Distinctiveness of Bean Landraces in Italy: the Case Study of the ‘Badda’ Bean
Abstract
:1. Introduction
2. Experimental Section
2.1. Plant Material and Morphological Analysis
| Accession type | Origin | Accession name | Accession code | Trial | HAB | |||
|---|---|---|---|---|---|---|---|---|
| Region | Province | Location | 1st | 2nd | ||||
| Landraces | Sicily | Palermo | Polizzi Generosa | ‘ Badda Bianco’ | BA4 | x | x | 2 |
| Palermo | Polizzi Generosa | ‘ Badda Bianco’ | BA7 | x | x | 2 | ||
| Palermo | Polizzi Generosa | ‘ Badda Nero’ | BA6 | x | x | 2 | ||
| Basilicata | Potenza | Rotonda | ‘ Castelluccisa’ | CA | x | 2 | ||
| Potenza | Rotonda | ‘ Zà Vincenza’ | ZV | x | 2 | |||
| Potenza | Sarconi | ‘ Monachella’ | MO | x | 2 | |||
| Potenza | Sarconi | ‘ O’Marozzo’ | MA | x | 2 | |||
| Campania | Salerno | Casalbuono | ‘ Munaciedd’ | MU | x | 2 | ||
| Salerno | Cilento | ‘ Tuvagliedd’ | TU | x | 2 | |||
| Salerno | Casalbuono | ‘ Panzariedd’ | PA | x | 2 | |||
| Latium | Rieti | Colle di Tora | ‘ A pisello’ | PH | x | 2 | ||
| Viterbo | Gradoli | ‘ Fagiolo del Purgatorio’ | PU | x | 1 | |||
| Marche | Pesaro e Urbino | Urbania | ‘ Borlotto rosso’ | AN8 | x | 2 | ||
| Pesaro e Urbino | Leccia | ‘ Borlotto’ | AN9 | x | 1 | |||
| Macerata | I Colli | ‘ Borlotto uova di quaglia’ | AN23 | x | x | 2 | ||
| Macerata | B.go S. Antonio | ‘ Monachello’ | AN39 | x | x | 2 | ||
| Commercial | Big borlotto | BB | x | 1 | ||||
| Clio | CL | x | x | 1 | ||||
| References | BAT93 | BAT | x | 1 | ||||
| JaloEEP558 | JA | x | 1 | |||||
2.2. Morphometric Analysis of Leaf Shape
2.3. Molecular Analyses
3. Results and Discussion
3.1. Preliminary Characterization of the ‘Badda’ Bean Landrace
| Trait | 1st trial | 2nd trial | ||
|---|---|---|---|---|
| Total | Degenerate primers | Non-degenerate primers | ||
| No. of informative primers | 12 | 28 | 11 | 17 |
| No. of bands | 138 | 194 | 69 | 125 |
| No. of bands per primer | 11.5 | 6.9 | 6.3 | 7.4 |
| No. of polymorphic bands | 93 (67.4) | 50 (25.8) | 20 (29.0) | 30 (24.0) |
