Decoding the Mitogenome of Takydromus intermedius: Insights into the Comparative Mitogenomics and Phylogenetic Relationships of Takydromus Lizards
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Mitogenome Sequencing, Assembly and Annotation
2.2.2. NCBI Database Sequence Acquisition
2.2.3. Analysis of Mitogenomic Characteristics
- (1)
- Repetitive Sequence Analysis
- (2)
- Nucleotide Composition and Divergence Analysis
- (3)
- Protein-Coding Gene Analysis
- (4)
- Genetic Distance Analysis
2.2.4. Molecular Phylogenetic Analysis
3. Results
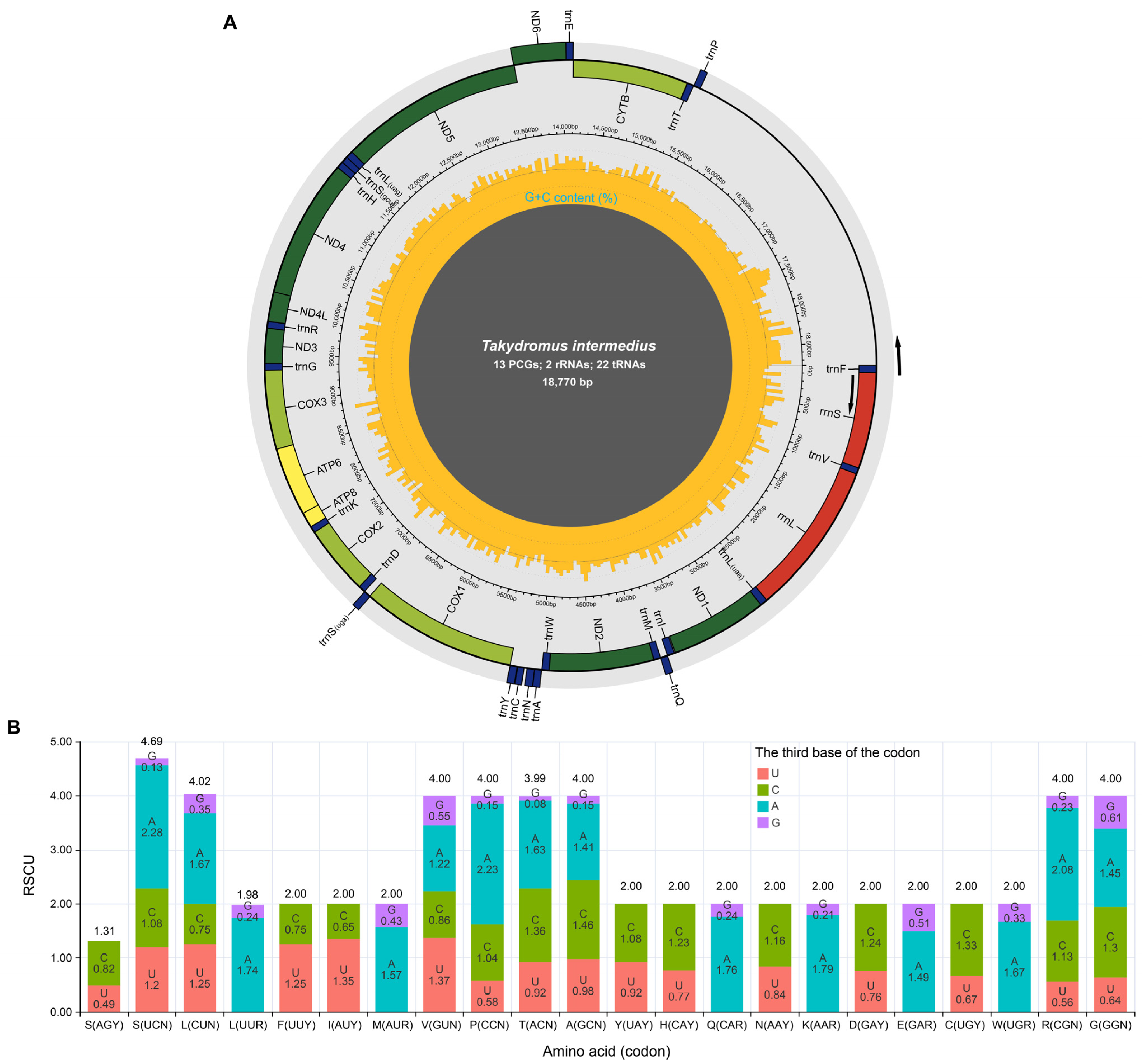
3.1. Characteristics of the Mitogenome of Takydromus intermedius
