Redefining Latrogastropoda Again and Searching for Its Sister Group in Hypsogastropoda (Gastropoda: Caenogastropoda)
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Methods
2.2. Data Set Assembly
2.3. Phylogenetic Analyses
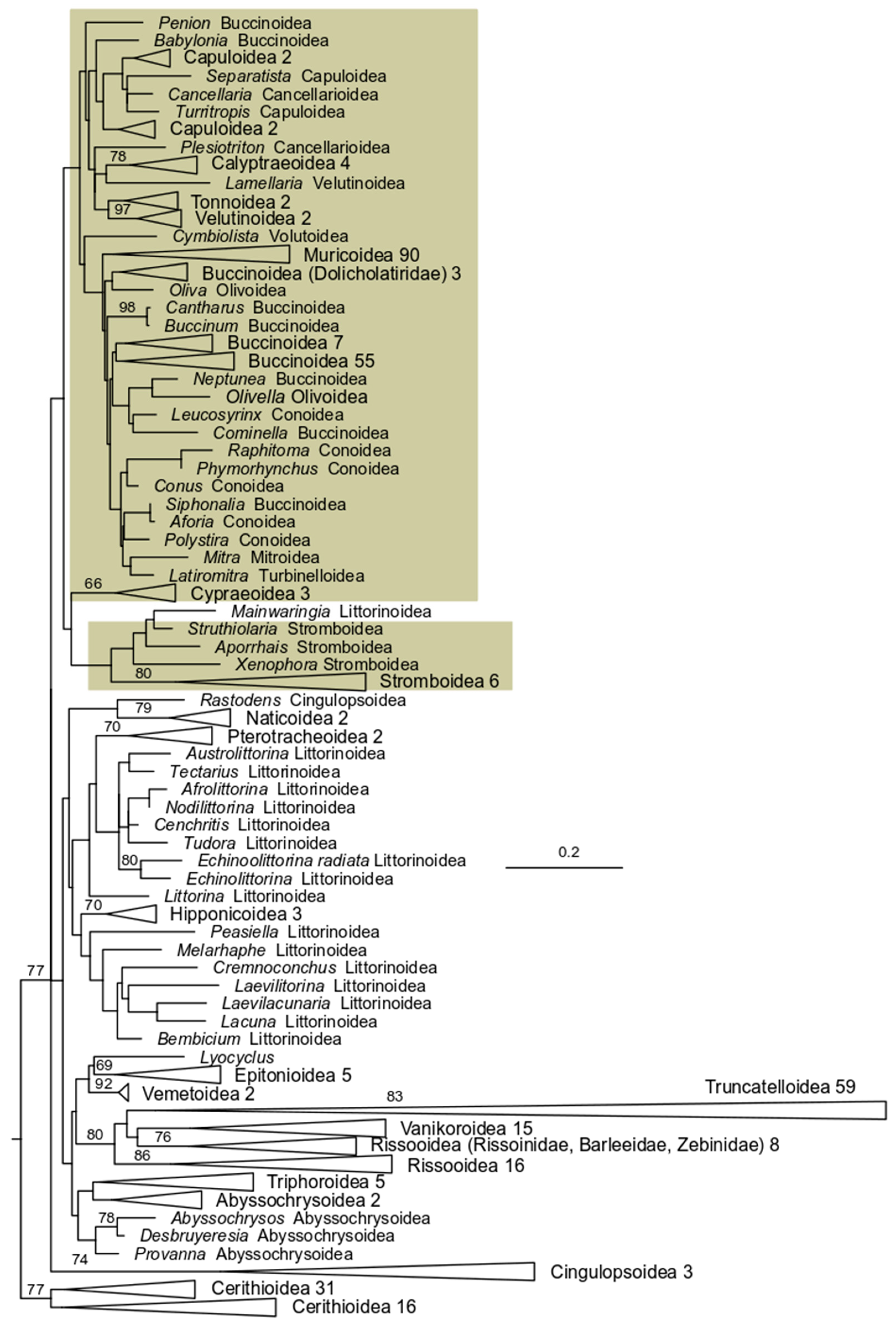
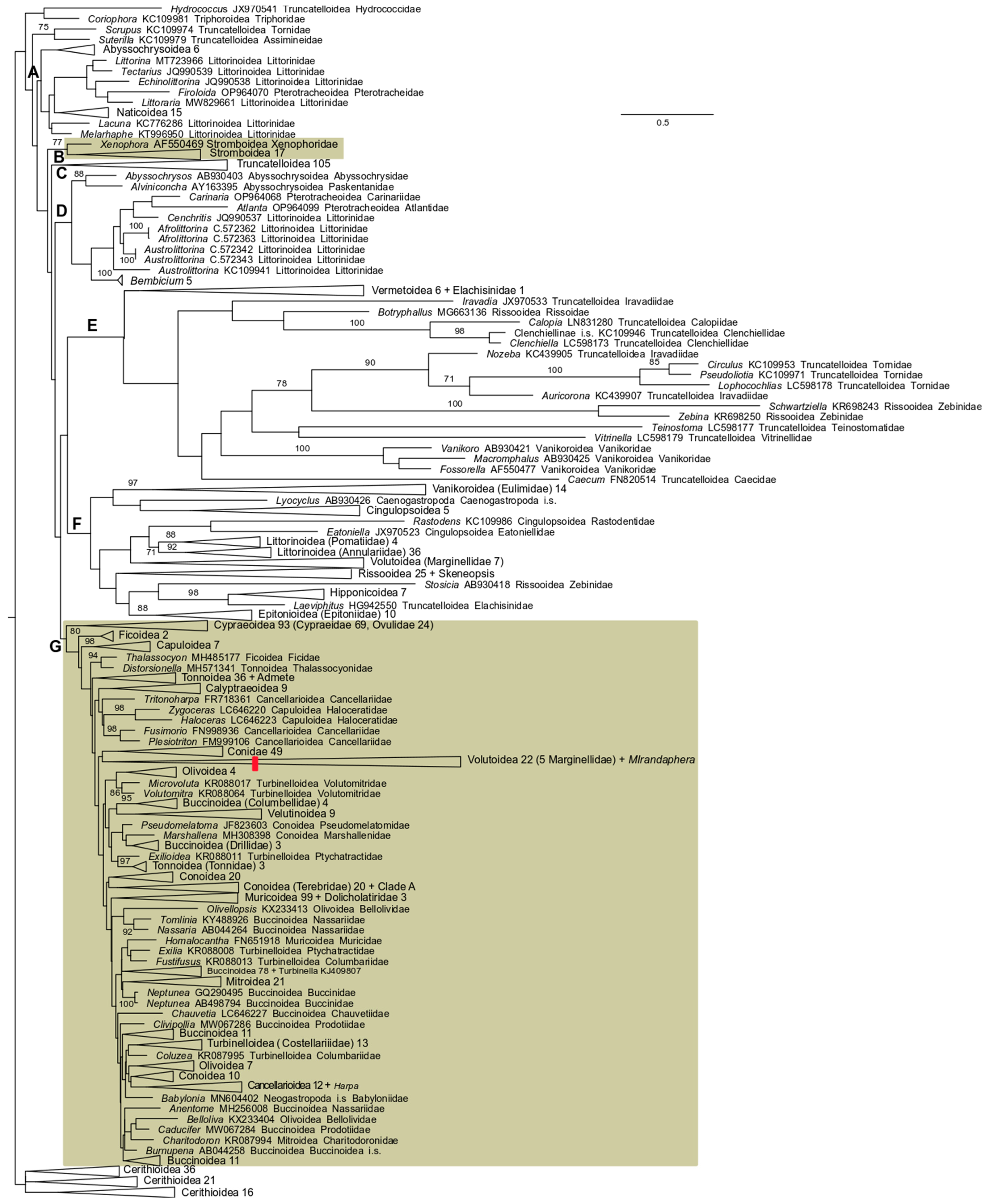
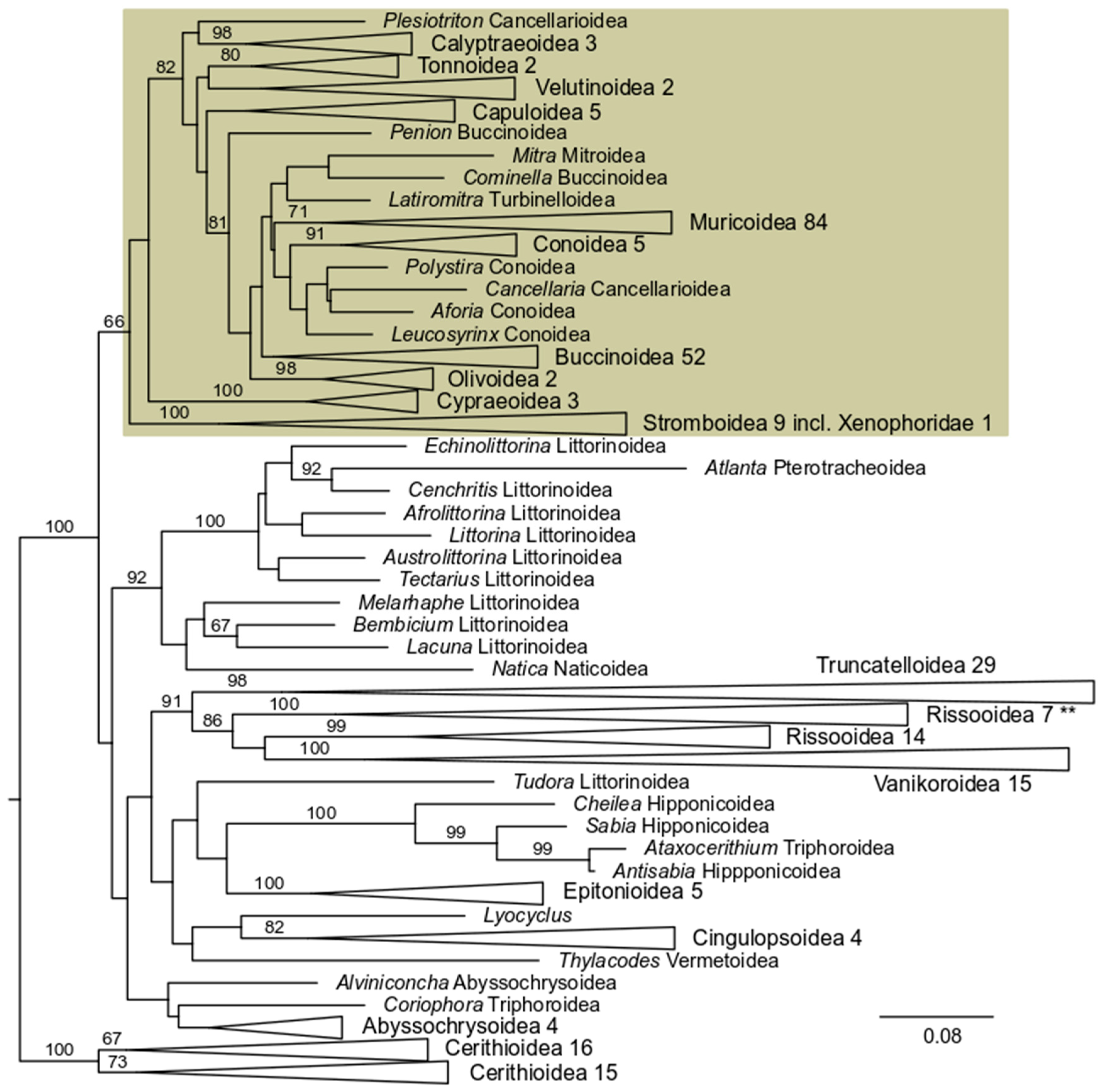
3. Results
3.1. Alignment Details
3.2. 28S rRNA Analysis
3.3. 16S rRNA Analysis
3.4. COI Analysis
3.5. 28 S rRNA + 16S rRNA Analysis
3.6. 28S rRNA + 16S rRNA + COI Analysis
3.7. GC/CG Motifs’ Occurrence
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bouchet, P.; Rocroi, J.-P.; Hausdorf, B.; Kaim, A.; Kano, Y.; Nützel, A.; Parkhaev, P.; Schrödl, M.; Strong, E.E. Revised classification, nomenclator and typification of gastropod and monoplacophoran families. Malacologia 2017, 61, 1–526. [Google Scholar] [CrossRef]
- Riedel, F. Ursprung Und Evolution Der “Höheren” Caenogastropoda. In Berliner Geowissenschaftliche Abhandlungen, Series E; Selbstverlag Fachbereich Geowissenschaften: Berlin, Germany, 2000; Volume 32, pp. 1–240, 21 pls. [Google Scholar]
- Simone, L.R.L. Phylogeny of the Caenogastropoda (Mollusca), based on comparative morphology. Arq. Zool. 2011, 42, 161–323. [Google Scholar] [CrossRef]
- Irwin, A.R.; Strong, E.E.; Kano, Y.; Harper, E.M.; Williams, S.T. Eight new mitogenomes clarify the phylogenetic relationships of Stromboidea within the caenogastropod phylogenetic framework. Mol. Phylogenet. Evol. 2021, 158, 107081. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Yang, M.; Duan, H.; Liao, X. The mitochondrial genome of Littoraria melanostoma reveals a phylogenetic relationship within Littorinimorpha. Diversity 2023, 15, 1005. [Google Scholar] [CrossRef]
- Ma, Y.; Zheng, B.; Li, J.; Meng, W.; Xu, K.; Ye, Y. Characterization of the complete mitochondrial genome of Desmaulus extinctorium (Littorinimorpha, Calyptraeoidea, Calyptraeidae) and molecular phylogeny of Littorinimorpha. PLoS ONE 2024, 19, e0301389. [Google Scholar] [CrossRef]
- Bouchet, P.; Rocroi, J.-P. Classification and nomenclator of gastropod families. Malacologia 2005, 47, 1–397. [Google Scholar]