| No. of polymorphic bands per primer | 7.8 | 1.8 | 1.8 | 1.8 |
| No. of private bands | 5 (3.6)* | 15 (7.7)** | 4 (5.8) | 11 (8.8) |
| No. of private bands per primer | 0.4 | 0.5 | 0.4 | 0.6 |
3.2. Morpho-Physiological Analysis
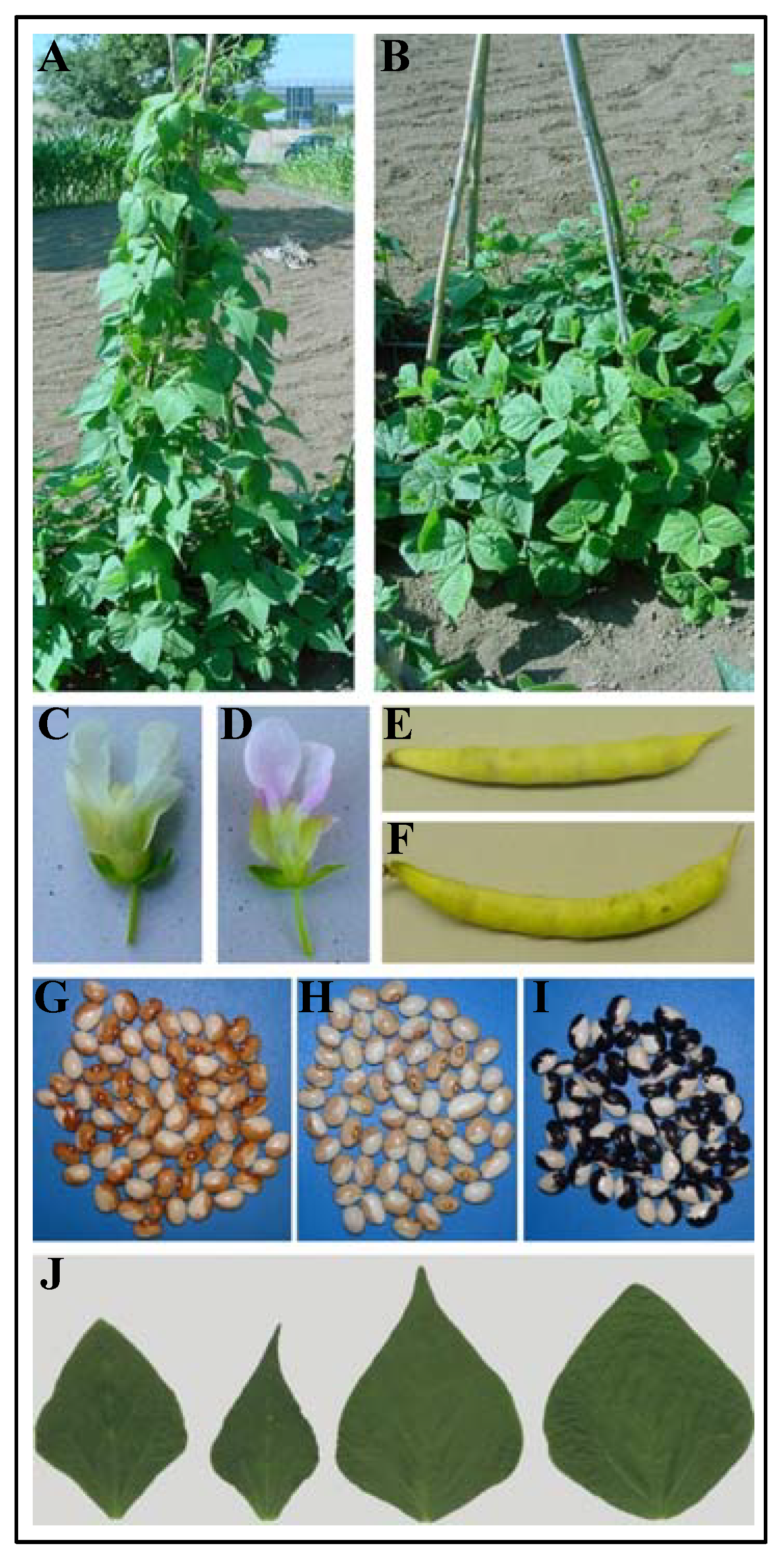
3.3. Morphometric Analysis of Leaf Shape
| 1st-level groups | 1st-level discriminant traits | 2nd-level groups | 2nd-level discriminant traits | Landraces | |||
|---|---|---|---|---|---|---|---|
| DLB | LST | LA | LSI | MH | |||
| ‘Symmetric leaflet’ | 0.48 (0.40–0.63) | 0.35 (0.28–0.50) | ‘Big leaflet’ | 33.6 (30.9–35.9) | 1.32 (1.28–1.38) | 8.6 (8.2–9.3) | AN8, MO, CA, PH |
| ‘Small leaflet’ | 25.1 (23.7–27.2) | 1.34 (1.32–1.40) | 7.6 (7.4–7.7) | BA4, AN39, ZV | |||
| ‘Asymmetric leaflet’ | 0.60 (0.53–0.70) | 0.26 (0.19–0.39) | ‘Big leaflet’ | 31.3 (29.9–33.6) | 1.36 (1.32–1.41) | 8.4 (8.2–8.7) | BA6, AN23, MA, MU |
| ‘Small leaflet’ | 27.5 (22.3–32.6) | 1.46 (1.39–1.54) | 8.4 (7.5–9.4) | BA7, PA, AN9, TU | |||
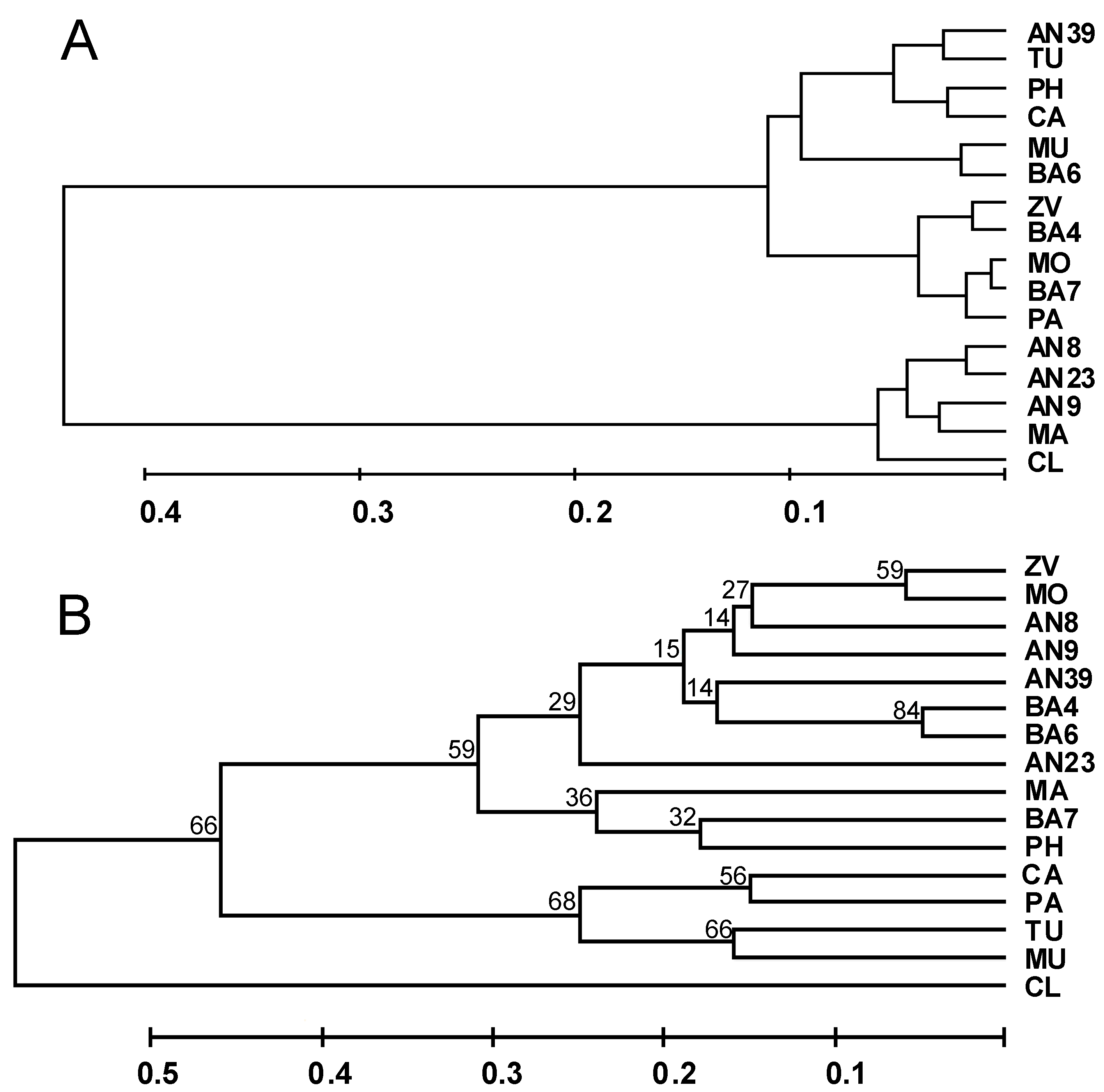
3.4. Molecular Analysis
4. Conclusions
Acknowledgements
References
- Gepts, P.; Bliss, F.A. Phaseolin variability among wild and cultivated common beans (Phaseolus vulgaris) from Colombia. Econ. Bot. 1986, 40, 469–478. [Google Scholar] [CrossRef]
- Gepts, P.; Bliss, F.A. Dissemination pathways of common bean (Phaseolus vulgaris, Fabaceae) deduced from phaseolin electrophoretic variability. II. Europe and Africa. Econ. Bot. 1988, 42, 86–104. [Google Scholar] [CrossRef]
- Papa, R.; Nanni, L.; Sicard, D.; Rau, D.; Attene, G. The evolution of genetic diversity in Phaseolus vulgaris L. In Darwin Harvest: New Approaches To The Origin, Evolution, and Conservation of Crops; Motley, T.J., Zerega, N., Cross, H., Eds.; Columbia University Press: New York, NY, USA, 2006; pp. 121–142. [Google Scholar]
- Zeven, A.C. The introduction of common bean (Phaseolus vulgaris L.) into Western Europe and the phenotypic variation of dry bean collected in The Netherlands in 1945. Euphytica 1997, 94, 319–328. [Google Scholar] [CrossRef]
