Abstract
The monitoring of algal communities has traditionally relied on optical microscopy. However, this technique is time-consuming and requires significant expertise to accurately identify species. In recent years, molecular techniques such as environmental DNA (eDNA) metabarcoding have facilitated the identification of algal communities. This study aims to compare both approaches for assessing planktonic microalgal communities in three areas of Lake Titicaca, using inverted light microscopy and eDNA metabarcoding. We found that the taxonomic composition obtained using the two methods differs significantly for Bacillariophyta, Chlorophyta, Charophyta, and Cyanobacteria, although genus- and order-level richness was similar across both approaches. A pronounced shift in species composition between techniques was revealed, with few shared genera and a high proportion of unassigned sequences (>50%) for Bacillariophyta. While microscopy provided more accurate estimates of microalgal density, metabarcoding revealed greater diversity, particularly among nanoplanktonic microalgae from the phyla Cryptophyta, Ochrophyta, Haptophyta, and Rhodophyta. To improve the accuracy and complementarity of these methodologies, it is essential to expand regional reference databases and work toward standardizing both approaches, allowing them to be used synergistically rather than independently.
1. Introduction
Lake Titicaca is one of the largest lakes in South America and has been recognized by the Ramsar Convention as a Wetland of International Importance [1]. However, anthropogenic activities are increasingly altering their biodiversity, which may in turn, affect the provision of essential ecosystem services such as water supply [2]. Despite growing concern, the effects of biodiversity loss on microorganisms such as algae remain poorly understood [3].
Water quality monitoring is essential for detecting and assessing deterioration in aquatic ecosystems [4]. In Peru, the Ministry of the Environment (MINAM) [5] regulates water quality using environmental quality standards (ECA), which set reference values for physical, chemical, and biological parameters to protect both human health and aquatic environments. Studies have found water quality issues in Lake Titicaca; Beltrán et al. noted low transparency, high conductivity, increased alkalinity, and significant fecal coliforms near Espinar Island due to untreated wastewater from Puno [6]. Similarly, Iquise identified poor water quality and reduced zooplankton diversity in Lake Titicaca’s Inner and Outer Bays [7].
When algae are suspended in a water column, they are referred to as phytoplankton. This group includes a diverse range of eukaryotic organisms—such as Bacillariophyta, Chlorophyta, and Charophyta—as well as prokaryotic Cyanobacteria. These microorganisms form the foundation of trophic networks in lakes, serving as the primary producers of aquatic ecosystems [8]. In addition, they provide a crucial food source for zooplankton [9]. Freshwater cyanobacteria and eukaryotic algae (hereafter referred to collectively as microalgae) respond strongly to environmental gradients [10] and have long been used as indicators of anthropogenic impacts on water quality [11]. Among them, cyanobacteria play a particularly critical role, as their transient blooms can occur in lakes across all trophic states [12]. They are widely recognized as key indicators of eutrophication in freshwater ecosystems [13]. However, certain cyanobacterial species can become harmful due to excessive growth and toxin production, resulting in harmful cyanobacterial blooms (CyanoHABs). These events pose significant threats to freshwater resources and public health, often requiring the implementation of monitoring, control, and management protocols [14].
The monitoring of algal communities has traditionally relied on optical microscopy, which enables taxonomic identification based on the morphology of these microorganisms. This method has been widely used for identifying and quantifying microalgae [15,16]. However, it requires considerable time and extensive expertise to accurately determine algal species based on morphological characteristics [17]. Furthermore, morphological identification presents limitations, as it does not allow for the detection of cryptic species [18]. Consequently, technical constraints persist, hindering the comprehensive analysis of microalgal assemblages [19]. Given the analytical challenges associated with traditional microscopy-based methods, molecular analyses provide a robust approach for identifying various aquatic species. Metabarcoding involves species composition analysis through the massive sequencing of a barcoding marker in an environmental sample [20]. This differs from metagenomics, defined as the direct sequencing of collective genomes in an environmental sample without amplifying specific genes [21]. Metabarcoding effectively overcomes the limitations of light microscopy by enabling rapid, high-throughput sample processing and facilitating the detection of rare, cryptic, and small-sized species at a relatively manageable cost [19]. However, despite its advantages, the reliability of metabarcoding remains insufficiently understood, and clear methodological guidelines for its application are still lacking [3].
To fully assess the potential of metabarcoding in estimating microbial diversity, it is essential to compare its results with morphology-based analyses. Several studies have conducted such comparisons, including Kim et al., who examined diatom communities in water samples from five estuaries in the Korean Peninsula [22]; Kulas et al., who evaluated the diversity and community structure of diatoms using morphological and molecular approaches along the Krka River (Croatia) [23]; Kutty et al., who evaluated a workflow that combines environmental DNA metabarcoding and morphological methods for diatom identification in six freshwater reservoirs in Singapore [24]; Mackeigan et al., who studied cyanobacteria from over 300 lakes in Canada [25]; Santi et al. who investigated microbial eukaryotes in Mediterranean coastal seawater mesocosms [26]; and Groendahl et al., who analyzed microalgae cultured in freshwater mesocosms at the University of Cologne, Germany [3]. Despite the significance of these comparisons for bioassessment programs and lake management, research in inland environments remains limited, particularly regarding Andean ecosystems and studies that directly compare barcoding with traditional microscopy-based identification of algae from identical samples. This study aims to contribute to this field by evaluating the strengths and limitations of both methodologies through a comparative analysis of planktonic microalgal communities in three areas of Lake Titicaca under contrasting environmental conditions (wet and dry seasons). The approaches employed inverted light microscopy and environmental DNA (eDNA) metabarcoding. Based on previous research, we hypothesize that molecular tools will offer a more comprehensive assessment of microalgal diversity by overcoming the inherent limitations of traditional morphometric techniques.