- Ponder, W.F.; Lindberg, D.R.; Ponder, J.M. Biology and Evolution of the Mollusca; CRC Press: Boca Raton, LA, USA, 2020; Volume 2. [Google Scholar]
- Ponder, W.F.; Colgan, D.J.; Healy, J.M.; Nützel, A.; Simone, L.R.L.; Strong, E.E. 2008. Caenogastropoda. In Phylogeny and Evolution of the Mollusca; Ponder, W.F., Lindberg, D.L., Eds.; University of California Press: Berkeley, CA, USA, 2008; pp. 331–383. [Google Scholar]
- Kantor, Y.I.; Fedosov, A.E.; Puillandre, N.; Bonillo, C.; Bouchet, P. Returning to the roots: Morphology, molecular phylogeny and classification of the Olivoidea (Gastropoda: Neogastropoda). Zool. J. Linn. Soc. 2017, 180, 493–541. [Google Scholar] [CrossRef]
- Kantor, Y.I.; Fedosov, A.E.; Kosyan, A.R.; Puillandre, N.; Sorokin, P.A.; Kano, Y.; Clark, R.; Bouchet, P. Molecular phylogeny and revised classification of the Buccinoidea (Neogastropoda). Zool. J. Linn. Soc. 2022, 194, 789–857. [Google Scholar] [CrossRef]
- Takano, T.; Warén, A.; Kano, Y. Phylogenetic position of the deep-sea snail family Haloceratidae and new insights into caenogastropod relationships. J. Molluscan Stud. 2022, 88, eyac012. [Google Scholar] [CrossRef]
- Irwin, A.R.; Bouchet, P.; Crame, J.A.; Harper, E.M.; Kronenberg, G.C.; Strong, E.E.; Williams, S.T. Molecular phylogenetics of the superfamily Stromboidea (Caenogastropoda): New insights from increased taxon sampling. Zool. Scr. 2024, 53, 818–838. [Google Scholar] [CrossRef]
- Fedosov, A.E.; Zaharias, P.; Lemarcis, T.; Modica, M.V.; Holford, M.; Oliverio, M.; Kantor, Y.I.; Puillandre, N. Phylogenomics of Neogastropoda: The backbone hidden in the bush. Syst. Biol. 2024, 73, 521–531. [Google Scholar] [CrossRef]
- Qi, L.; Zhang, N.; Xu, B.; Xu, Q.; Han, X.; Kong, L.; Li, Q. Increased microgastropoda sampling give new insights into the phylogenetic relationships of Littorinoidea (Littorinimorpha). Mol. Phylogenet. Evol. 2024, 199, 108139. [Google Scholar] [CrossRef]
- Qu, J.; Yang, W.; Teng, X.; Xu, L.; Zhang, D.; Xing, Z.; Wang, S.; Liu, X.; Wang, L.; Wang, X. Gene characterization and phylogenetic analysis of four mitochondrial genomes in Caenogastropoda. Acta Oceanol. Sin. 2024, 43, 137–150. [Google Scholar] [CrossRef]
