Phylogenetic Relationships of Dianthus (Caryophyllaceae) Species Found in South Africa
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling and Collection of Plant Material
2.2. Choosing Outgroups
2.3. DNA Extraction, Amplification and Sequence Alignment
2.4. Data Analyses
2.5. Molecular Phylogenetic Analyses
2.6. Distribution Maps Analyses
3. Results
3.1. Statistics of Molecular Data
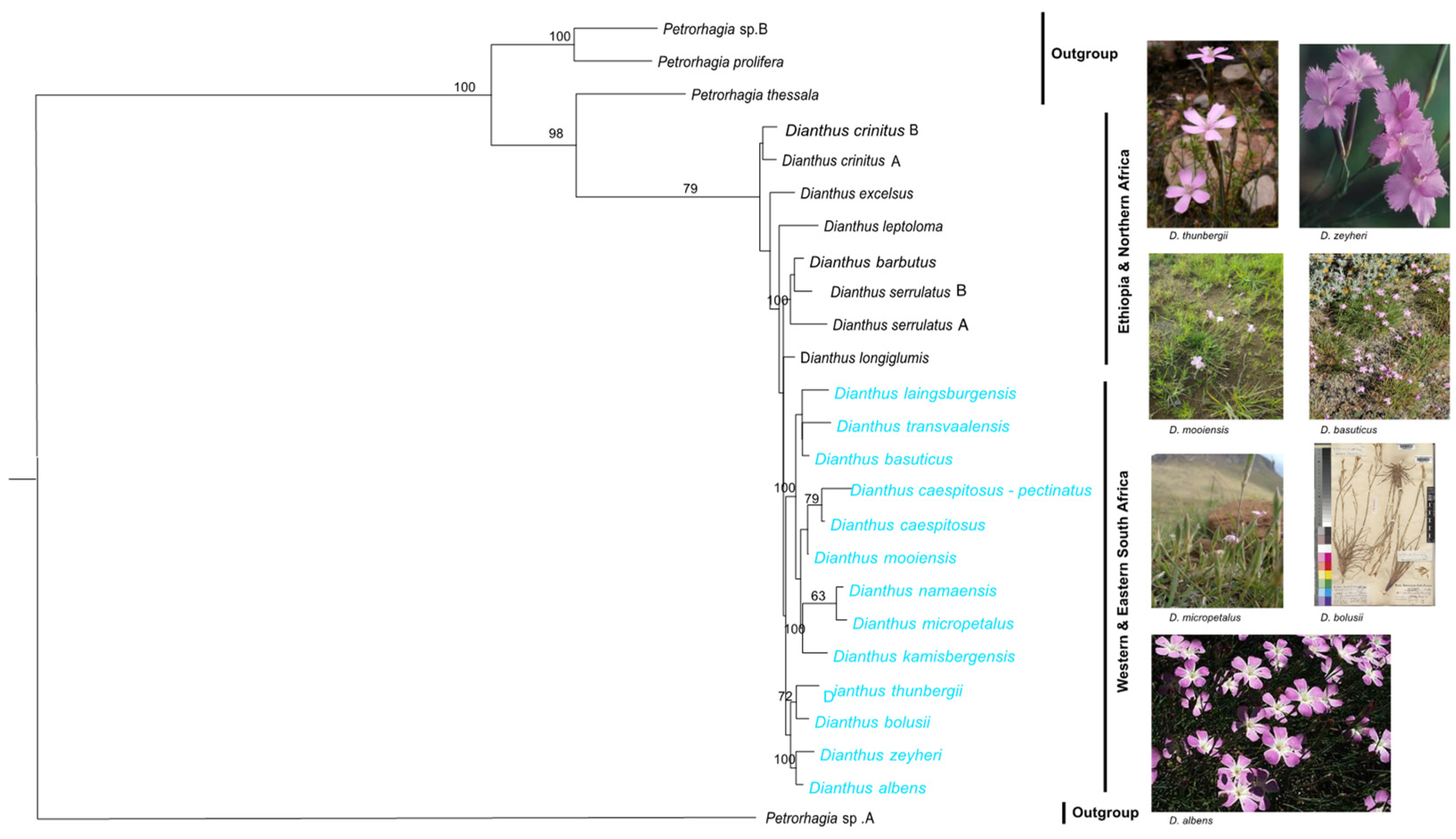
3.2. Phylogenetic Analyses of All the Dianthus Species in This Study
3.3. Phylogenetic Analysis and Relationships Among South African Dianthus
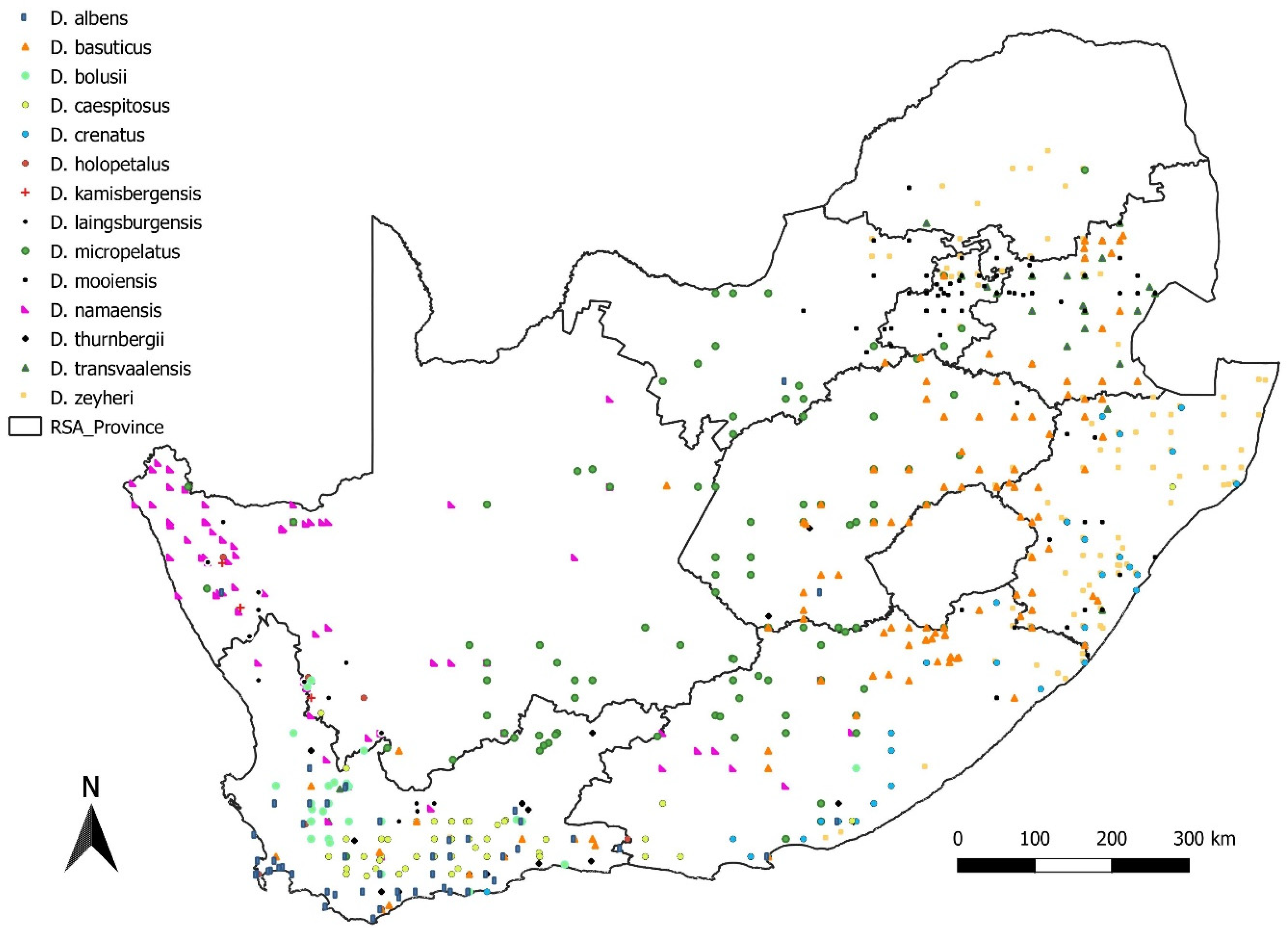
3.4. Dianthus Distribution in South Africa
4. Discussion
4.1. Phylogenetic Incongruence and Topological Conflicts
4.2. Phylogenetic Relationships Within Dianthus
4.3. Geographical Distribution of Dianthus in South Africa
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fassou, G.; Korotkova, N.; Nersesyan, A.; Koch, M.A.; Dimopoulos, P.; Borsch, T. Taxonomy of Dianthus (Caryophyllaceae): Overall phylogenetic relationships and assessment of species diversity based on a first comprehensive checklist of the genus. PhytoKeys 2022, 196, 91–214. [Google Scholar] [CrossRef] [PubMed]
- Pooley, E. A Field Guide to Wildflowers of KwaZulu-Natal and the Eastern Region, 1st ed.; Natal Flora Publications Trust: Durban, South Africa, 1998. [Google Scholar]
- Valente, L.M.; Savolainen, V.; Vargas, P. Unparalleled rates of species diversification in Europe. Proc. R. Soc. B Biol. Sci. 2010, 277, 1489–1497. [Google Scholar] [CrossRef]
- Constantinidis, T. Dianthus haematocalyx subsp. phitosianus (Caryophyllaceae), a New Serpentine Endemic from Greece. Phyton 1999, 39, 277–291. [Google Scholar]
