Molecular Polymorphisms in Tunisian Pomegranate (Punica granatum L.) as Revealed by RAPD Fingerprints
Abstract
:1. Introduction
2. Results and Discussion
2.1. Primers and Resolving power (Rp)
| Primer | Sequence (5’−3’) | Amplified bands | ||
|---|---|---|---|---|
| Total | Polymorphic | Rp | ||
| OPA 04 | AATCGGGCTG | 3 | 3 | 1.333 |
| OPA 19 | CAAACGTCGG | 10 | 8 | 4.832 |
| OPH 07 | CAAACGTCGG | 11 | 9 | 4.666 |
| OPH 08 | GAAACACCCC | 5 | 4 | 2.000 |
| Total | 29 | 24 | 12.831 | |
2.2. Relationships between Cultivars
| GR2 | 0 | |||||||||||
| GR1 | 0.292 | 0 | ||||||||||
| BL1 | 0.042 | 0.333 | 0 | |||||||||
| ZG1 | 0.292 | 0.500 | 0.333 | 0 | ||||||||
| MZ3 | 0.417 | 0.292 | 0.458 | 0.375 | 0 | |||||||
| TN6 | 0.292 | 0.167 | 0.250 | 0.417 | 0.208 | 0 | ||||||
| TN1 | 0.208 | 0.333 | 0.167 | 0.333 | 0.458 | 0.250 | 0 | |||||
| JB3 | 0.208 | 0.250 | 0.250 | 0.250 | 0.292 | 0.167 | 0.167 | 0 | ||||
| MZ1 | 0.208 | 0.500 | 0.167 | 0.417 | 0.625 | 0.417 | 0.250 | 0.333 | 0 | |||
| GB9 | 0.667 | 0.458 | 0.708 | 0.375 | 0.250 | 0.458 | 0.542 | 0.458 | 0.792 | 0 | ||
| GB8 | 0.625 | 0.583 | 0.667 | 0.333 | 0.292 | 0.500 | 0.583 | 0.417 | 0.667 | 0.292 | 0 | |
| CH3 | 0.458 | 0.250 | 0.417 | 0.667 | 0.375 | 0.250 | 0.333 | 0.417 | 0.583 | 0.375 | 0.667 | 0 |
| Cultivars | GR2 | GR1 | BL1 | ZG1 | MZ3 | TN6 | TN1 | JB3 | MZ1 | GB9 | GB8 | CH3 |
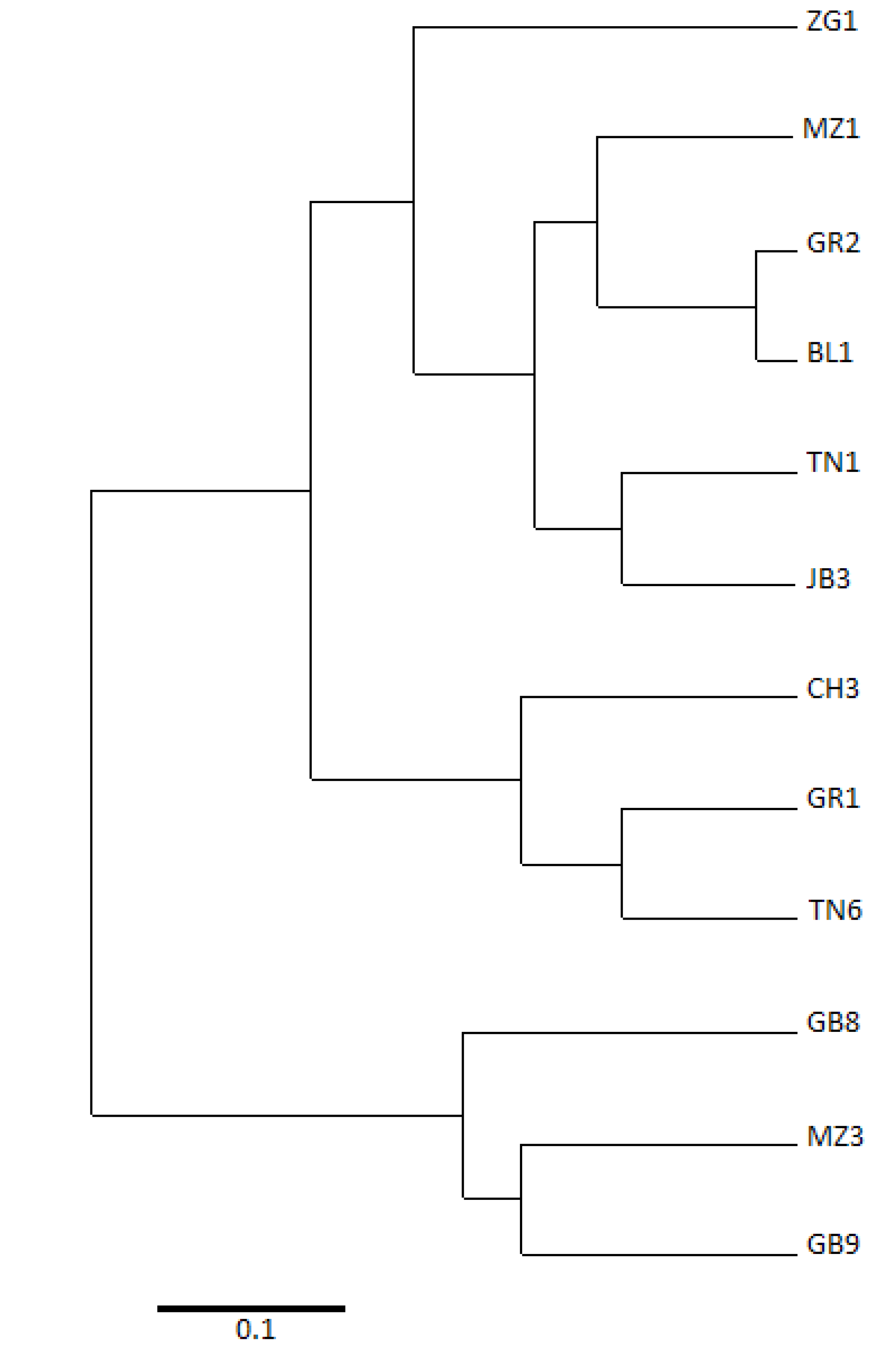
3. Experimental Section
3.1. Plant Material
| Cultivar | Label | Origin | Main characteristics | |||
| Taste | Rind color | Arils color | ||||
| Gabsi 8 | GB8 | Kettana | Sweet | Red | Redish pink | |
| Gabsi 9 | GB9 | Zerkine II | Sweet | Red | Redish pink | |
| Tounsi 1 | TN1 | Testour | Sweet | Red | Dark red | |
| Tounsi 6 | TN6 | Tozeur | Sweet | Pale red | Red | |
| Chelfi 3 | CH3 | Testour | Sweet | Red | Redish pink | |
| Mezzi 1 | MZ1 | Tozeur | Sour | Pale red | Dark red | |
| Mezzi 3 | MZ3 | Tozeur | Sour | Pale red | Redish pink | |
| Jebali 3 | JB3 | El Alia | Sweet | Dark red | Redish pink | |
| Garoussi 1 | GR1 | Mareth | Sweet | Red | White pink | |
| Garoussi 2 | GR2 | Chott Mariam | Sour | Pale red | Red | |