- Wang, H.; He, X.; Chen, C.; Gao, K.; Dai, Y.; Sun, J. New insights into the phylogeny of Neogastropoda aided by draft genome sequencing of a volutid snail. Zool. Scr. 2024, 53, 805–817. [Google Scholar] [CrossRef]
- Colgan, D.J.; Ponder, W.F.; Beacham, E.; Macaranas, J. Molecular phylogenetics of Caenogastropoda (Gastropoda: Mollusca). Mol. Phylogenet. Evol. 2007, 42, 717–737. [Google Scholar] [CrossRef] [PubMed]
- Osca, D.; Templado, J.; Zardoya, R. Caenogastropod mitogenomics. Mol. Phylogenet. Evol. 2015, 93, 118–128. [Google Scholar] [CrossRef]
- Santos, C.A.; Bezerra, F.O.; Andrade, S.C. Littoraria flava (Gastropoda: Littorinidae) mitogenome: Phylogenetic considerations within the Caenogastropoda and evidence of microscale local adaptation. Mar. Biol. 2015, 169, 121. [Google Scholar]
- Li, F.; Li, W.; Zhang, Y.; Wang, A.; Liu, C.; Gu, Z.; Yang, Y. The molecular phylogeny of Caenogastropoda (Mollusca, Gastropoda) based on mitochondrial genomes and nuclear genes. Gene 2024, 928, 148790. [Google Scholar] [CrossRef] [PubMed]
- Strong, E.E. Refining molluscan characters: Morphology, character coding and a phylogeny of the Caenogastropoda. Zool. J. Linn. Soc. 2003, 137, 447–554. [Google Scholar] [CrossRef]
- Goulding, T.C.; Strong, E.E.; Quattrini, A.M. Target-capture probes for phylogenomics of the Caenogastropoda. Mol. Ecol. Resour. 2023, 23, 1372–1388. [Google Scholar] [CrossRef]
- Mindell, D.P.; Honeycutt, R.L. Ribosomal RNA in vertebrates: Evolution and phylogenetic applications. Ann. Rev. Ecol. Syst. 1990, 21, 541–566. [Google Scholar] [CrossRef]
- Fontanilla, I.K.; Naggs, F.; Wade, C.M. Molecular phylogeny of the Achatinoidea (Mollusca: Gastropoda). Mol. Phylogenet. Evol. 2017, 114, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Li, Q.; Kong, L. Additional gene data and increased sampling give new insights into the phylogenetic relationships of Neogastropoda, within the caenogastropod phylogenetic framework. Mol. Phylogenet. Evol. 2011, 61, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Takano, T.; Kano, Y. Molecular phylogenetic investigations of the relationships of the echinoderm-parasite family Eulimidae within Hypsogastropoda (Mollusca). Mol. Phylogenet. Evol. 2014, 79, 258–269. [Google Scholar] [CrossRef]