- Negri, V.; Tosti, N. Phaseolus genetic diversity maintained on-farm in central Italy. Genet. Res. Crop Evol. 2002, 49, 511–520. [Google Scholar] [CrossRef]
- Santalla, M.; Rodino, A.P.; De Ron, A.M. Allozyme evidence supporting southwestern Europa as a secondary center of genetic diversity for common bean. Theor. Appl. Genet. 2002, 104, 934–944. [Google Scholar] [CrossRef]
- Piergiovanni, A.R.; Laghetti, G. The common bean landraces from Basilicata (Southern Italy): an example of integrated approach applied to genetic resources management. Genet. Res. Crop Evol. 1999, 46, 47–52. [Google Scholar] [CrossRef]
- Piergiovanni, A.R.; Taranto, G.; Losavio, F.; Pignone, D. Common bean (Phaseolus vulgaris L.) landraces from Abruzzo and Lazio regions (Central Italy). Genet. Res. Crop Evol. 2006, 53, 313–322. [Google Scholar] [CrossRef]
- Lioi, L.; Piergiovanni, A.R.; Pignone, D.; Puglisi, S.; Santantonio, M.; Sonnante, G. Genetic diversity of some surviving on-farm Italian common bean (Phaseolus vulgaris L.) landraces. Plant Breed. 2005, 124, 576–581. [Google Scholar] [CrossRef]
- Sicard, D.; Nanni, L.; Porfidi, O.; Bulfon, D.; Papa, R. Genetic diversity of Phaseolus vulgaris L. and P. coccineus L. landraces in Central Italy. Plant Breed. 2005, 124, 464–472. [Google Scholar] [CrossRef]
- Santangelo, E.; Mazzucato, A.; Picarella, M.E.; Mosconi, P.; Lioi, L.; Soressi, G.P. Caratterizzazione del ‘Fagiolo del Purgatorio’ di Gradoli (VT). Italus Hortus 2006, 13, 496–502. [Google Scholar]
- Foschiani, A.; Miceli, F.; Vischi, M. Assessing diversity in common bean (Phaseolus vulgaris L.) accessions at phenotype and molecular level: a preliminary approach. Genet. Res. Crop Evol. 2009, 56, 445–453. [Google Scholar] [CrossRef]
- Santalla, M.; De Ron, A.M.; Voysest, O. European bean market classes. In Catalogue of Bean Genetic Resources; Amurrio, M., Santalla, M., De Ron, A.M., Eds.; PHASELIEU-FAIRPL97-3463, Misión Biológica de Galicia (CSIC): Pontevedra, Spain, 2001; pp. 79–94. [Google Scholar]