| Zaghouani 1 | ZG1 | Zaghouan | Sweet | Dark red | Redish pink | |
| Bellahi 1 | BL1 | Tozeur | Sweet | Red green | White pink | |
3.2. DNA Extraction
3.3. Primers and PCR Assays
3.4. Data Analysis
4. Conclusions
References
- Evreinoff, V.A. Le grenadier. Fruits d'Outre-Mer 1949, 4, 161–170. [Google Scholar]
- Tous, J.; Ferguson, L. Mediterranean fruits. In Progress in New Crops; Janick, J., Ed.; ASHS Press: Arlington, VA, USA, 1996; pp. 416–430. [Google Scholar] [Green Version]
- Mars, M. Ressources génétiques du grenadier (Punica granatum L.) en Tunisie : prospection, conservation et analyse de la diversité.; Thèse de Doctorat d’Etat, Université El Manar, Faculté des Sciences de Tunis: Tunis, Tunisia, 2001. [Google Scholar] [Green Version]
- Mars, M.; Marrakchi, M. Conservation et valorisation des ressources génétiques du grenadier (Punica granatum L.) en Tunisie. Plant Genet. Resour. Newslett. 1998, 114, 35–39. [Google Scholar]
- Mars, M.; Marrakchi, M. Diversity of pomegranate (Punica granatum L.) germplasm in Tunisia. Genet. Resour. Crop Ev. 1999, 46, 461–467. [Google Scholar] [CrossRef]
- Baht, K.V.; Jarret, R.L.; Rana, P.S. DNA profiling of banana and plantain cultivars using random amplified polymorphic DNA (RAPD) and restriction fragment length polymorphism (RFLP) markers. Electrophoresis 1995, 16, 1736–1745. [Google Scholar] [CrossRef]
- Onguso, J.M.; Kahangi, E.M.; Ndiritu, D.W.; Mizutani, F. Genetic characterisation of cultivated bananas and plantains in Kenya by RAPD markers. Sci. Hort. 1999, 99, 9–20. [Google Scholar]
- Sarkhosh, A.; Zamani, Z.; Fatahi, R.; Ebadi, A. RAPD markers reveal polymorphism among some Iranian pomegranate (Punica granatum L.) genotypes. Sci. Hort. 2006, 111, 24–29. [Google Scholar] [CrossRef]
- Durgaç, C.; Ozgen, M.; Simsek, O.; Kaçar, Y.K.; Kyga, Y.; Celebi, S.; Gunduz, K.; Serce, S. Molecular and pomological diversity among pomegranate (Punica granatum L.) cultivars in Eastern Mediterranean region of Turkey. Afr. J. Biotechnol. 2008, 7, 1294–1301. [Google Scholar]