- Machkour-M’Rabet, S.; Hanes, M.M.; Martínez-Noguez, J.J.; Cruz-Medina, J.; García-De León, F.J. The queen conch mitogenome: Intra- and interspecific mitogenomic variability in Strombidae and phylogenetic considerations within the Hypsogastropoda. Sci. Rep. 2021, 11, 11972. [Google Scholar] [CrossRef] [PubMed]
- Simone, L.R.L. Comparative morphological study of representatives of the three families of the Stromboidea and the Xenophoroidea (Mollusca, Caenogastropoda), with an assessment of their phylogeny. Arq. Zool. 2005, 37, 141–267. [Google Scholar] [CrossRef]
- Fassio, G.; Stefani, M.; Russini, V.; Buge, B.; Bouchet, P.; Treneman, N.; Malaquias, M.A.E.; Schiaparelli, S.; Modica, M.V.; Oliverio, M. Neither slugs nor snails: A molecular reappraisal of the gastropod family Velutinidae. Zool. J. Linn. Soc. 2023, 197, 924–964. [Google Scholar] [CrossRef]
- Criscione, F.; Ponder, W.F. A phylogenetic analysis of rissooidean and cingulopsoidean families (Gastropoda: Caenogastropoda). Mol. Phylogenet. Evol. 2013, 66, 1075–1082. [Google Scholar] [CrossRef]
- Criscione, F.; Ponder, W.F.; Köhler, F.; Takano, T.; Kano, Y. A molecular phylogeny of Rissoidae (Caenogastropoda: Rissooidea) allows testing the diagnostic utility of morphological traits. Zool. J. Linn. Soc. 2016, 179, 23–40. [Google Scholar] [CrossRef]
- Mcarthur, A.G.; Koop, B.F. Partial 28S rDNA sequences and the antiquity of the hydrothermal vent endemic gastropods. Mol. Phylogenet. Evol. 1999, 13, 255–274. [Google Scholar] [CrossRef]
- Colgan, D.J.; Ponder, W.F.; Beacham, E.; Macaranas, J.M. Molecular phylogenetic studies of Gastropoda based on six gene segments representing coding or non-coding and mitochondrial or nuclear DNA. Molluscan Res. 2003, 23, 123–148. [Google Scholar] [CrossRef]
- Colgan, D.J.; Ponder, W.F.; Eggler, P.E. Gastropod evolutionary rates and phylogenetic relationships assessed using partial 28S rDNA and histone H3 sequences. Zool. Scr. 2000, 29, 29–63. [Google Scholar] [CrossRef]
- Palumbi, S.R.; Martin, A.; Romano, S.; McMillan, W.O.; Stice, L.; Grabowski, G. The Simple Fool’s Guide to PCR; Department of Zoology and Kewalo Marine Laboratory, University of Hawai’i: Honolulu, HI, USA, 1991. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- MolluscaBase Editors. MolluscaBase. Available online: https://www.molluscabase.org (accessed on 25 June 2025).