2. Materials and Methods
2.1. Study Area
Lake Titicaca, located in South America, spans southern Peru and northwestern Bolivia, between 14°05′ and 16°50′ north and 68°10′ and 71°05′ west. In Perú, it lies entirely within the department of Puno and has a surface water availability of 9877 hm3. Water samples were collected from Lake Titicaca in 2021 during the wet season (April) and dry season (September). Fourteen sampling sites were selected across three distinct areas of Lake Titicaca, namely Puno Bay (BA), Major Lake (MA), and Minor Lake (MI) (Figure 1). These same sampling sites were used for molecular analyses.
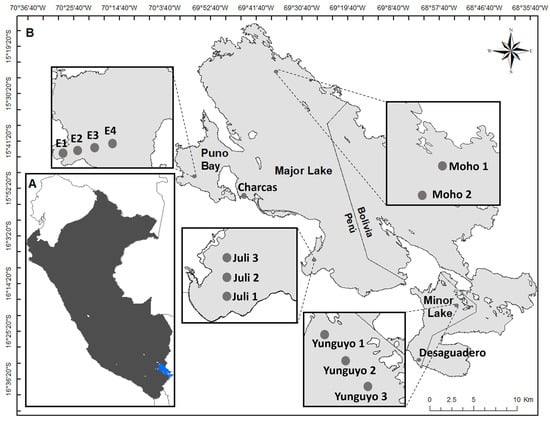
Figure 1.
(A) Map of Lake Titicaca (blue) in Peru (black). (B) Sampling sites in Puno Bay (BA), Major Lake (MA), and Minor Lake (MI) of Lake Titicaca.
2.2. Sampling
For phytoplankton community sampling, 250 mL of surface water (0.5 m depth) was collected using a Niskin bottle and fixed with 4% formaldehyde [27]. Additionally, 40 L of surface water was filtered through a plankton net with a 20-micron mesh opening. The resulting filtrate was transferred into 250 mL airtight bottles and directly fixed with 4% formaldehyde. In addition, in situ surface-level measurements of dissolved oxygen (DO), temperature (TE), hydrogen ion potential (pH), and electrical conductivity (EC) were carried out using a portable multiparameter device (HandyLab 680, Mainz city, Germany) (see Supplementary Table S1). A total of 28 net samples and 28 water samples were obtained for qualitative and quantitative phytoplankton analysis. Biological samples were stored at 4 °C and subsequently transported to the Molecular Ecology and Biodiversity Laboratory of the Faculty of Biological Sciences at UNMSM. In parallel, for molecular analysis, 1 L of surface water (0.5 m depth) was collected using a Niskin bottle and sterile vials, then filtered with a portable pump through a 0.22 µm Sterivex filter. The 28 filters designated for molecular analysis were packed in a refrigerated cooler for transport to Lima.
2.3. DNA Metabarcoding
Total DNA was isolated from the Sterivex filter using the NucleoMag DNA/RNA (MachereyNagel, Düren, Germany) Water kit, following the manufacturer’s protocol. Briefly, the process began with cell lysis using Buffer C1, followed by vortexing. The lysate was then transferred into a new tube for DNA binding to NucleoMag B-Beads using MWA2 Buffer. The tube was placed in a magnetic separator for five minutes, ensuring complete bead binding to the magnets. This was followed by successive washes with MWA3 and MWA4 Buffers, concluding with DNA elution. Amplification, library construction, and sequencing of the samples were conducted by a commercial service at Jonah Venture (Boulder city, NV, USA).
The sequencing began with the amplification of a fragment of ~410 bp of the 23S rRNA gene from each sample using PCR with the primers p23SrV_f1 and p23Sr1V_r1 [28]. This gene has been used as a universal plastid marker to identify multiple algal lineages, including both prokaryotic and eukaryotic algae [28]. Both forward and reverse primers contained a 5′ adapter sequence to facilitate subsequent indexing and Illumina sequencing. Each 40 µL PCR reaction was prepared according to the specifications of the Promega PCR Master Mix (Promega, Madison, WI, USA), which included 12.5 µL of Master Mix, 0.5 µM of each primer, 1.0 µL of genomic DNA (gDNA), and 10.5 µL of DNase/RNase-free water.
PCR amplification was carried out under the following thermal cycling conditions: an initial denaturation at 94 °C for 3 min, followed by 40 cycles of 94 °C for 30 s, 55 °C for 45 s, and 72 °C for 1 min, with a final extension at 72 °C for 10 min. To verify amplicon size and PCR efficiency, 5 µL of each reaction product was visualized on 2% agarose gel. Subsequently, PCR products were cleaned by incubating the amplicons with Exonuclease I and Shrimp Alkaline Phosphatase (Exo1/SAP) at 37 °C for 30 min, followed by enzyme inactivation at 95 °C for 5 min. For barcode indexing, a second round of PCR was performed to assign a unique 12-nucleotide index sequence to each sample. This indexing PCR included the Promega PCR Master Mix, 0.5 µM of each index primer, and 2 µL of the cleaned PCR amplicon as the DNA template. The thermal profile consisted of an initial denaturation at 95 °C for 3 min, followed by 8 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s.
For library normalization and pooling, 25 µL of each indexed PCR amplicon was cleaned and normalized using the SequalPrep Normalization Plate Kit (Life Technologies, Carlsbad, CA, USA), following the manufacturer’s protocol. Subsequently, 5 µL of each normalized sample was pooled into a single library. Final pooled libraries were submitted for sequencing on an Illumina NovaSeq 6000 platform (San Diego, CA, USA) at the Texas A&M Agrilife Genomics and Bioinformatics Sequencing Core Facility using the SP Reagent Kit v1.5 (500 cycles).
Raw sequence data was demultiplexed using pheniqs v2.1.0 [29], enforcing strict sample barcode index matching (i.e., no errors). Cutadapt v3.4 [30] was used to remove PCR primers from forward and reverse reads, discarding read pairs where one or both primers were not found in the expected location (5′) with an error rate <0.15. Raw sequences were processed using the DADA2 v1.16 package [31] in R v.4.4.3 [32]. Sequences were filtered by quality using standard filtering parameters. Error rates were estimated, and the ASV inference algorithm was applied to all filtered and trimmed sequence data. Subsequently, a consensus of the forward and reverse reads was performed. Chimeric sequences (“artifacts”) were then removed using the removeBimeraDenovo function. Sequences were compared against the SILVA database (https://www.arb-silva.de, accessed on 3 June 2024) [33] and ugreen-db (http://microgreen-23sdatabase.ea.inra.fr/, accessed on 3 June 2024) [34], which contains taxonomic information on bacteria, archaea, and eukaryotes. Taxonomy was assigned using DADA2’s assign Taxonomy function, which applies a naive Bayesian classifier up to genus level, and the assign Species function, which assigns a binomial genus–species name only on an exact identity match (100%).
