | Christmas Island | 44 | 35 | 0.983 ± 0.011 | 3.63 ± 1.83 | 15.70 ± 7.14 |
| C. joculator | Cocos Islands | 41 | 37 | 0.995 ± 0.007 | 9.99 ± 4.91 | 41.65 ± 18.45 |
| C. joculator | Christmas and Cocos Islands | 85 | 66 | 0.991 ± 0.004 | 6.92 ± 0.34 | 30.16 ± 13.30 |
| C. joculator | Group 1 (Cocos Islands) | 10 | 10 | 1 ± 0.045 | 3.34 ± 1.85 | 14.49 ± 7.10 |
| C. joculator | Group 2 (Christmas and Cocos Islands) | 75 | 56 | 0.988 ± 0.005 | 3.66 ± 1.83 | 15.83 ± 7.14 |
3.2. Widespread Species C. bispinosa and C. flavicauda
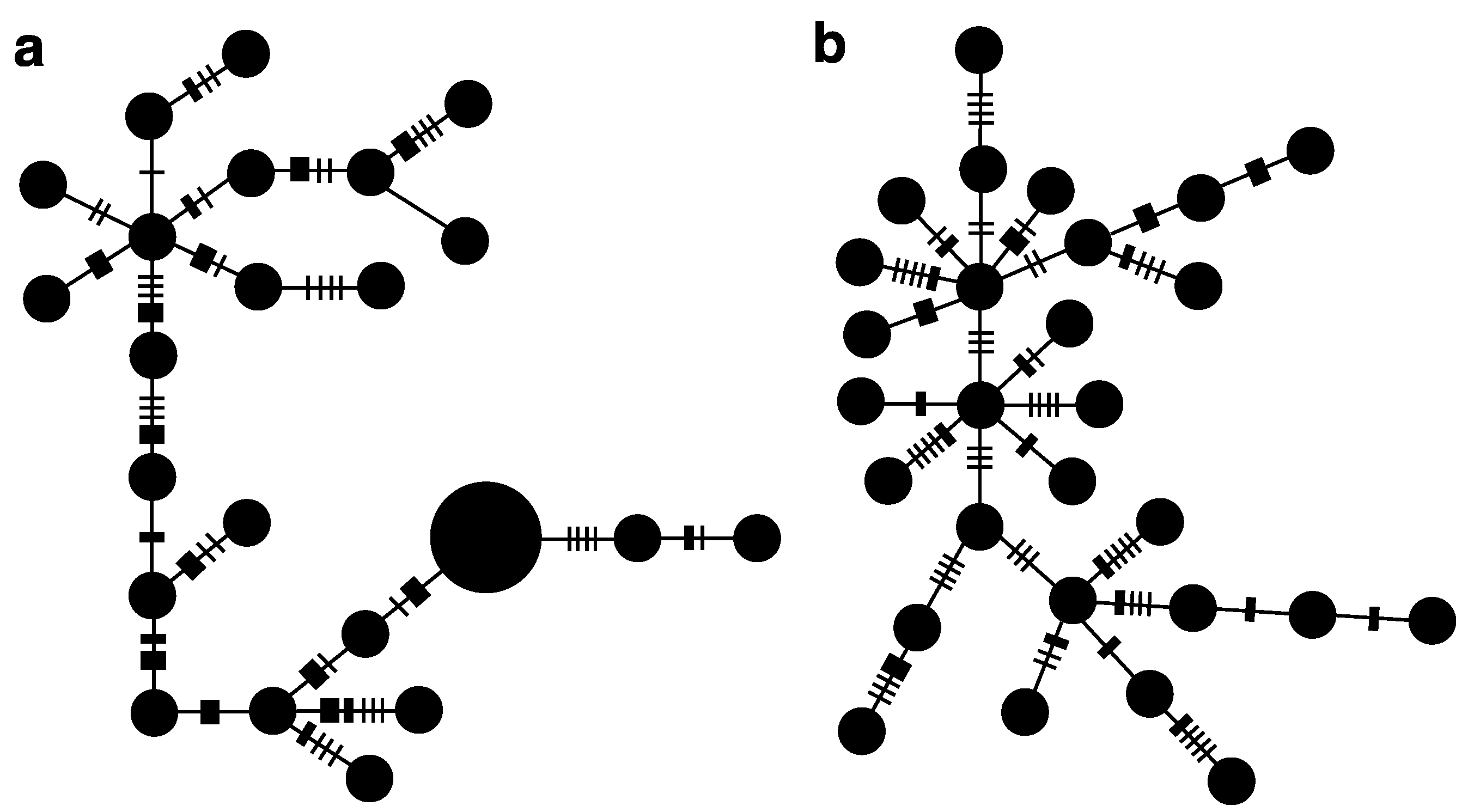
4. Discussion
5. Conclusions
Acknowledgments
References and Notes
- Frankham, R. Do island populations have less genetic variation than mainland populations? Heredity 1997, 78, 311–327. [Google Scholar]
- Frankham, R. Inbreeding and extinction: Island populations. Conserv. Biol. 1998, 12, 665–675. [Google Scholar] [CrossRef]
- Whittaker, R.J. Island Biogeography: Ecology, Evolution, and Conservation; Oxford University Press: Oxford, UK, 1998. [Google Scholar]
- Pimm, S.L. The Balance of Nature: Ecological Issues in the Conservation of Species and Communities; University of Chicago Press: Chicago, IL, USA, 1991. [Google Scholar]
- Gaston, K.J. Rarity; Chapman & Hall: London, UK, 1994. [Google Scholar]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- McNeely, J.A.; Miller, K.R.; Reid, W.V.; Mittermeier, R.A.; Werner, T.B. Conserving the World's Biological Diversity; IUCN: Gland, Switzerland, 1990. [Google Scholar]
- Gaston, K.J.; Blackburn, T.M.; Lawton, J.H. Interspecific abundance-range size relationships: An appraisal of mechanisms. J. Anim. Ecol. 1997, 66, 579–601. [Google Scholar] [CrossRef]
- Frankham, R. Relationship of genetic variation to population size in wildlife. Conserv. Biol. 1996, 10, 1500–1508. [Google Scholar]
- Diamond, J. “Normal” extinctions of isolated populations. In Extinctions; Nitecki, M.H., Ed.; University of Chicago Press: Chicago, IL, USA, 1984; pp. 191–246. [Google Scholar]
- Randall, J.E. Zoogeography of shore fishes of the Indo-Pacific region. Zool. Stud. 1998, 37, 227–268. [Google Scholar]
- Robertson, D.R. Population maintenance among tropical reef fishes: Inferences from small-island endemics. Proc. Natl. Acad. Sci. USA 2001, 98, 5667–5670. [Google Scholar] [CrossRef]
- Hughes, T.P.; Bellwood, D.R.; Connolly, S.R. Biodiversity hotspots, centres of endemicity, and the conservation of coral reefs. Ecol. Lett. 2002, 5, 775–784. [Google Scholar] [CrossRef]
- Jones, G.P.; Caley, M.J.; Munday, P.L. Rarity in coral reef fish communities. In Coral Reef Fishes: Dynamics and Diversity in a Complex Ecosystem; Sale, P.F., Ed.; Academic Press: San Diego, CA, USA, 2002; pp. 81–101. [Google Scholar]
- Roberts, C.M.; Hawkins, J.P. Extinction risk in the sea. Trends Ecol. Evol. 1999, 14, 241–246. [Google Scholar] [CrossRef]