- Germshuizen, Z.; Meyer, N.L.; Steenkamp, Y.; Keith, M. (Eds.) A Checklist of South African Plants. Southern African Botanical Diversity Network; Report No. 41; SABONET, South African National Biodiversity Institute: Pretoria, South Africa, 2006. [Google Scholar]
- Hooper, S.S. Dianthus L. (Caryophyllaceae). Flora Zambesiaca 1961, 1, 355–358. [Google Scholar]
- Hooper, S.S. The genus Dianthus in Central and South Africa. Hooker’s Icones Plantarum. 1959, 37, 1–58. [Google Scholar]
- Goldblatt, P.; Manning, J. Cape Plants: A conspectus of the Cape Flora of South Africa; National Botanical Institute of South Africa: Pretoria, South Africa, 2000. [Google Scholar]
- Govaerts, R. World Checklist of Seed Plants Database in ACCESS D; Missouri Botanical Garden: St. Louis, MO, USA, 2000. [Google Scholar]
- Raimondo, D.; Von Staden, L.; Foden, W.; Victor, J.E.; Helme, N.A.; Turner, R.C.; Kamundi, D.A.; Manama, P.A. (Eds.) Red List of South African Plants; Strelitzia 25; South African National Biodiversity Institute: Pretoria, South Africa, 2009. [Google Scholar]
- Burtt Davy, J. A Revision of the South African Species of Dianthus. Kew Bull. Misc. Inf. 1922, 7, 209–223. [Google Scholar]
- Sonder, O.W. Dianthus L. (Caryophylleae). Flora Capensis 1860, 1, 122–124. [Google Scholar]
- Thiers, B. Index Herbariorium: A Global Directory of Public Herbaria and Associated Staff. New York Botanical Garden’s Virtual Herbarium. 2025. Available online: https://sweetgum.nybg.org/science/ih/ (accessed on 31 January 2025).
- Fior, S.; Karis, P.O.; Casazza, G.; Minuto, L.; Sala, F. Molecular phylogeny of the Caryophyllaceae (Caryophyllales) inferred from chloroplast matK and nuclear rDNA its sequences. Am. J. Bot. 2006, 93, 399–411. [Google Scholar] [CrossRef]