2.4. Microscopy-Based Analysis
Quantitative analysis was conducted using an inverted microscope, LEICA Microsystems (DMIL LED) (Wetzlar, Germany), following the methodology of Utermöhl [16] according to APHA (2012) [35] standards. Water samples were gently homogenized before placing subsamples of 10 and 25 mL for sedimentation over 32 and 64 h, respectively. To count highly abundant species and those smaller than 20 µm, an 86.75 mm2 area was examined at 400× magnification. Organisms larger than 20 µm and less abundant were counted across the entire chamber at 200× magnification. The results were expressed in cells/L. This qualitative analysis using net samples provided an overview of the microalgae composition before carrying out a detailed quantitative analysis [36] and allowed us to take microphotographs of the most abundant species using a Nikon-brand phase-contrast trinocular microscope equipped with a camera and software TOUPVIEW (4.11.19728 (serie 4.11)) (model Eclipse SI) (Tokyo, Japan). Net samples were processed using various staining techniques to enhance the identification of chlorophytes, cyanobacteria, and dinoflagellates. Additionally, diatom shells were cleaned to facilitate species identification prior to enumeration, following the Utermöhl methodology [16].
2.5. Statistical Analyses
For microscopic data, microalgal abundance was quantified as cell density (cells/L) for each taxonomic group. In contrast, metabarcoding data were represented by the relative abundance of each amplicon sequence variant (ASV). Cluster analyses based on Bray–Curtis similarity were performed using PRIMER v6 software [37] applying a log(x+1) transformation to both ASV abundance (molecular data) and taxon density (microscopy data) for 14 samples (N = 14). To compare the composition of microalgal assemblages between the two methods, relative abundance matrices were analyzed separately for each sampling area, Puno Bay (BA), Major Lake (MA), and Minor Lake (MI), across both wet (WS) and dry (DS) seasons.
All statistical analyses were conducted in R version 4.0.2 [32]. To evaluate significant differences between molecular and microscopy-derived data, we compared proportions across the three study areas (BA, MA, and MI) at both the phylum and order taxonomic levels. Additionally, taxonomic richness at the order and genus levels was compared between methods using Student’s t-tests, implemented via the PAST 4 software package.
To minimize noise and potential artifacts—such as sequencing errors in metabarcoding or misidentifications in microscopy—genera representing less than (<2%) of the total relative abundance across the entire study area were excluded from both datasets.
3. Results and Discussion
3.1. Spatial Variability
Overall, cluster analyses based on both ASV data (metabarcoding) and abundance data (microscopy) revealed three main clusters, which corresponded closely to the geographic locations of the sampling sites (BA, MA, and MI) in both seasons. Using the molecular approach, sample E4-BA consistently clustered with MA in both the wet and dry seasons, likely due to its greater similarity with MA sites, particularly the dominance of the diatom Eunotia and the ochrophyte Nannochloropsis (Figure 2). In contrast, the microscopy-based analysis showed that the Charcas site did not group with any other cluster during the dry season, primarily due to the high relative abundance of the chlorophyte Chlamydomonadales (Figure 3).
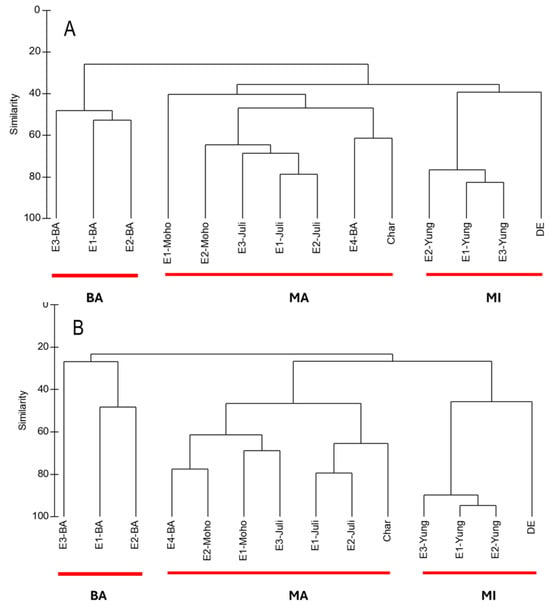
Figure 2.
(A) Cluster analysis of microalgae detected with molecular technique in 14 sampling sites in wet season (WS) based on Bray–Curtis similarities calculated from log-transformed ASV data [log(X+1)]. (B) Cluster analysis of microalgae detected with metabarcoding in 14 sampling sites in dry season (DS) based on Bray–Curtis similarities calculated from log-transformed ASV data [log(X+1)].
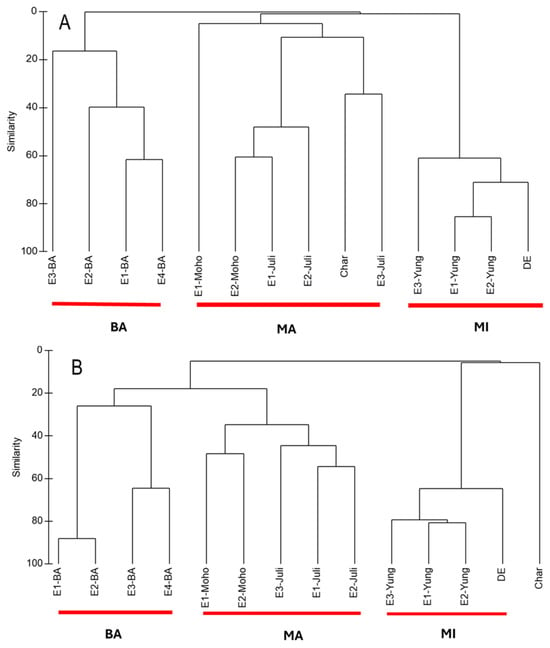
Figure 3.