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL X Windows interface: Flexible strategiesfor multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Katoh, K.; Asimenos, G.; Toh, H. Multiple alignment of DNA sequences with MAFFT. In Bioinformatics for DNA Sequence Analysis; Posada, D., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; pp. 39–64. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program forWindows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 312–313. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 2008, 75, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2016, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. Figtree Version 1.4.3. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 18 November 2016).
- Pattengale, N.D.; Alipour, M.; Bininda-Emonds, O.R.P.; Moret, B.M.E.; Stamatakis, A. How many bootstrap replicates arenecessary? J. Comput. Biol. 2010, 17, 337–354. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2016, 35, 518–522. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J. Tracer 1.3. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 31 August 2023).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ponder, W.F. The anatomy and relationships of Elachisina Dall (Gastropoda: Rissoacea). J. Molluscan Stud. 1985, 51, 23–34. [Google Scholar] [CrossRef]
- Xia, X.; Lemey, P. Assessing substitution saturation with DAMBE. In The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny, 2nd ed.; Lemey, P., Salemi, M., Vandamme, A.-M., Eds.; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Wenz, W. Gastropoda. In Handbuch der Palaozoologie; Schindewolf, O.H., Ed.; Bornträger: Berlin, Germany, 1938; Volume 6, pp. 831–999. [Google Scholar]
- Fedosov, A.E.; Caballer Gutierrez, M.; Buge, B.; Sorokin, P.V.; Puillandre, N.; Bouchet, P. Mapping the missing branch on the neogastropod tree of life: Molecular phylogeny of marginelliform gastropods. J. Molluscan Stud. 2019, 85, 439–451. [Google Scholar] [CrossRef]
- Harasewych, M.G.; Sei, M.; Oleinik, A.; Uribe, J.E. The complete mitochondrial genome of Voluta musica Linnaeus, 1758 (Neogastropoda: Volutidae: Volutinae). Nautilus 2024, 138, 1–7. [Google Scholar]