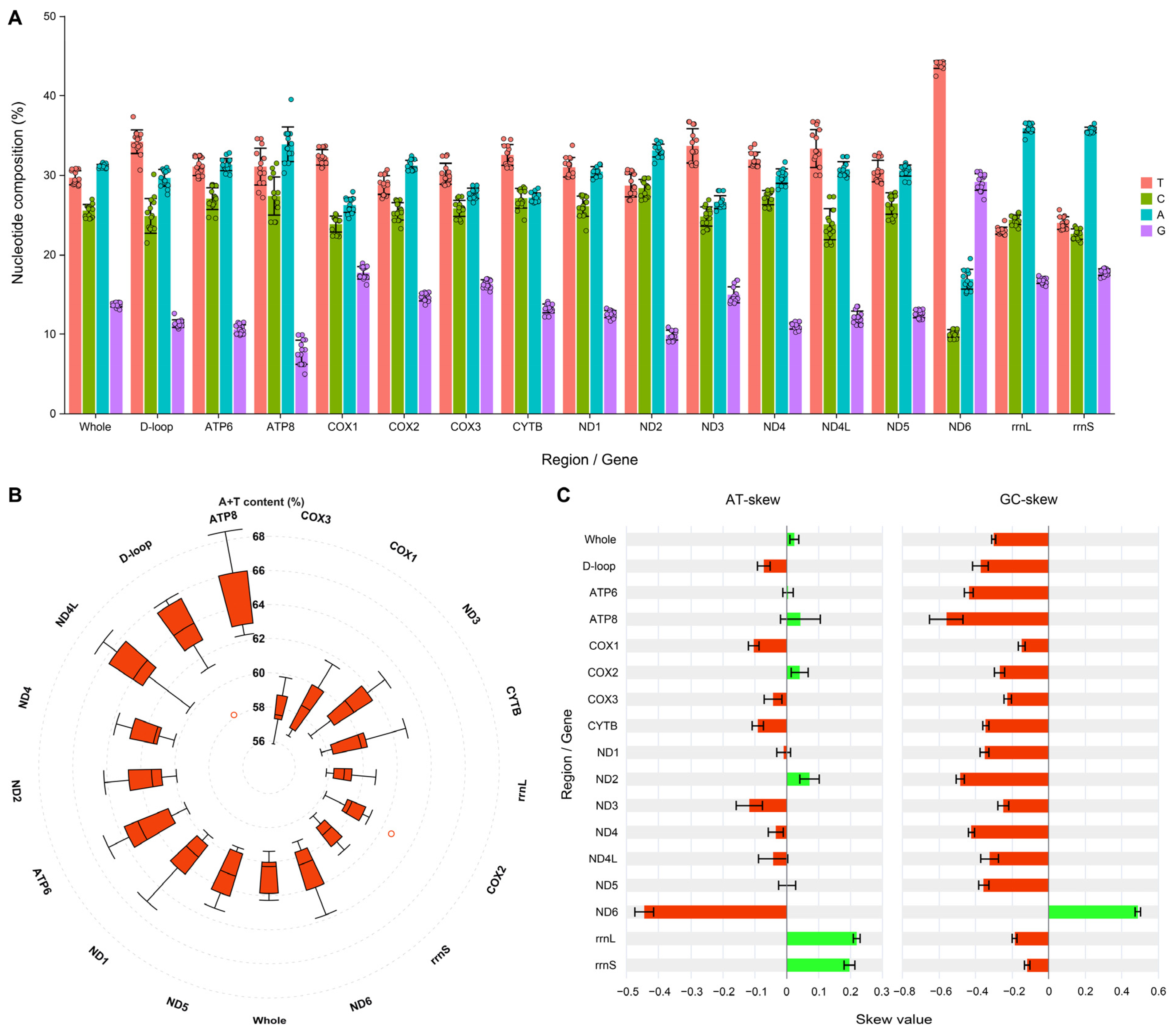
3.2. Nucleotide Composition and Skewness of the Mitogenomes Within the Genus Takydromus
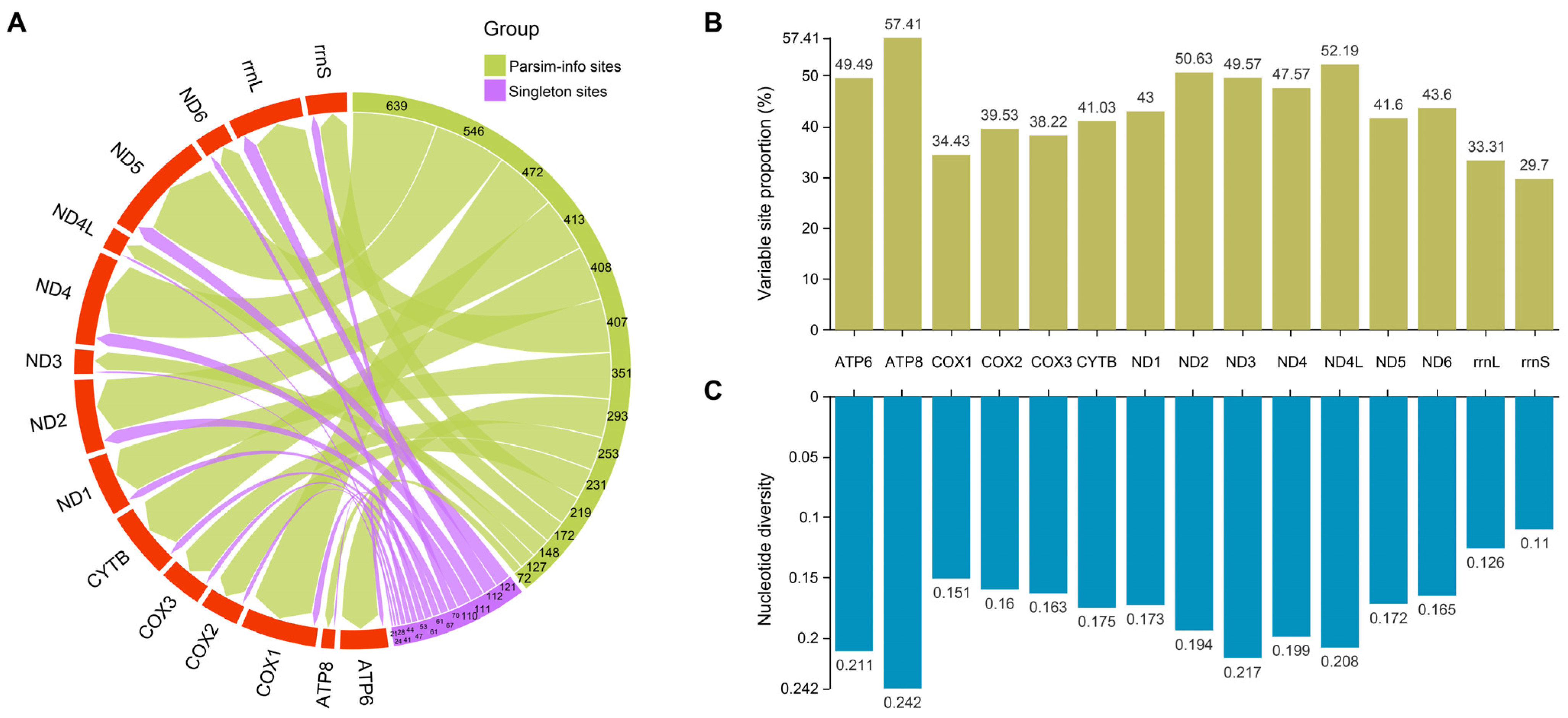
3.3. Nucleotide Variation and Diversity of the Mitogenomes Within the Genus Takydromus
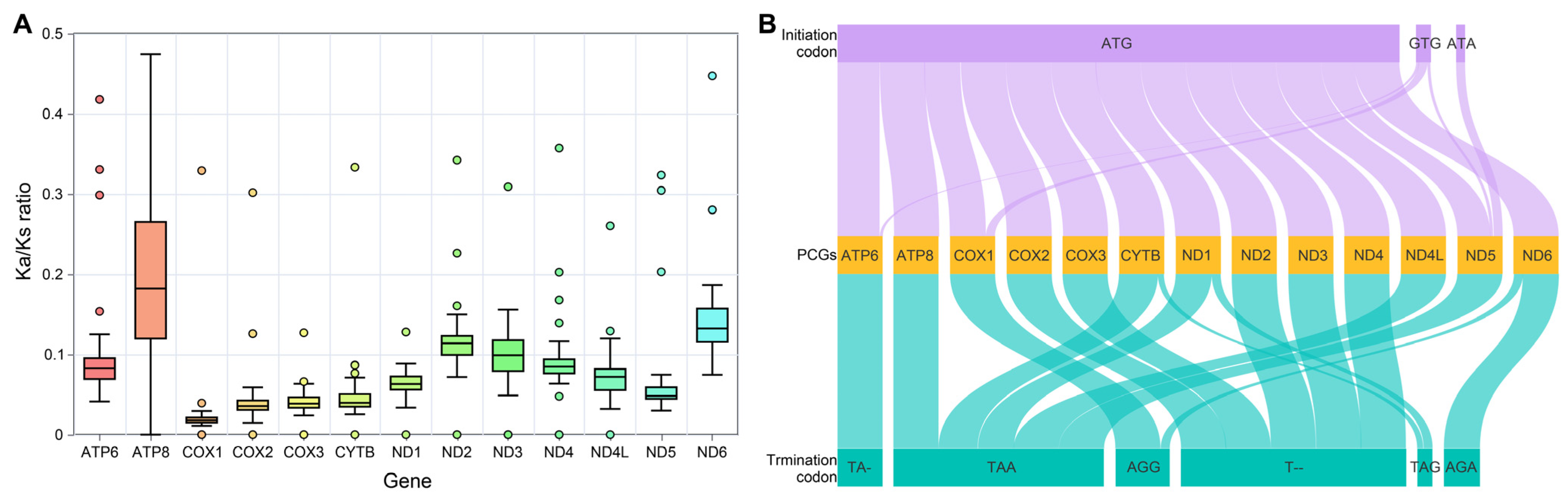
3.4. Evolutionary Pressure and Codon Usage of Mitochondrial Protein-Coding Genes Within the Genus Takydromus
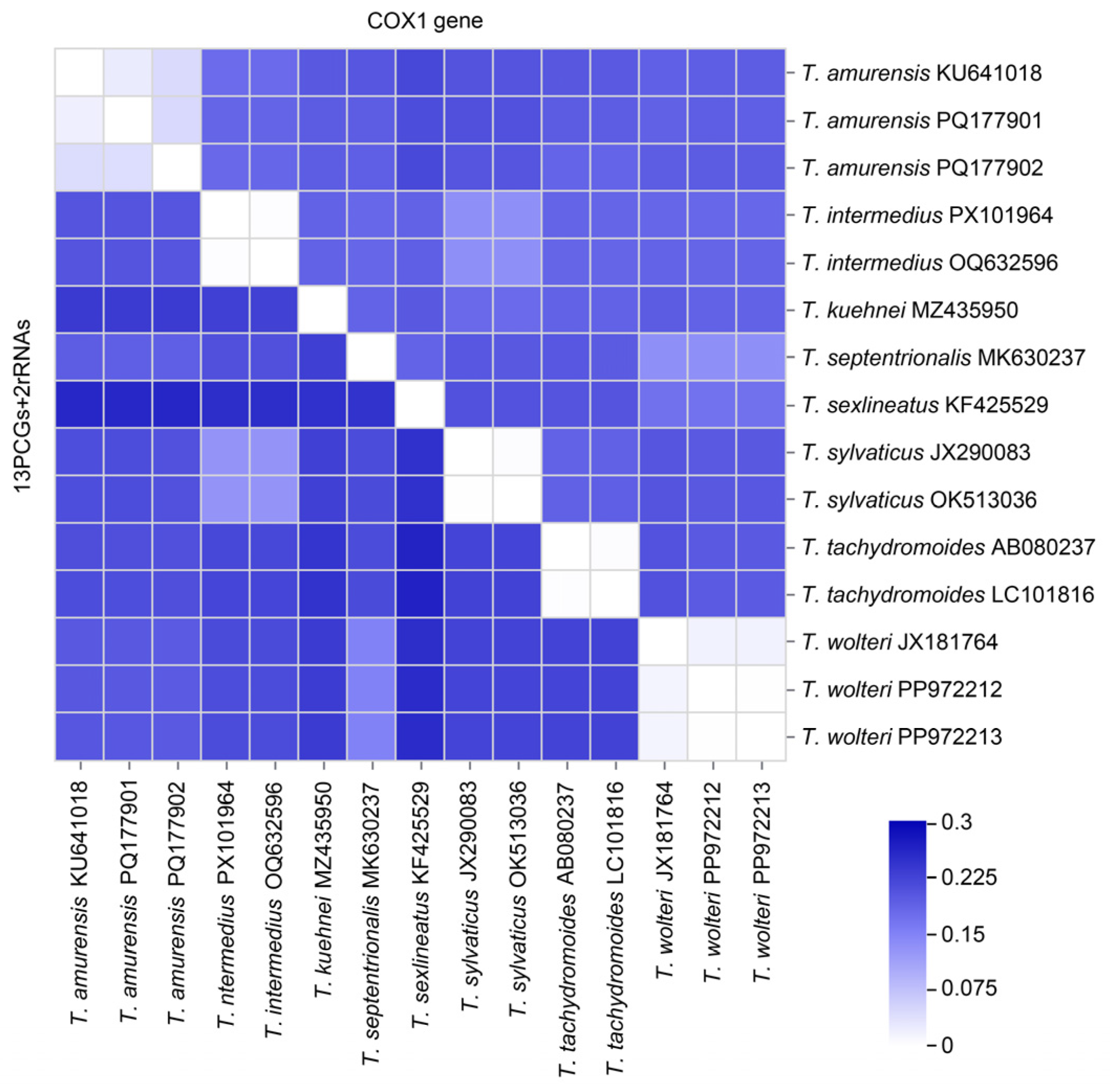
3.5. Interspecific Genetic Distances Within the Genus Takydromus
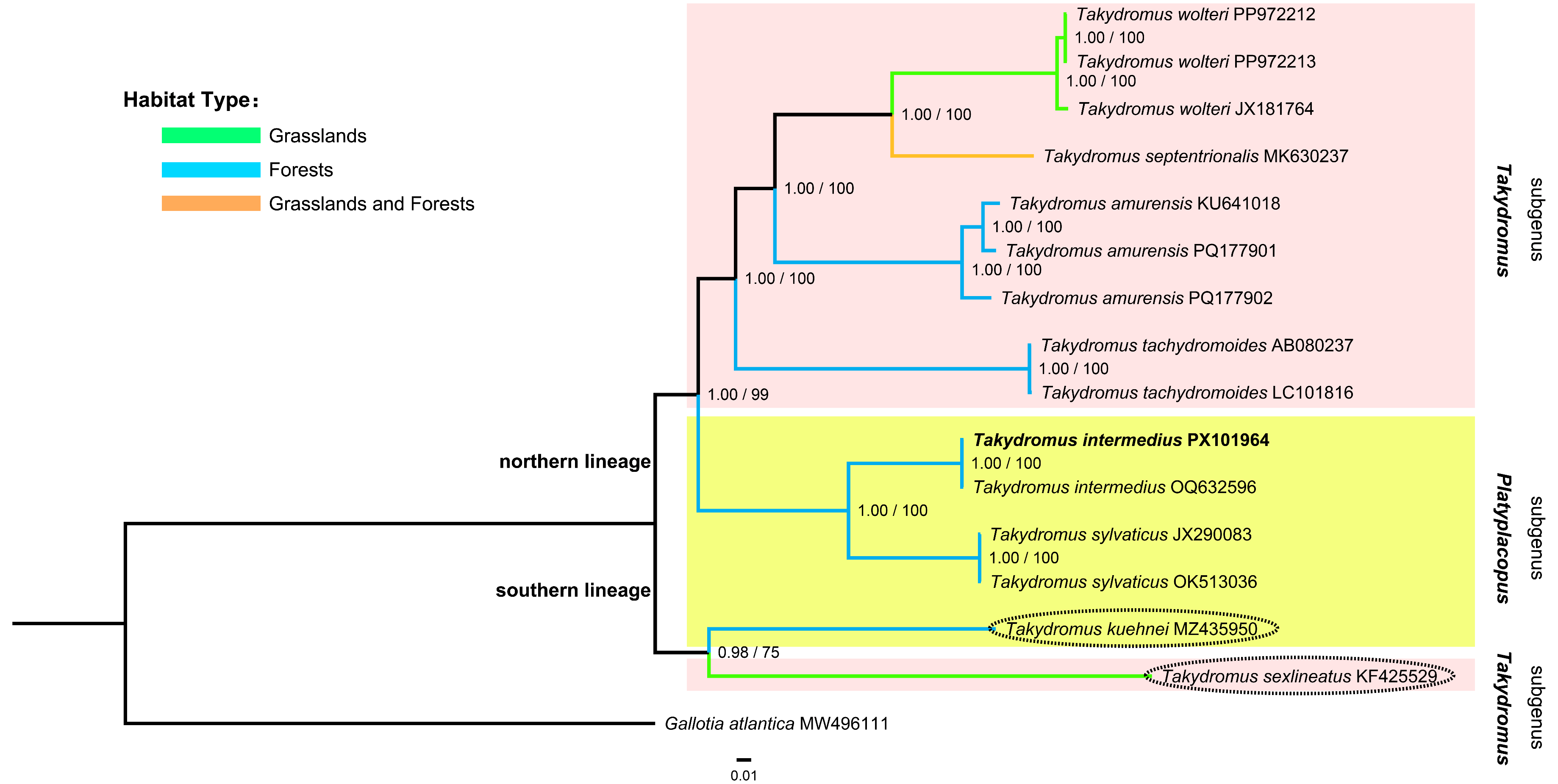
3.6. Phylogenetic Relationships Within the Genus Takydromus
4. Discussion