(A) Cluster analysis of microalgae detected with microscopy technique in 14 samples sites in wet season (WS) based on Bray–Curtis similarities calculated from log-transformed ASV data [log(X+1)]. (B) Cluster analysis of microalgae detected with microscopy in 14 sampling sites in dry season (DS) based on Bray–Curtis similarities calculated from log-transformed ASV data [log(X+1)].
3.2. Molecular and Microscopy-Based Approaches
A total of 54 and 45 taxa were detected using the molecular approach during the wet and dry seasons, respectively, encompassing nine taxonomic divisions. Most taxa were identified at the genus level, with only one ASV assigned to the species level. Overall, the molecular technique revealed a dominance of chlorophytes, cyanobacteria, and diatoms. In contrast, the microscopy-based approach recorded 76 and 83 taxa during the wet and dry seasons, respectively, belonging to five and six divisions. The majority of these taxa were identified at the species level. Across the study area, a clear dominance of diatoms and chlorophytes was consistently observed (Table 1) (Supplemental Tables S2–S10).

Table 1.
Phytoplankton composition (%) by phylum with molecular technique and microscopy technique in wet season (WS) and dry season (DS) of 2021.
3.3. Comparing Both Approaches
Chi-squared estimates, evaluated at the 0.1% significance level (p < 0.001), were used to compare the results obtained from the molecular and microscopy-based methods. The analyses revealed significant differences in the proportional representation of microalgae between the two approaches across the three areas (BA, MA, and MI) for each phylum (Table 2) and each order (Supplemental Table S11). These discrepancies were also evident when comparing the relative abundances derived from metabarcoding and microscopy for both phyla and orders. However, a high level of concordance was observed in terms of the number of detected orders and genera. In other words, microalgal richness at the order and genus levels was comparable between the microscopy and DNA metabarcoding methods (Table 3).

Table 2.
Chi-squared test estimates comparing the proportions of microalgal phyla detected using the molecular (mol) and microscopy (mic) techniques during both the wet season (WS) and dry season (DS) of 2021. Analyses were conducted for each of the three study sites, Puno Bay (BA), Lake Major (MA), and Lake Minor (MI).

Table 3.
Chi-squared test estimates comparing the number of microalgal taxa at the order and genus levels detected using molecular and microscopy techniques. The analysis evaluates the similarity in taxonomic richness between both approaches across all sampling sites.
In the case of diatoms (Bacillariophyta), only two genera—Cyclotella and Thalassiosira—were detected by both methods. Using the molecular technique, the majority of diatom sequences were classified as “Unclassified” (>50%), meaning they could not be assigned to known taxa. In contrast, microscopy allowed for more precise taxonomic identification, with most diatoms identified at the species level. For example, Aulacoseira granulata accounted for over 70% of the diatom community in BA and MA, while Fragilaria crotonensis represented more than 49% in MI during both seasons (Figure 4 and Figure 5A,B). These findings align with those of Kim et al. [22], who reported superior species-level resolution using microscopy compared to metabarcoding. One key reason for the lack of overlap in taxa, particularly at lower taxonomic levels, is the incompleteness of reference barcode databases. Freshwater diatoms are notably underrepresented in these libraries compared to other taxonomic groups such as fish, Trichoptera, or vascular plants [38]. Another possible explanation for these differences is that non-living diatom frustules, although still morphologically identifiable, may contain only highly degraded eDNA, which prevents their assignment to a specific taxon [39]. Water samples also exhibit an inherent bias toward certain diatom groups. According to Kutty et al. [24], increasing the sample volume can enhance taxon representation and improve congruence between identification methods.
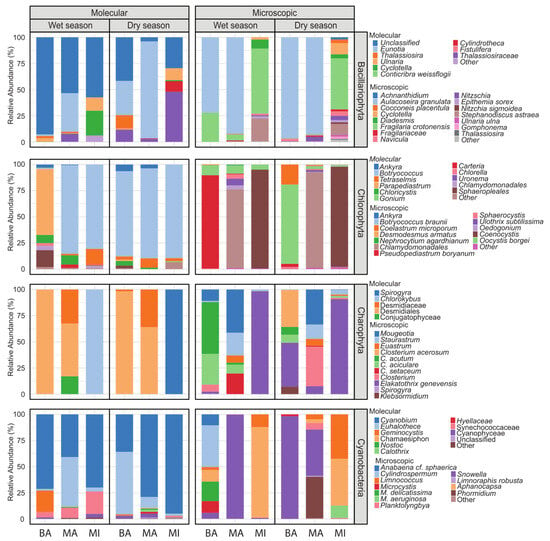
Figure 4.
Composition of Bacillariophyta, Chlorophyta, Charophyta, and Cyanobacteria communities at the genus level for each sample analyzed in Bahía de Puno (BA), Lago Major (MA), and Lago Minor (MI) using molecular and microscopy analyses, showing the relative abundance of the genera detected in the wet season (WS) and dry season (DS) of 2021.