- Ponder, W.F. The origin and evolution of the Neogastropoda. Malacologia 1974, 12, 295–338. [Google Scholar]
- Modica, M.V.; Bouchet, P.; Cruaud, C.; Utge, J.; Oliverio, M. Molecular phylogeny of the nutmeg shells (Neogastropoda, Cancellariidae). Mol. Phylogenet. Evol. 2011, 59, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.G.; Dyal, P.; Williams, S.T. A global molecular phylogeny of 147 periwinkle species (Gastropoda, Littorininae). Zool. Scr. 2012, 41, 125–136. [Google Scholar] [CrossRef]
- Reid, D.G. Mainwaringia Nevill, 1885, a littorinid genus from Asiatic mangrove forests, and a case of protandrous hermaphroditism. J. Molluscan Stud. 1986, 52, 225–242. [Google Scholar] [CrossRef]
- Reid, D.G. Systematic revision of the Recent species of Peasiella Nevill, 1885 (Gastropoda: Littorinidae), with notes on the fossil species. Nautilus 1989, 103, 43–69. [Google Scholar]
- Reid, D.G.; Mak, Y.M. Additions and corrections to the taxonomy of the genus Peasiella Nevill, 1885 (Gastropoda: Littorinidae). Naut. 1998, 112, 6–33. [Google Scholar]
- Simone, L.R.L. Convergence with naticids: Phenotypic phylogenetic study on some Antarctic littorinoideans, with description of the zerotulid new genus Pseudonatica, and its presence in Brazil (Mollusca, Caenogastropoda). J. Mar. Biol. Assoc. UK 2018, 98, 1365–1381. [Google Scholar] [CrossRef]
- Ponder, W.F.; Fukuda, H.; Hallan, A. A review of the family Clenchiellidae (Mollusca: Caenogastropoda: Truncatelloidea). Zootaxa 2014, 3872, 101–153. [Google Scholar] [CrossRef] [PubMed]
| Taxon | Composition | Reference |
|---|---|---|
| Latrogastropoda | Calyptraeoidea a, Cassoidea (= Tonnoidea), Cypraeoidea, Ficoidea, Lamellarioidea b, Laubierinoidea c, Naticoidea, Neogastropoda | [2] |
| Latrogastropoda | Calyptraeoidea, Cypraeoidea (including Velutinidae), Ficoidea, Neogastropoda, Stromboidea, Tonnoidea, Xenophoroidea | [1] |
| Latrogastopoda | Calyptraeoidea, Capuloidea, Cypraeoidea, Ficoidea, Stromboidea (includes Xenophoridae), Tonnoidea, Velutinoidea and Neogastropoda (Buccinoidea Rafinesque, 1815, Cancellarioidea, Forbes & Hanley, 1851, Conoidea Fleming, 1822, Mitroidea Swainson, 1831, Muricoidea Rafinesque, 1815, Olivoidea Latreille, 1825, Turbinelloidea Swainson, 1835, and Volutoidea Rafinesque, 1815) | Redefined here |