- Dulvy, N.K.; Sadovy, Y.; Reynolds, J.D. Extinction vulnerability in marine populations. Fish Fish. 2003, 4, 25–64. [Google Scholar] [CrossRef]
- Bellwood, D.R.; Hughes, T.P.; Folke, C.; Nyström, M. Confronting the coral reef crisis. Nature 2004, 429, 827–833. [Google Scholar] [CrossRef]
- Munday, P.L.; Leis, J.M.; Lough, J.M.; Paris, C.B.; Kingsford, M.J.; Berumen, M.L.; Lambrechts, J. Climate change and coral reef connectivity. Coral Reefs 2009, 28, 379–395. [Google Scholar] [CrossRef]
- Grant, W.A.S.; Bowen, B.W. Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. J. Hered. 1998, 89, 415–426. [Google Scholar]
- Craig, M.T.; Eble, J.A.; Bowen, B.W.; Robertson, D.R. High genetic connectivity across the Indian and Pacific Oceans in the reef fish Myripristis berndti (Holocentridae). Mar. Ecol. Prog. Ser. 2007, 334, 245–254. [Google Scholar] [CrossRef]
- Klanten, O.S.; Choat, J.H.; Van Herwerden, L. Extreme genetic diversity and temporal rather than spatial partitioning in a widely distributed coral reef fish. Mar. Biol. 2007, 150, 659–670. [Google Scholar]
- Horne, J.B.; Van Herwerden, L.; Choat, J.H.; Robertson, D.R. High population connectivity across the Indo-Pacific: Congruent lack of phylogeographic structure in three reef fish congeners. Mol. Phylogenet. Evol. 2008, 49, 629–638. [Google Scholar]
- Gaither, M.R.; Toonen, R.J.; Robertson, D.R.; Planes, S.; Bowen, B.W. Genetic evaluation of marine biogeographical barriers: Perspectives from two widespread Indo-Pacific snappers (Lutjanus kasmira and Lutjanus fulvus). J. Biogeogr. 2010, 37, 133–147. [Google Scholar]
- Briggs, J.C. Marine zoogeography; McGraw-Hill: New York, NY, USA, 1974. [Google Scholar]
- Allen, G.R.; Steene, R.C.; Allen, M. A guide to angelfishes & butterflyfishes; Odyssey Publishing/Tropical Reef Research: Perth, Australia, 1998. [Google Scholar]
- Eble, J.A.; Toonen, R.J.; Bowen, B.W. Endemism and dispersal: Comparative phylogeography of three surgeonfishes across the hawaiian archipelago. Mar. Biol. 2009, 156, 689–698. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular cloning: A laboratory manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Lee, W.J.; Conroy, J.; Howell, W.H.; Kocher, T.D. Structure and evolution of teleost mitochondrial control regions. J. Mol. Evol. 1995, 41, 54–66. [Google Scholar]
- Hall, T. Bioedit: Biological Sequence Alignment; Ibis biosciences: Carlsbad, CA, USA, 2007. [Google Scholar]
- Schneider, S.; Roessli, D.; Excoffier, L. Arlequin: A Software for Population Genetics Data Analysis; Genetics and Biometry Laboratory, University of Geneva: Geneva, Switzerland, 2000. [Google Scholar]
- Rohlf, F.J. Algorithm 76. Hierarchical clustering using the minimum spanning tree. Comput. J. 1973, 16, 93–95. [Google Scholar]