- Palumbi, S.R. Nucleic acids II: The Polymerase Chain Reaction. Molecular Systematics; Hillis, D.M., Moritz, C., Mable, B.K., Eds.; Sinauer Associates: Sunderland, MA, USA, 1996; pp. 205–247. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, D.G.M., Sninsky, J., White, T., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef]
- Wicke, S.; Quandt, D. Universal primers for the amplification of the plastid trnK/matK region in land plants. An. Del Jard. Bot. De Madr. 2009, 66, 285–288. [Google Scholar] [CrossRef]
- Schäferhoff, B.; Müller, K.F.; Borsch, T. Caryophyllales phylogenetics: Disentangling Phytolaccaceae and Molluginaceae and description of Microteaceae as a new isolated family. Willdenowia 2009, 39, 209–228. [Google Scholar] [CrossRef]
- Swofford, D.L.; Sullivan, J. Phylogeny inference based on parsimony and other methods using PAUP. Phylogenetic Handb. A Pract. Approach DNA Protein Phylogeny Cáp 2003, 7, 160–206. [Google Scholar]
- Fitch, W.M. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Biol. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Felsenstein, J. Phylogenies and the comparative method. Am. Nat. 1985, 125, 1–15. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef]
- Sugiura, N. Further analysts of the data by Akaike’s information criterion and the finite corrections: Further analysts of the data by Akaike’s. Commun. Stat. -Theory Methods 1978, 7, 13–26. [Google Scholar] [CrossRef]
- South African National Biodiversity Institute. Botanical Database of Southern Africa (BODATSA). 2016. Available online: https://posa.sanbi.org/sanbi/Explore (accessed on 16 February 2025).