| Siphonogastropoda | Cypraeoidea (including Velutinidae), Neogastropoda, Tonnoidea (including Ficidae) | [3] |
| Littorinimorpha | Hypsogastropoda except Neogastropoda | [7] |
| Rissoidina (= “Rissoiform clade” of [1]) | Rissooidea Gray, 1840, Truncatelloidea Gray, 1840 and Vanikoroidea Gray, 1840 | [8] |
| “Asiphonate group” | Cingulopsoidea Fretter & Patil, 1958, Epitoniodea Berry, 1910 (1812), Vanikoroidea, Hipponicoidea Troschel, 1861, Littorinoidea Children, 1834, Naticoidea, Pterotracheoidea Rafinesque, 1814, Rissooidea, Triphoroidea Gray, 1847, Truncatelloidea and Vermetoidea Rafinesque, 1815 | [9] |
| “Siphonate group” | Calyptraeoidea, Cypraeoidea, Stromboidea, Tonnoidea, Xenophoridae and the neogastropod superfamilies Buccinoidea, Cancellarioidea, Conoidea, Mitroidea, Muricoidea and Volutoidea | [9] |
| Taxon | Latrogastropoda sensu [2] | Latrogastropoda sensu [1] | Siphonogastropoda [3] | “Siphonate” Clade [9] | Latrogastropoda as Here Redefined |
|---|---|---|---|---|---|
| Calyptraeidae | Yes | Yes | No | Yes | Yes |
| Capulidae | Yes | No | No | ? | Yes |
| Cypraeoidea | Yes | Yes | Yes | Yes | Yes |
| Ficoidea | Yes | Yes | Yes | ? | Yes |
| Hipponicoidea | Yes | No | No | Yes | Yes |
| Neogastropoda | Yes | Yes | Yes | Yes | Yes |
| Naticoidea | Yes | No | No | No | No |
| Stromboidea | No | Yes | No | Yes | Yes |
| Tonnoidea | Yes | Yes | Yes | Yes | Yes |
| Velutinoidea | Yes | Yes | Yes | ? | Yes |
| Gene | Primer Name | Primer Sequence | Reference |
|---|---|---|---|
| 28S rRNA | 28S D1F | ACCCSCTGAAYTTAAGCAT | [33] |
| 28S D1R | AACTCTCTCMTTCARAGTTC | [33] | |
| 28SAF | GACCCGAAAGATGGTGAACTAT | [34] | |
| 28SARC | TTTTGGTAAGCAGAACTGGCGCT | Reverse complement of 28SAR | |
| 28S D6 | CAACTAGCCCTTAAAATGGATGG | [35] | |
| 28S D6FC | CCATCCATTTTAAGGGCTAGTTG | Reverse complement of 28SD6 | |
| 28S D6R | AMAGAAAAGARAACTCTYCC | [35] | |
| 28SBF | GGGAGTTTGACTGGGGCGGTACA | [35] | |
| 28SBR | TGGGTGAACAATCCAACGCTTGG | [35] | |
| 16S rRNA | 16Sar | CGCCTGTTTATCAAAAACAT | [36] |
| 16Sbr | CCGGTCTGAACTCAGATCACGT | [36] | |