- Nei, M. Molecular evolutionary genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar]
- Zwickl, D.J. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. Thesis, The University of Texas, Austin, 2006. [Google Scholar]
- Swofford, D.L. PAUP*: Phylogenetic analysis using parsimony; Sinauer Associates: Sunderland, MA, USA, 2003; Information confirmed correct. [Google Scholar]
- Bay, L.; Choat, J.H.; Herwerden, L.; Robertson, D.R. High genetic diversities and complex genetic structure in an Indo-Pacific tropical reef fish (Chlorurus sordidus): Evidence of an unstable evolutionary past? Mar. Biol. 2004, 144, 757–767. [Google Scholar] [CrossRef]
- Bowen, B.W.; Muss, A.; Rocha, L.A.; Grant, W.S. Shallow mtdna coalescence in atlantic pygmy angelfishes (genus Centropyge) indicates a recent invasion from the indian ocean. J. Hered. 2006, 97, 1–12. [Google Scholar]
- Hickey, A.J.R.; Lavery, S.D.; Hannan, D.A.; Baker, C.S.; Clements, K.D. New zealand triplefin fishes (family tripterygiidae): Contrasting population structure and mtDNA diversity within a marine species flock. Mol. Ecol. 2009, 18, 680–696. [Google Scholar]
- Winters, K.L.; van Herwerden, L.; Choat, J.H.; Robertson, D. Phylogeography of the indo-pacific parrotfish Scarus psittacus: Isolation generates distinctive peripheral populations in two oceans. Mar. Biol. 2010, 157, 1679–1691. [Google Scholar] [CrossRef]
- Hamrick, J.; Godt, M. Allozyme diversity in plant species. In Plant Population Genetics, Breeding, and Genetic Resources; Brown, A., Clegg, M., Kahler, A., Weir, B., Eds.; Sinauer: Sunderland, UK, 1989; pp. 43–63. [Google Scholar]
- Hobbs, J.-P.A.; Jones, G.P.; Munday, P.L. Extinction risk in endemic marine fishes. Conserv. Biol. 2011, 25, 1053–1055. [Google Scholar] [CrossRef]
- Hobbs, J.-P.A.; Jones, G.; Munday, P. Rarity and extinction risk in coral reef angelfishes on isolated islands: Interrelationships among abundance, geographic range size and specialisation. Coral Reefs 2010, 29, 1–11. [Google Scholar] [CrossRef]
- Lewis, P.O.; Crawford, D.J. Pleistocene refugium endemics exhibit greater allozymic diversity than widespread congeners in the genus Polygonella (Polygonaceae). Am. J. Bot. 1995, 141–149. [Google Scholar]
- Aleksić, J.M.; Geburek, T. Mitochondrial DNA reveals complex genetic structuring in a stenoendemic conifer Picea omorika [(panč.) purk.] caused by its long persistence within the refugial Balkan region. Plant System. Evol. 2010, 285, 1–11. [Google Scholar] [CrossRef]
- Voris, H.K. Maps of pleistocene sea levels in southeast asia: Shorelines, river systems and time durations. J. Biogeogr. 2000, 27, 1153–1167. [Google Scholar] [CrossRef]
- Froese, R.; Pauly, D. Fishbase. International centre for living aquatic resource management, manila. Available online: http://www.fishbase.org (accessed on 20 January 2013).