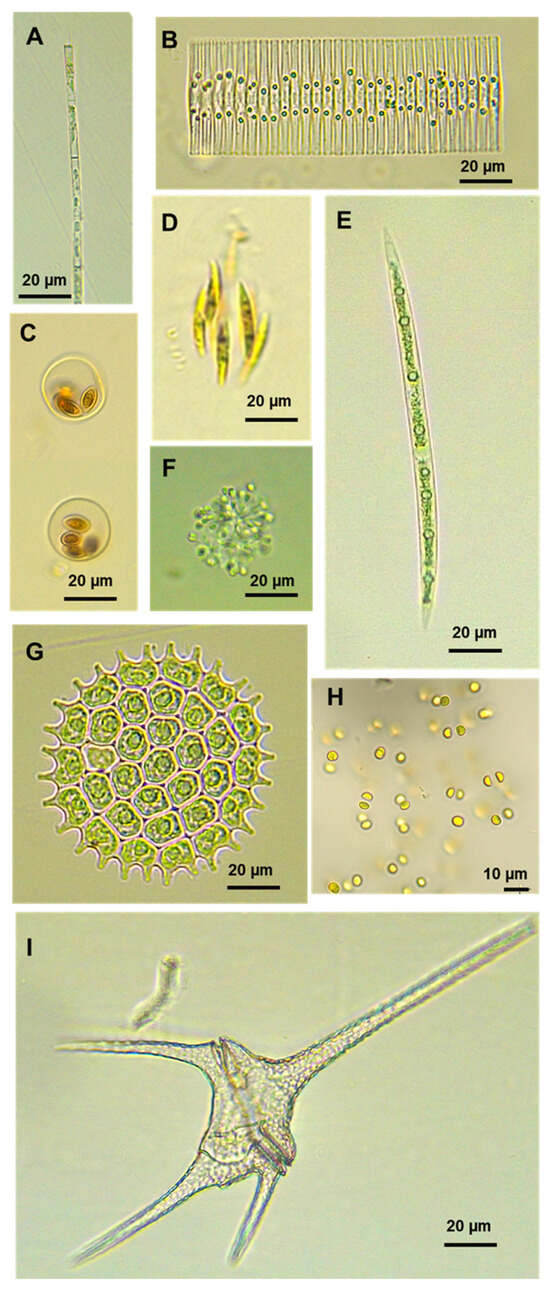
Figure 5.
Micrographs of some microalgae identified using optical microscopy in Lake Titicaca. (A) Aulacoseira granulata, (B) Fragilaria crotonensis, (C) Oocystis borgei, (D) Elakatothrix genevensis, (E) Closterium sp., (F) Snowella sp., (G) Pseudopediastrum boryanum, (H) Limnococcus sp., and (I) Ceratium hirundinella.
In Chlorophyta (chlorophytes), only two genera, Ankyra and Botryococcus, were commonly identified by both methods. In the molecular analysis, Parapediastrum was dominant in Puno Bay (BA), accounting for over 60% of chlorophyte reads, while Botryococcus dominated in Lake Major (MA) and Lake Minor (MI), exceeding 80% of reads. In contrast, microscopy enabled more detailed species-level identification, with taxa such as Pseudopediastrum boryanum (89.2%) and Oocystis borgei (75.9%) (Figure 5C,G) being prevalent in BA during the wet and dry seasons, respectively; Chlamydomonadales were dominant (>75%) in MA and Coenocystis in MI throughout both seasons. Microscopy revealed greater species richness overall, while several genera identified through metabarcoding were not detected using traditional methods. These discrepancies observed in chlorophytes, as well as in diatoms and other algal groups that will be analyzed later, can be partly attributed to differences in the volume of sample processed, with 25/10 mL for microscopy (using the inverted method) versus 1000 mL for metabarcoding. Such a variation complicates direct comparisons. Similar findings were reported by MacKeigan et al. [25], who used 2–10 mL for microscopy and 500 mL for metabarcoding. Furthermore, the units of analysis differ substantially—with individual cells for microscopy versus ASV sequences for metabarcoding—making direct comparison inherently challenging [40,41].
For charophycean algae (Charophyta), only one genus, Spirogyra, was identified using both methodologies. The molecular technique primarily detected taxa at higher taxonomic levels, including the order Desmidiales and family Desmidiaceae, with only two genera—Chlorokybus and Spirogyra—identified in MI during the wet and dry seasons, respectively. In contrast, microscopy allows for the best identification at the species/genus level. For example, Closterium acutum (49.6%) was dominant in BA; Mougeotia (41.4%) and Elakatothrix genevensis (98.3%) were prevalent in MA and MI, respectively, during the wet season. During the dry season, Closterium acerosum (35.1%) was recorded in BA, Mougeotia (33.5%) and Closterium (37.9%) in MA (Figure 5E), and Elakatothrix genevensis (90.7%) in MI (Figure 4 and Figure 5D). The discovery of Chlorokybus, a terrestrial alga, is notable. Its genetic material may have reached Lake Titicaca via runoff, rain, wind, or surrounding soil and vegetation. This possibility has been supported by the study of Bozdogan et al. [42], who evaluated the potential of eDNA present in pond water to reflect nearby terrestrial plant communities. The authors concluded that the plant taxa detected in the water’s eDNA were generally consistent with the adjacent terrestrial plant ecosystem.
For cyanobacteria, no taxa were identified using both methodologies. The molecular approach consistently detected the genus Cyanobium (ranging from 35% to 95%) across both seasons, as well as Euhalothece (47% and 59%) in MA and BA, respectively. In contrast, the microscopy-based approach identified several different genera, such as Snowella (99.7% and 98% in MA and BA, respectively) and Aphanocapsa (>45% in MI) (Figure 4). These discrepancies are consistent with the findings of MacKeigan et al. [25], who compared planktonic cyanobacterial assemblages using inverted light microscopy and 16S rRNA metabarcoding. The key difference reported was the detection of picocyanobacteria via the molecular technique. Cyanobium is a freshwater picocyanobacterium (0.2 to 2 μm) that is often overlooked in optical microscopy due to its minute size. Euhalothece, while not strictly a picocyanobacterium, also presents size and morphological variability depending on environmental conditions [43], typically measuring between 2.0 and 6.0 μm in width [44]. These small dimensions make it challenging to detect and identify using traditional microscopy. Overall, our results underscore the difficulty of accurately identifying cyanobacteria using light microscopy, largely due to their diminutive cell size and the prevalence of cryptic species [45]. An additional noteworthy finding from the molecular data is Euhalothece, typically found in hypersaline environments [44]. Although a BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi accessed on 27 July 2025) search of this ASV suggests greater similarity to Synechocystis or another Cyanophyceae, curated databases, like Silva, identified it as Euhalothece (record AY584507), which BLAST also links closely to Synechocystis. This underscores a key limitation of molecular methods: incomplete reference databases hinder reliable taxonomic assignment at lower levels for many freshwater algae, leading to unclassified or misclassified taxa [26].