4.1. Structural and Evolutionary Features of the Takydromus Mitogenome
4.2. Nucleotide Diversity and Selective Pressure of the Takydromus Mitogenome
4.3. Phylogenetic Relationships Among the Takydromus Lizards
4.4. Taxonomic Implications for the Genus Takydromus
4.5. Ecological Implications for the Genus Takydromus
4.6. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ota, H.; Honda, M.; Chen, S.-L.; Hikida, T.; Panha, S.; Oh, H.-S.; Matsui, M. Phylogenetic relationships, taxonomy, character evolution and biogeography of the lacertid lizards of the genus Takydromus (Reptilia: Squamata): A molecular perspective. Biol. J. Linn. Soc. 2002, 76, 493–509. [Google Scholar] [CrossRef][Green Version]
- Arnold, E.N. Interrelationships and evolution of the east Asian grass lizards, Takydromus (Squamata: Lacertidae). Zool. J. Linn. Soc. 1997, 119, 267–296. [Google Scholar] [CrossRef]
- Arnold, E.N.; Arribas, O.; Carranza, S. Systematics of the Palaearctic and Oriental lizard tribe Lacertini (Squamata: Lacertidae: Lacertinae), with descriptions of eight new genera. Zootaxa 2007, 1430, 1–86. [Google Scholar] [CrossRef]
- Uetz, P.; Freed, P.; Aguilar, R.; Reyes, F.; Kudera, J.; Hošek, J. The Reptile Database. Available online: http://www.reptile-database.org (accessed on 20 September 2025).