- Lioi, L. Geographical variation of phaseolin patterns in an old world collection of Phaseolus vulgaris. Seed Sci. Technol. 1989, 17, 317–324. [Google Scholar]
- Sánchez, E.; Sifres, A.; Casañas, F.; Nuez, F. The endangered future of organoleptically prestigious European landraces: Ganxet bean (Phaseolus vulgaris L.) as an example of a crop originating in the Americas. Genet. Res. Crop Evol. 2008, 55, 45–52. [Google Scholar] [CrossRef]
- Marotti, I.; Bonetti, A.; Minelli, M.; Catizone, P.; Dinelli, G. Characterization of some Italian common bean (Phaseolus vulgaris L.) landraces by RAPD, semi-random and ISSR molecular markers. Genet. Res. Crop Evol. 2007, 54, 175–188. [Google Scholar] [CrossRef]
- Maciel, F.L.; Echeverrigaray, S.; Gerald, L.T.S.; Grazziotin, F.G. Genetic relationships and diversity among Brazilian cultivars and landraces of common beans (Phaseolus vulgaris L.) revealed by AFLP markers. Genet. Res. Crop Evol. 2003, 50, 887–893. [Google Scholar] [CrossRef]
- Masi, P.; Logozzo, G.; Donini, P.; Spagnoletti-Zeuli, P. Analysis of genetic structure in widely distributed common bean landraces with different plant growth habits using SSR and AFLP markers. Crop Sci. 2009, 49, 187–199. [Google Scholar] [CrossRef]
- Galván, M.Z.; Bornet, B.; Balatti, B.A.; Branchard, M. Inter simple sequence repeat (ISSR) markers as a tool for the assessment of both genetic diversity and gene pool origin in common bean (Phaseolus vulgaris L.). Euphytica 2003, 132, 297–301. [Google Scholar] [CrossRef]
- Gonzalez, A.; Wong, A.; Delgado Salinas, A.; Papa, R.; Gepts, P. Assessment of inter simple sequence repeat markers to differentiate sympatric wild and domesticated populations of common bean (Phaseolus vulgaris L.). Crop Sci. 2005, 45, 606–615. [Google Scholar] [CrossRef]
- Zizumbo-Villarreal, D.; Colunga-GarcíaMarín, P.; de la Cruz, E.P.; Delgado-Valerio, P.; Gepts, P. Population structure and evolutionary dynamics of wild-weedy-domesticated complexes of common bean in a Mesoamerican region. Crop Sci. 2005, 45, 1073–1083. [Google Scholar] [CrossRef]