In the case of euglenoids (Euglenozoa), only the genus Phacus was detected in Puno Bay (BA) using microscopy, while the molecular approach identified Eutreptia, Colacium, and Euglena. The absence of these taxa in microscopic observations may be attributed to the use of formaldehyde as a fixative. Formaldehyde is not suitable for the fixation of naked algal cells, as the cell shape is distorted and flagella are lost [15], for example, in euglenoids. As a result, certain soft-bodied or unarmored algal forms may deteriorate or go undetected under microscopy following preservation [46]. In contrast, two dinoflagellate species—Ceratium hirundinella (Figure 5I) and Peridinium cinctum—were identified using microscopy, while none were detected via metabarcoding. Although dinoflagellates have been extensively studied [26], their taxonomic classification primarily relies on distinctive morphological characteristics, such as the number, shape, and arrangement of thecal plates [47]. However, their molecular representation in databases remains limited [26]. This limitation underscores one of the primary challenges of DNA metabarcoding. Improving these databases by incorporating DNA sequences from local species in Lake Titicaca could significantly enhance the accuracy and resolution of molecular identifications in future studies.
Using only the metabarcoding approach, four additional phyla were detected—Cryptophyta, Ochrophyta, Haptophyta, and Rhodophyta—represented by the following genera: Plagioselmis, Rhodomonas (5–10 µm in diameter), Nannochloropsis (4.0–5.1 µm in diameter), Trachydiscus (2–6 µm in diameter), Ochromonas (8–10 µm in length), Chrysochromulina (3–13 µm in length), and Porphyridium (5–10 µm in length) (Table 4). These nanoplanktonic microalgae are challenging to detect using microscopy due to their minute size and morphological features that are either indistinct or invisible under a light microscope. For instance, the haptophyte Chrysochromulina possesses minute organic scales that are not discernible with light microscopy and can only be observed using transmission electron microscopy [48].

Table 4.
Phylum composition detected only with molecular techniques in the wet season (WS) and dry season (DS) of 2021.
4. Conclusions
The composition of the microalgal community differed significantly between the molecular and morphology-based approaches, likely due to the incompleteness of the reference databases, disparities in sample volume, and the use of chemical fixatives. Metabarcoding revealed a greater abundance of nanoplanktonic microalgae and picocyanobacteria in Lake Titicaca, taxa that are often underrepresented or undetected in microscopy-based assessments. The molecular monitoring of algae in the lake will still need to be complemented by microscopic analysis, despite its high cost, due to the large number of unclassified (“Unclassified”) matches associated with diatom barcodes. The findings indicate the need to improve regional reference databases and standardize both methodologies. This would enable them to complement each other and provide a more thorough and precise assessment of microalgal diversity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d17080560/s1. Table S1. Physicochemical parameters: dissolved oxygen (DO), temperature (TE), hydrogen ion potential (pH) y electrical conductivity (EC), per site and season of 2021. Wet season (WS) and dry season (DS). Table S2. Number of ASVs assigned to the Genus taxonomic level for the Phylum Bacillariophyta per site and season of 2021. Wet season (WS) and dry season (DS). Table S3. Number of ASVs assigned to the Genus taxonomic level for the Phylum Chlorophyta per sampling site and season of 2021. Wet season (WS) and dry season (DS). Table S4. Number of ASVs assigned to the Genus taxonomic level for the Phylum Charophyta per sampling site and season of 2021. Wet season (WS) and dry season (DS). Table S5. Number of ASVs assigned to the Genus taxonomic level for the Phylum Cyanobacteria per sampling site and season of 2021. Wet season (WS) and dry season (DS). Table S6. Number of ASVs assigned to the Genus taxonomic level for the Phylum Cryptophyta Euglenozoa, Ochrophyta, Haptophyta y Rhodophyta per sampling site and season of 2021. Wet season (WS) and dry season (DS). Table S7. Abundance (cells/L) of the species/genus of the Phylum Bacillariophyta per sampling site and season of 2021. Table S8. Abundance (cells/L) of the species/genus of the Phylum Chlorophyta per sampling site and season of 2021. Table S9. Abundance (cells/L) of the species/genus of the Phylum Charophyta per sampling site and season of 2021. Wet season (WS) and dry season (DS). Table S10. Abundance (cells/L) of the species/genus of the Phylum Cyanobacteria, Miozoa y Euglenozoa per sampling site and season of 2021. Wet season (WS) and dry season (DS). Table S11. Chi-square test estimates comparing the proportions of microalgal community orders identified using molecular (mol) and microscopy (mic) techniques during both the wet season (WS) and dry season (DS) of 2021. Analyses were performed for each of the three sites: Puno Bay (BA), Lake Mayor (MA), and Lake Minor (MI).
Author Contributions
Conceptualization, M.B.; methodology, M.B. and J.L.R.; writing—original draft preparation, M.B. and J.L.R.; writing—review and editing, M.B. and J.L.R.; contributed to the writing of the manuscript, M.B.; funding acquisition M.B. All authors have read and agreed to the published version of the manuscript.
Funding
FONDECYT for the funding granted through the project “DNA metabarcoding and morphological identification of microalgae and cyanobacteria: biological indicators to evaluate water quality in Lake Titicaca”, contract 130-2020-FONDECYT.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The raw sequence reads obtained in this study have been deposited in the NCBI SRA (BioProject PRJNA1281193).