- Wendel, J.F.; Doyle, J.J. Phylogenetic incongruence: Window into genome history and molecular evolution. In Molecular Systematics of Plants II: DNA Sequencing; Soltis, D.E., Soltis, P.S., Doyle, J.J., Eds.; Kluwer Academics Publishers: Norwell, MA, USA, 1998; pp. 265–296. [Google Scholar]
- Álvarez, I.; Wendel, J.F. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenetics Evol. 2003, 29, 417–434. [Google Scholar] [CrossRef]
- Hershkovitz, M.A.; Zimmer, E.A. Ribosomal DNA evidence and disjunctions of western American Portulacaceae. Mol. Phylogenetics Evol. 1999, 15, 419–439. [Google Scholar] [CrossRef]
- Soltis, D.E.; Kuzoff, R.K.; Mort, M.E.; Zanis, M.; Fishbein, M.; Hufford, L.; Koontz, J.; Arroyo, M.K. Elucidating deep-level phylogenetic relationships in Saxifragaceae using sequences for six chloroplast and nuclear DNA regions. Ann. Mo. Bot. Gard. 2001, 88, 669–693. [Google Scholar] [CrossRef]
- Madhani, H.; Rabeler, R.; Pirani, A.; Oxelman, B.; Heubl, G.; Zarre, S. Untangling phylogenetic patterns and taxonomic confusion in tribe Caryophylleae (Caryophyllaceae) with special focus on generic boundaries. Taxon 2018, 67, 83–112. [Google Scholar] [CrossRef]
- Vlok, J.; Schutte-Vlok, A.L. Plants of the Klein Karoo; Umdaus Press: Hatfield, UK, 2010. [Google Scholar]


| Locus | Primer | Reference |
|---|---|---|
| ITS | ||
| ITS5 | GGA AGT AAA AGT CGTAAC AAG G | [16] |
| ITS4 | TCCTCCGCT TAT TGATAT GC | [16] |
| trnH-psbA | ||
| trnH | TGA TCC ACT TGG CTA CCG CC | [17] |
| psbA | GCT AAC CTT GGT ATG GAA GT | [17] |
| trnK-matK | ||
| trnK-F | GGG TTG CTA ACT CAA TGGTAG AG – | [18] |
| CARYmatK1440R | AKC GTA AAT GAG AGG ATT G | [19] |
| trnK-psbA | ||
| trnK | GGG TTG CTA ACT CAA TGGTAG AG – | [17] |
| psbA | GCT AAC CTT GGT ATG GAA GT | [17] |
| Gene Regions | No. of Characters | No. of Taxa | Characters Constant | Parsimony Uninformative | Parsimony Informative | Missing | jMODELTEST AIC | Number of Trees per Run |
|---|---|---|---|---|---|---|---|---|
| ITS | 659 | 95 | 181 | 361 | 117 | >5% | F81+I+G | |
| trnH-psbA | 258 | 93 | 183 | 39 | 36 | >5% | TN93+I+G | |
| trnK-matK | 1376 | 95 | 1252 | 72 | 52 | >5% | GTR+I+G | |
| trnK-psbA | 772 | 91 | 668 | 83 | 21 | >5% | HKY85+I+G | |
| Combined regions | 3065 | 98 | 2284 | 555 | 226 | >5% | - | 20,001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mnxati, S.; Mankga, L. Phylogenetic Relationships of Dianthus (Caryophyllaceae) Species Found in South Africa. Diversity 2025, 17, 202. https://doi.org/10.3390/d17030202
Mnxati S, Mankga L. Phylogenetic Relationships of Dianthus (Caryophyllaceae) Species Found in South Africa. Diversity. 2025; 17(3):202. https://doi.org/10.3390/d17030202
Chicago/Turabian StyleMnxati, Sifiso, and Ledile Mankga. 2025. "Phylogenetic Relationships of Dianthus (Caryophyllaceae) Species Found in South Africa" Diversity 17, no. 3: 202. https://doi.org/10.3390/d17030202
APA StyleMnxati, S., & Mankga, L. (2025). Phylogenetic Relationships of Dianthus (Caryophyllaceae) Species Found in South Africa. Diversity, 17(3), 202. https://doi.org/10.3390/d17030202