| COI | 1490 | GGTCAACAAATCATAAAGATATTGG | [37] |
| 2198 | TAAACTTCAGGGTGACCAAAAAATCA | [37] |
| Data Set Metric | 28S rRNA | 16S rRNA | COI | 28S rRNA + 16S rRNA | 28S rRNA + 16S rRNA + COI |
|---|---|---|---|---|---|
| No. of sequences | 390 | 936 | 996 | 304 | 236 |
| Aligned length | 2603 | 714 | 658 | 3068 | 3793 |
| Parsimony informative | 866 | 519 | 400 | 1212 | 1456 |
| Log likelihood | −52,389.25 | −136,063.82 | −198,409.51 | −98,740.44 | −131,235.3164 |
| Bootstrap replicates (RAxML) | 450 | 450 | 400 | 350 | 350 |
| Motif or Taxon | Number of Species |
| CG | |
| Vetigastropoda | 9 |
| Heterobranchia | 260 |
| Neritimorpha | 2 |
| Peltospiridae | 2 |
| Architaenioglossa | 6 |
| Campaniloidea | 1 |
| Cerithioidea | 6 |
| Hypsogastropoda | |
| Cypraeoidea | 1 |
| Tonnoidea | 2 |
| Stromboidea (1 Xenophoridae) | 8 |
| Calyptraeoidea | 2 |
| Neogastropoda | 50 |
| GC | |
| Abyssochrysoidea | 2 |
| Cingulopsoidea | 1 |
| Epitonioidea | 2 |
| Littorinidae | 8 |
| Hipponicoidea | 1 |
| Naticidae | 3 |
| Pterotracheidae | 1 |
| Rissooidea | 1 |
| Triphoroidea | 3 |
| Truncatelloidea | 6 |
| Vanikoroidea (2 Eulimidae) a | 2 |
| Vermetoidea | 1 |
| Unknown | |
| Latrogastropoda | |
| Capuloidea | Not available |
| Ficoidea | Not available |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colgan, D.J.; Ponder, W.F. Redefining Latrogastropoda Again and Searching for Its Sister Group in Hypsogastropoda (Gastropoda: Caenogastropoda). Diversity 2025, 17, 524. https://doi.org/10.3390/d17080524
Colgan DJ, Ponder WF. Redefining Latrogastropoda Again and Searching for Its Sister Group in Hypsogastropoda (Gastropoda: Caenogastropoda). Diversity. 2025; 17(8):524. https://doi.org/10.3390/d17080524
Chicago/Turabian StyleColgan, Donald J., and Winston F. Ponder. 2025. "Redefining Latrogastropoda Again and Searching for Its Sister Group in Hypsogastropoda (Gastropoda: Caenogastropoda)" Diversity 17, no. 8: 524. https://doi.org/10.3390/d17080524
APA StyleColgan, D. J., & Ponder, W. F. (2025). Redefining Latrogastropoda Again and Searching for Its Sister Group in Hypsogastropoda (Gastropoda: Caenogastropoda). Diversity, 17(8), 524. https://doi.org/10.3390/d17080524