Acknowledgments
We thank Anderson Villanueva and Leonardo Mendoza for their collaboration in the sampling and sample analysis process.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| BA | Puno Bay |
| MA | Major Lake |
| MI | Minor Lake |
| DS | Dry season |
| WS | Wet season |
References
- MINAM. Línea Base Ambiental de la Cuenca del Lago Titicaca; Ministerio del Ambiente: Lima, Peru, 2013. [Google Scholar]
- Balvanera, P.; Pfisterer, A.B.; Buchmann, N.; He, J.S.; Nakashizuka, T.; Raffaelli, D. Quantifying the evidence for biodiversity effects on ecosystem functioning and services. Ecol. Lett. 2006, 9, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Groendahl, S.; Kahlert, M.; Fink, P. The best of both worlds: A combined approach for analyzing microalgal diversity via metabarcoding and morphology-based methods. PLoS ONE 2017, 12, e0172808. [Google Scholar] [CrossRef]
- Wondmagegn, T.; Mengistou, S. Effects of anthropogenic activities on macroinvertebrate assemblages in the littoral zone of Lake Hawassa, a tropical Rift Valley Lake in Ethiopia. Lakes Reserv. Res. Manag. 2020, 25, 61–71. [Google Scholar] [CrossRef]
- MINAN. Aprueban estándares de calidad ambiental (ECA) para agua y establecen disposiciones complementarias. El Peru. 2017, 6–9. [Google Scholar]
- Beltrán, D.F.; Palomino, R.P.; Moreno, E.G.; Peralta, C.G.; Montesinos-Tubée, D.B. Calidad de agua de la bahía interior de Puno, lago Titicaca durante el verano del 2011. Rev. Peru. Biol. 2015, 22, 335–340. [Google Scholar] [CrossRef]
- Iquise, S.R. Variación Espacial del Zooplancton en Tres Épocas en Relación a Factores Fisicoquímicos del Agua en la Bahía Interior y Exterior del Lago Titicaca, Puno. Bachelor’s Thesis, Universidad Nacional del Altiplano, Puno, Peru, 2017. Available online: http://repositorio.unap.edu.pe/handle/20.500.14082/7062 (accessed on 15 April 2025).
- Oviedo, A.I.H. Variación espacial y temporal de la diversidad y abundancia del fitoplancton del lago de Yojoa en un año hidrológico 2014–2015. Rev. Cienc. Tecnol. 2017, 19, 40–77. [Google Scholar] [CrossRef]
- Mariano-Astocondor, M. Composición y estructura de la comunidad fitoplanctónica en la laguna Tranca Grande (Junín, Perú). Rev. Peru. Biol. 2001, 8, 114–124. [Google Scholar] [CrossRef]
- Rimet, F. Recent views on river pollution and diatoms. Hydrobiology 2012, 683, 1–24. [Google Scholar] [CrossRef]
- Stevenson, R.J.; Pan, Y.; Van Dam, H. Assessing environmental conditions in rivers and streams with diatoms. In The Diatoms: Applications for the Environmental and Earth Sciences; Cambridge University Press: Cambridge, UK, 2010; pp. 57–85. [Google Scholar]
- Paerl, H.W.; Paul, V.J. Climate change: Links to global expansion of harmful cyanobacteria. Water Res. 2012, 46, 1349–1363. [Google Scholar] [CrossRef]
- Reynolds, C.S. The Ecology Freshwataer Phytoplankton; Cambridge University Press: Cambridge, UK, 1984; p. 390. [Google Scholar]
- Chorus, I.; Bartram, J. Toxic Cyanobacteria in Water: A Guide to Public Health Significance, Monitoring and Management; Spon/Chapman & Hall: London, UK, 1999. [Google Scholar]
- Karlson, B.; Cusack, C.; Bresnan, E. Microscopic and molecular methods for quantitative phytoplankton analysis. In Intergovernmental Oceanographic Commission, Manuals and Guides; UNESCO: Paris, France, 2010. [Google Scholar] [CrossRef]
- Utermohl, H. Zur vervollkommnung der cuantitativo del fitoplancton metódico. Int. Ver. Theor. Angew. Limnol. Mitteilungen 1958, 9, 1–38. [Google Scholar] [CrossRef]
- Lindeque, P.K.; Hay, S.J.; Heath, M.R.; Ingvarsdottir, A. Integrating conventional microscopy and molecular analysis to analyse the abundance and distribution of four Calanus congeners in the North Atlantic. J. Plankton Res. 2006, 28, 221–238. [Google Scholar] [CrossRef][Green Version]
- Chen, G.; Hare, M.P. Cryptic ecological diversification of a planktonic estuarine copepod, Acartia tonsa. Mol. Ecol. 2008, 17, 1451–1468. [Google Scholar] [CrossRef]
- Abad, D.; Albaina, A.; Aguirre, M.; Laza-Martínez, A.; Uriarte, I.; Iriarte, A.; Villate, F.; Estonba, A. Is metabarcoding suitable for estuarine plankton monitoring? A comparative study with microscopy. Mar. Biol. 2016, 163, 149–150. [Google Scholar] [CrossRef]
- Gojobori, T.; Wada, T.; Kobayashi, T.; Mineta, K. Marine Metagenomics: Technological Aspects and Applications; Springer Nature Singapore Pte Ltd.: Singapore, 2019. [Google Scholar]
- Streit, W.R.; Daniel, R. Metagenomics: Methods and Protocols; Spring: New York, NY, USA, 2010. [Google Scholar]
- Kim, Y.S.; Yun, H.S.; Lee, J.H.; Lee, K.L.; Choi, J.S.; Won, D.H.; Kim, Y.J.; Kim, H.S.; Yoon, H.S. Comparison of Metabarcoding and Microscopy Methodologies to Analyze Diatom Communities in Five Estuaries Along the Southern Coast of the Korean Peninsula. Microb. Ecol. 2024, 87, 95. [Google Scholar] [CrossRef]
- Kulaš, A.; Udovič, M.G.; Tapolczai, K.; Žutinić, P.; Orlić, S.; Levkov, Z. Diatom eDNA metabarcoding and morphological methods for bioassessment of karstic river. Sci. Total Environ. 2022, 829, 154536. [Google Scholar] [CrossRef]
- Kutty, S.N.; Loh, R.K.; Bannister, W.; Taylor, D. Evaluation of a diatom eDNA-based technique for assessing water quality variations in tropical lakes and reservoirs. Ecol. Indic. 2022, 141, 109108. [Google Scholar] [CrossRef]
- MacKeigan, P.W.; Garner, R.E.; Monchamp, M.È.; Walsh, D.A.; Onana, V.E.; Kraemer, S.A.; Pick, F.R.; Beisner, B.E.; Agbeti, M.D.; da Costa, N.B.; et al. Comparing microscopy and DNA metabarcoding techniques for identifying cyanobacteria assemblages across hundreds of lakes. Harmful Algae Mar. 2022, 113, 102187. [Google Scholar] [CrossRef]
- Santi, I.; Kasapidis, P.; Karakassis, I.; Pitta, P.A. Comparison of DNA Metabarcoding and Microscopy Methodologies for the Study of Aquatic Microbial Eukaryotes. Diversity 2021, 13, 180. [Google Scholar] [CrossRef]
- Samanez, I.; Rimarachin Ching, V.; Palma, C.; Maestre, J.; Ortega, H.; Correa, V.; Hidalgo, M. Métodos de Colecta, Identificación y Análisis de Comunidades Biológicas: Plancton, Perifiton, Bentos (Macroinvertebrados) y Necton (Peces) en Aguas Continentales del Perú; Ministerio del Ambiente: Lima, Peru, 2014. [Google Scholar]
- Sherwood, A.R.; Presting, G.G. Universal primers amplify a 23S rDNA plastid marker in eukaryotic algae and cyanobacteria. J. Phycol. 2007, 43, 605–608. [Google Scholar] [CrossRef]
- Galanti, L.; Shasha, D.; Gunsalus, K.C. Pheniqs 2.0: Accurate, high-performance Bayesian decoding and confidence estimation for combinatorial barcode indexing. BMC Bioinform. 2021, 22, 359. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. Available online: https://journal.embnet.org/index.php/embnetjournal/article/view/200 (accessed on 1 April 2025). [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. Dada2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.r-project.org/ (accessed on 1 April 2025).