- Sonnante, G.; Pignone, D. Assessment of genetic variation in a collection of lentil using molecular tools. Euphytica 2001, 120, 301–307. [Google Scholar] [CrossRef]
- Schachl, R.; De la Rosa, L. Characterisation of Phaseolus accessions. In Handbook on Evaluation of Phaseolus Germplasm; De la Cuadra, C., De Ron, A.M., Schachl, R., Eds.; PHASELIEU-FAIRPL97-3463, Misión Biológica de Galicia (CSIC): Pontevedra, Spain, 2001; pp. 29–43. [Google Scholar]
- SAS Institute Inc. SAS Guide for Personal Computers, 9th ed.; SAS Institute: Cary, NC, USA, 2002. [Google Scholar]
- Gower, J.C. A general coefficient of similarity and some of its properties. Biometrics 1971, 27, 623–637. [Google Scholar] [CrossRef]
- Brewer, M.T.; Lang, L.; Fujimura, K.; Dujmovic, N.; Gray, S.; van der Knaap, E. Development of a controlled vocabulary and software application to analyze fruit shape variation in tomato and other plant species. Plant Physiol. 2006, 141, 15–25. [Google Scholar] [CrossRef]
- Gonzalo, M.J.; Brewer, M.T.; Anderson, C.; Sullivan, D.; Gray, S.; van der Knaap, E. Tomato fruit shape analysis using morphometric and morphology attributes implemented in Tomato Analyzer software program. J. Amer. Soc. Hort. Sci. 2009, 134, 77–87. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Nei, M.; Tajima, F.; Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J. Mol. Evol. 1983, 91, 153–170. [Google Scholar]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy; W.H. Freeman: San Francisco, CA, USA, 1973. [Google Scholar]
- Van de Peer, Y.; De Wachter, Y. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Applic. Biosci. 1994, 10, 569–570. [Google Scholar]
- Lioi, L. Geographical variation of phaseolin patterns in an Old World collection of Phaseolus vulgaris. Seed Sci. Technol. 1989, 17, 317–324. [Google Scholar]
- Limongelli, G.; Laghetti, G.; Perrino, P.; Piergiovanni, A.R. Variation of seed storage proteins in landraces of common bean (Phaseolus vulgaris L.) from Basilicata, Southern Italy. Euphytica 1995, 92, 393–399. [Google Scholar] [CrossRef]