- Torres-Díaz, C.; Ruiz, E.; González, F.; Fuentes, G.; Cavieres, L.A. Genetic diversity in Nothofagus alessandrii (Fagaceae), an endangered endemic tree species of the coastal Maulino forest of central Chile. Ann. Bot. 2007, 100, 75–82. [Google Scholar] [CrossRef]
- Fatemi, M.; Gross, C.L. Life on the edge-high levels of genetic diversity in a cliff population of Bertya ingramii are attributed to B. rosmarinifolia (Euphorbiaceae). Biol. Conserv. 2009, 142, 1461–1468. [Google Scholar] [CrossRef]
- Zidana, H.; Turner, G.F.; van Oosterhout, C.; Hänfling, B. Elevated mtDNA diversity in introduced populations of Cynotilapia afra (günther 1894) in Lake Malawi National Park is evidence for multiple source populations and hybridization. Mol. Ecol. 2009, 18, 4380–4389. [Google Scholar]
- Bay, L.K.; Caley, M.J. Greater genetic diversity in spatially restricted coral reef fishes suggests secondary contact among differentiated lineages. Diversity 2011, 3, 483–502. [Google Scholar]
- Hobbs, J.-P.A.; Salmond, J.K. Cohabitation of Indian and Pacific Ocean species at Christmas and Cocos (Keeling) Islands. Coral Reefs 2008, 27, 933–933. [Google Scholar] [CrossRef]
- Hobbs, J.-P.A.; Frisch, A.J.; Allen, G.R.; van Herwerden, L. Marine hybrid hotspot at Indo-Pacific biogeographic border. Biol. Lett. 2009, 5, 258–261. [Google Scholar]
- Schultz, J.K.; Pyle, R.L.; DeMartini, E.; Bowen, B.W. Genetic connectivity among color morphs and pacific archipelagos for the flame angelfish, Centropyge loriculus. Mar. Biol. 2007, 151, 167–175. [Google Scholar] [CrossRef]
- Brothers, E.B.; Thresher, R.E. Pelagic duration, dispersal, and the distribution of Indo-Pacific coral reef fishes. In The ecology of coral reefs; Reaka, M.L., Ed.; NOAA: Washington, DC, USA, 1985; Volume 1, pp. 53–70. [Google Scholar]
- Stobutzki, I.C.; Bellwood, D.R. Sustained swimming abilities of the late pelagic stages of coral reef fishes. Mar. Ecol. Prog. Ser. 1997, 149, 35–41. [Google Scholar] [CrossRef]
- McMillan, W.O.; Palumbi, S.R. Rapid rate of control-region evolution in Pacific butterflyfishes (Chaetodontidae). J. Mol. Evol. 1997, 45, 473–484. [Google Scholar] [CrossRef]
- Bellwood, D.R.; Herwerden, L.; Konow, N. Evolution and biogeography of marine angelfishes (pisces: Pomacanthidae). Mol. Phylo. Evol. 2004, 33, 140–155. [Google Scholar] [CrossRef]
- Johannesson, K.; André, C. Life on the margin: Genetic isolation and diversity loss in a peripheral marine ecosystem, the Baltic sea. Mol. Ecol. 2006, 15, 2013–2029. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hobbs, J.-P.A.; Van Herwerden, L.; Jerry, D.R.; Jones, G.P.; Munday, P.L. High Genetic Diversity in Geographically Remote Populations of Endemic and Widespread Coral Reef Angelfishes (genus: Centropyge). Diversity 2013, 5, 39-50. https://doi.org/10.3390/d5010039
Hobbs J-PA, Van Herwerden L, Jerry DR, Jones GP, Munday PL. High Genetic Diversity in Geographically Remote Populations of Endemic and Widespread Coral Reef Angelfishes (genus: Centropyge). Diversity. 2013; 5(1):39-50. https://doi.org/10.3390/d5010039
Chicago/Turabian StyleHobbs, Jean-Paul A., Lynne Van Herwerden, Dean R. Jerry, Geoffrey P. Jones, and Philip L. Munday. 2013. "High Genetic Diversity in Geographically Remote Populations of Endemic and Widespread Coral Reef Angelfishes (genus: Centropyge)" Diversity 5, no. 1: 39-50. https://doi.org/10.3390/d5010039
APA StyleHobbs, J.-P. A., Van Herwerden, L., Jerry, D. R., Jones, G. P., & Munday, P. L. (2013). High Genetic Diversity in Geographically Remote Populations of Endemic and Widespread Coral Reef Angelfishes (genus: Centropyge). Diversity, 5(1), 39-50. https://doi.org/10.3390/d5010039