- Sheidai, M.; Saneghi, A.; Shahreiyari, Z.H.; Noormohammadi, Z.; Farahanei, F.; Tabatabaei-Ardakanei, S. RAPD and cytogenetic study of some pomegranate (Punica granatum L.) cultivars. Caryologia 2008, 6, 68–73. [Google Scholar]
- Yuan, Z.; Yin, Y.; Qu, J.; Zhu, L.; Li, Y. Population genetic diversity in Chinese pomegranate (Punica granatum L.) cultivars revealed by fluorescent-AFLP markers. J. Genet. Genomics 2007, 34, 1061–1071. [Google Scholar]
- Jbir, R.; Hasnaoui, N.; Mars, M.; Marrakchi, M.; Trifi, M. Characterization of Tunisian pomegranate (Punica granatum L.) cultivars using amplified fragment length polymorphism analysis. Sci. Hort. 2008, 115, 231–237. [Google Scholar] [CrossRef]
- Melgarejo, P.; Martinez, J.J.; Hernandez, F.; Martinez, R.; Legua, P.; Oncina, R.; Martinez-Murcia, A. Cultivar identification using 18S-28S rDNA intergenic spacer-RFLP in pomegranate (Punica granatum L.). Sci. Hort. 2009, 120, 500–503. [Google Scholar] [CrossRef]
- Landry, B.; Li, R.; Cheung, W.; Granger, R. Phylogeny analysis of 25 apple root stocks using RAPD markers and tactical gene targging. Theor. Appl. Genet. 1994, 89, 847–852. [Google Scholar]
- Qu, X.P.; Lu, J.; Lamikanra, O. Genetic diversity in Muscadine and American bunch grapes based on randomly amplified polymorphic DNA (RAPD) analysis. J. Am. Soc. Hort. Sci. 1996, 121, 1020–1023. [Google Scholar]
- Ronning, C.M.; Schnell, R.J.; Gazit, S. Use of random amplified polymorphic DNA (RAPDs) to identify annona cultivars. J. Am. Soc. Hort. Sci. 1995, 120, 726–729. [Google Scholar]
- Ben Abdallah, A.; Stiti, K.; Lepoivre, P.; Du Jardin, P. Identification de cultivars de palmier dattier (Phoenix dactylifera L.) par l’amplification aléatoire d’ADN (RAPD). Cah. Étud. Recher. Francoph./Agric. 2000, 9, 103–107. [Google Scholar]
- Chatti, K.; Salhi-Hannachi, A.; Mars, M.; Marrakchi, M.; Trifi, M. Genetic diversity and phylogenic relationships in Tunisian fig (Ficus carica L.) cultivars mediated by RAPD. Biol. Tunis. 2004, 1, 1–4. [Google Scholar]
- Ebrahimi, R.; Zamani, Z.; Kashi, A. Genetic diversity evaluation of wild Persian shallot (Allium hirtifolium Boiss.) using morphological and RAPD markers. Sci. Hort. 2009, 119, 345–351. [Google Scholar] [CrossRef]