- Singh, S.P.; Gepts, P.; Debouck, D.G. Races of common bean (Phaseolus vulgaris, Fabaceae). Econ. Bot. 1991, 45, 379–396. [Google Scholar] [CrossRef]
- Piergiovanni, A.R.; Lioi, L. La conservazione on-farm: importanza della caratterizzazione e valutazione degli agro-ecotipi di fagiolo. Agroindustria 2007, 6, 29–36. [Google Scholar]
- Venora, G.; Grillo, O.; Ravalli, C.; Cremonini, R. Identification of Italian landraces of bean (Phaseolus vulgaris L.) using an image analysis system. Sci. Horticult. 2009, 121, 410–418. [Google Scholar] [CrossRef]
Appendices
| Primer type | Primer code | Primer sequence | Ta (°C) |
|---|---|---|---|
| Degenerate* | ISSR 1 | (GACA)3 RG | 42 |
| ISSR 2 | (GACAC)2 | 42 | |
| ISSR 3 | (GA)8 RG | 42 | |
| ISSR 4 | YR (GACA)3 | 42 | |
| ISSR 5 | (ACTG)3 RG | 42 | |
| ISSR 6 | (GACA)3 RT | 42 | |
| ISSR 7 | (AG)8 YG | 56 | |
| ISSR 8 | (CA)8 RY | 56 | |
| ISSR 9 | (AC)8 YA | 56 | |
| ISSR10 | (GA)8 YT | 56 | |
| ISSR11 | (GT)8 YC | 56 | |
| ISSR12 | BDB (CA)7 | 56 | |
| Non-degenerate | ISSR 15 | GTGC(AC)7 | 56 |
| ISSR 16 | GGGC(AC)7 | 56 | |
| ISSR 17 | CAGC(CT)7 | 56 | |
| ISSR 19 | CCTGC(AC)7 | 56 | |
| ISSR 32 | (AGC)4C | 56 | |
| ISSR 35 | (AGC)4GA | 56 | |
| ISSR 38 | (AGC)4AT | 56 | |
| ISSR 39 | (AGC)4AC | 56 | |
| ISSR 50 | CCA(GCT)4 | 56 | |
| ISSR 51 | GCACC(CA)9 | 56 | |
| ISSR 52 | GGCAC(CA)9 | 56 | |
| ISSR 53 | CGCAA(CA )9 | 56 | |
| ISSR 54 | GGCTA(CA)9 | 56 | |
| ISSR 55 | CCATC(CA)9 | 56 | |
| ISSR 56 | GCTAC(CA)9 | 56 | |
| ISSR 58 | CGAAC(CA)9 | 56 | |
| ISSR 59 | GGCCA(GCT)6 | 56 | |
| ISSR 60 | GCCAC(GCT)6 | 56 |
| Accession code | FLO* | SET* | HAR* | PLE* | PWI* | FCO | PBP | PBS | PCU | PCO | SCO | STY | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (das) | (das) | (das) | (cm) | (cm) | |||||||||||||
| BA 4 | 83 | ab | 93 | a | 150 | a | 11.4 | b | 1.2 | d | 1 | 1 | 1 | 3 | 3 | 4 | 2 |
| BA 7 | 89 | a | 92 | a | 150 | a | 12.3 | b | 1.5 | bcd | 1 | 1 | 1 | 3 | 5 | 3 | 2 |
| BA 6 | 81 | ab | 85 | ab | 150 | a | 13.9 | b | 1.7 | abc | 1 | 1 | 2 | 1 | 4 | 2 | 2 |