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Pablo, T.; Pablo, Y.; Jörg, Y.; Frank, P.; Glöckner, O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Djemiel, C.; Plassard, D.; Terrat, S. µgreen-db: A reference database for the 23S rRNA gene of eukaryotic plastids and cyanobacteria. Sci. Rep. 2020, 10, 5915. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Waste Water, 22nd ed.; American Public Health Association: Washington, DC, USA, 2012. [Google Scholar]
- UNE-EN 15204; Calidad del Agua Guía Para el Recuento de Fitoplancton por Microscopía Invertida (Técnica Utermöhl). Asociación Española de Normalización y Certificación (AENOR): Madrid, Spain, 2007; p. 44.
- Clarke, K.; Goley, R. Plymout Routines in Multivariate Ecological Research v5. User Manual/Tutorial; PRIMER-E LTDA: Auckland, New Zealand, 2001; p. 91. [Google Scholar]
- Weigand, H.; Beermann, A.J.; Čiampor, F.; Costa, F.O.; Csabai, Z.; Duarte, S.; Geiger, M.F.; Grabowski, M.; Rimet, F.; Rulik, B.; et al. DNA barcode reference libraries for the monitoring of aquatic biota in Europe: Gap-analysis and recommendations for future work. Sci. Total Environ. 2019, 678, 499–524. [Google Scholar] [CrossRef]
- Kang, W.; Anslan, S.; Börner, N.; Schwarz, A.; Schmidt, R.; Künzel, S.; Rioual, P.; Echeverría-Galindo, P.; Vences, M.; Wang, J.; et al. Diatom metabarcoding and microscopic analyses from sediment samples at Lake Nam Co, Tibet: The effect of sample-size and bioinformatics on the identified communities. Ecol. Indic. 2021, 121, 107070. [Google Scholar] [CrossRef]
- Pawlowski, J.; Kelly-Quinn, M.; Altermatt, F.; Apoth’eloz-Perret-Gentil, L.; Beja, P.; Boggero, A.; Borja, A.; Bouchez, A.; Cordier, T.; Domaizon, I.; et al. The future of biotic indices in the ecogenomic era: Integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci. Total Environ. 2018, 11, 96. [Google Scholar] [CrossRef]
- Vuorio, K.; Mäki, A.; Salmi, P.; Aalto, S.L.; Tiirola, M. Consistency of targeted metatranscriptomics and morphological characterization of phytoplankton communities. Front. Microbiol. 2020, 11, 96. [Google Scholar] [CrossRef]
- Bozdogan, D.; Takizawa, S.; Furukori, N.; Homma, K.; Abe, H.; Sakio, H.; Harada, N.; Suzuki, K. Pond Water eDNA Reflects Broad Consistency with Surrounding Terrestrial Plant Ecosystems. Biology 2025, 14, 62. [Google Scholar] [CrossRef]
- Bhatt, H.H.; Pasricha, R.; Upasani, V.N. Aislamiento y caracterización de una cianobacteria halófila, Euhalothece SLVH01, del lago salado de Sambhar, India. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 215–224. [Google Scholar] [CrossRef]
- Mogany, T.; Swalaha, F.M.; Allam, M.; Mtshali, P.S.; Ismail, A.; Kumari, S.; Bux, F. Caracterización fenotípica y genotípica de una cianobacteria unicelular hipersalina autóctona única, Euhalothece sp. nov. Microbiol. Res. 2018, 211, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Sciuto, K.; Moro, I. Cyanobacteria: The bright and dark sides of a charming group. Biodivers. Conserv. 2015, 24, 711–738. [Google Scholar] [CrossRef]
- CEN/TC 230; Water Quality—Guidance on Quantitative and Qualitative Sampling of Marine Phytoplankton. iTeh Standards: San Francisco, CA, USA, 2005; p. 26.
- Boltovskoy, A. Taxonomía y Morfología de los Dinoflagelados: Métodos de Trabajo; Alveal, K., Ferrario, M.E., Oliveira, C., Sar, E., Eds.; Universidad de Concepción: Concepción, Chile, 1995; pp. 55–82. [Google Scholar]
- Bicudo, C.E.M.; Menezes, M. Gêneros de Algas de Águas Continentais do BRASIL: Chave Para Identificação e Descrições, 3rd ed.; RiMa: São Carlos, Brazi, 2017; p. 572. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).