- Salhi-Hannachi, A.; Chatti, K.; Mars, M.; Marrakchi, M.; Trifi, M. Comparative analysis of genetic diversity in two collections of fig cultivars based on random amplified polymorphic DNA and inter simple sequence repeats fingerprints. Genet. Resour. Crop Ev. 2005, 52, 563–573. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar] [Green Version]
- Williams, J.G.K.; Kubelik, A.R.; Livak, J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef]
- Gilbert, J.E.; Lewis, E.V.; Wilkinson, M.J.; Caligari, P.D.S. Developing an appropriate strategy to assess genetic variability in plant germplasm collections. Theor. Appl. Genet. 1999, 98, 1125–1131. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.H. Mathematical models for studying genetic variation in terms of restriction endonucleases. P. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef]
- Felsenstein, J. PHYLIP (Phylogeny Interference Package) version 3,5 c; Department of Genetics, University of Washington: Seattle, WA, USA, 1995. [Google Scholar] [Green Version]
- Page, R.D.M. Tree View: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996, 12, 357–358. [Google Scholar]
© 2010 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hasnaoui, N.; Mars, M.; Chibani, J.; Trifi, M. Molecular Polymorphisms in Tunisian Pomegranate (Punica granatum L.) as Revealed by RAPD Fingerprints. Diversity 2010, 2, 107-114. https://doi.org/10.3390/d2010107
Hasnaoui N, Mars M, Chibani J, Trifi M. Molecular Polymorphisms in Tunisian Pomegranate (Punica granatum L.) as Revealed by RAPD Fingerprints. Diversity. 2010; 2(1):107-114. https://doi.org/10.3390/d2010107
Chicago/Turabian StyleHasnaoui, Néjib, Messaoud Mars, Jemni Chibani, and Mokhtar Trifi. 2010. "Molecular Polymorphisms in Tunisian Pomegranate (Punica granatum L.) as Revealed by RAPD Fingerprints" Diversity 2, no. 1: 107-114. https://doi.org/10.3390/d2010107
APA StyleHasnaoui, N., Mars, M., Chibani, J., & Trifi, M. (2010). Molecular Polymorphisms in Tunisian Pomegranate (Punica granatum L.) as Revealed by RAPD Fingerprints. Diversity, 2(1), 107-114. https://doi.org/10.3390/d2010107