| CA | 87 | ab | 93 | a | 150 | a | 10.9 | b | 1.6 | bcd | 1 | 1 | 3 | 2 | 3 | 3 | 2 |
| ZV | 87 | ab | 93 | a | 150 | a | 13.1 | b | 1.7 | abc | 2 | 1 | 1 | 3 | 4 | 4 | 2 |
| MO | 85 | ab | 92 | a | 140 | ab | 13.3 | b | 1.7 | abc | 1 | 1 | 1 | 3 | 3 | 3 | 2 |
| MA | 66 | cd | 71 | bc | 129 | b | 11.8 | b | 1.4 | cd | 2 | 1 | 1 | 3 | 1 | 6 | 3 |
| MU | 59 | d | 64 | cd | 129 | b | 14 | b | 1.7 | abc | 1 | 2 | 2 | 1 | 4 | 3 | 2 |
| TU | 62 | d | 68 | cd | 150 | a | 11.5 | b | 1.5 | d | 1 | 1 | 2 | 2 | 2 | 5 | 2 |
| PA | 76 | bc | 79 | ab | 140 | ab | 13.3 | b | 1.5 | bcd | 1 | 1 | 1 | 1 | 1 | 3 | 2 |
| PH | 64 | cd | 80 | ab | 150 | a | 11.5 | b | 1.5 | bcd | 1 | 1 | 3 | 2 | 4 | 1 | 1 |
| AN8 | 55 | d | 64 | cd | 93 | c | 17.3 | a | 1.5 | bcd | 2 | 1 | 3 | 2 | 1 | 6 | 3 |
| AN9 | 43 | e | 54 | d | 93 | c | 10.7 | b | 1.7 | abc | 2 | 1 | 1 | 3 | 3 | 6 | 3 |
| AN23 | 55 | d | 64 | cd | 93 | c | 10.9 | b | 2 | a | 2 | 1 | 2 | 2 | 4 | 6 | 3 |
| AN39 | 59 | d | 65 | cd | 93 | c | 11 | b | 1.4 | cd | 1 | 1 | 1 | 2 | 1 | 2 | 2 |
| CL | 43 | e | 54 | c | 93 | c | 13.8 | b | 1.4 | cd | 3 | 2 | 1 | 2 | 1 | 6 | 3 |
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Paniconi, G.; Gianfilippi, F.; Mosconi, P.; Mazzucato, A. Distinctiveness of Bean Landraces in Italy: the Case Study of the ‘Badda’ Bean. Diversity 2010, 2, 701-716. https://doi.org/10.3390/d2050701
Paniconi G, Gianfilippi F, Mosconi P, Mazzucato A. Distinctiveness of Bean Landraces in Italy: the Case Study of the ‘Badda’ Bean. Diversity. 2010; 2(5):701-716. https://doi.org/10.3390/d2050701
Chicago/Turabian StylePaniconi, Giulia, Federica Gianfilippi, Pietro Mosconi, and Andrea Mazzucato. 2010. "Distinctiveness of Bean Landraces in Italy: the Case Study of the ‘Badda’ Bean" Diversity 2, no. 5: 701-716. https://doi.org/10.3390/d2050701
APA StylePaniconi, G., Gianfilippi, F., Mosconi, P., & Mazzucato, A. (2010). Distinctiveness of Bean Landraces in Italy: the Case Study of the ‘Badda’ Bean. Diversity, 2(5), 701-716. https://doi.org/10.3390/d2050701