- Wang, Y.-Y.; Gong, S.-P.; Liu, P.; Wang, X. A new species of the genus Takydromus (Squamata: Lacertidae) from Tianjingshan Forestry Station, northern Guangdong, China. Zootaxa 2017, 4338, 441–458. [Google Scholar] [CrossRef][Green Version]
- Wang, J.; Lyu, Z.-T.; Yang, C.-Y.; Li, Y.-L.; Wang, Y.-Y. A new species of the genus Takydromus (Squamata, Lacertidae) from southwestern Guangdong, China. Zookeys 2019, 871, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Hu, Y.-H.; Chen, J.; Zhong, J.; Ji, X. A new species of the genus Takydromus (Squamata: Lacertidae) from northeastern Guangxi, China. Animals 2024, 14, 1402. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Ji, X.; Wang, Y.; Rao, D.; Huang, S.; Wang, Y.; Song, Z.; Guo, X.; Jiang, J. An annotated list of lizards (Sauria: Squamata) recorded from the People’s Republic of China. Asian Herpetol. Res. 2022, 13, 64–74. [Google Scholar]
- Sheremetyeva, I.N.; Popova, A.A. Evolution of East Asian forest kingdom has increased the number of Asian grass lizard (Takydromus) species. Biol. Bull. 2025, 52, 188. [Google Scholar] [CrossRef]
- Guo, K.; Zhong, J.; Xie, F.; Zhu, L.; Qu, Y.-F.; Ji, X. Climate warming will increase chances of hybridization and introgression between two Takydromus lizards (Lacertidae). Ecol. Evol. 2021, 11, 8573–8584. [Google Scholar] [CrossRef]
- Boulenger, G.A. A revision of the lizards of the genus Tachydromus. Mem. Asiat. Soc. Bengal 1917, 5, 207–235. [Google Scholar]
- Pope, C.H. The Reptiles of China: Turtles, Crocodilians, Snakes, Lizards; American Museum of Natural History: New York, NY, USA, 1935. [Google Scholar]
- Arnold, E.N. Towards a phylogeny and biogeography of the Lacertidae: Relationships within an Old-World family of lizards derived from morphology. Bull. Br. Mus. Nat. Hist. (Zool.) 1989, 55, 209–257. [Google Scholar]
- Lin, S.-M.; Chen, C.A.; Lue, K.-Y. Molecular phylogeny and biogeography of the grass lizards genus Takydromus (Reptilia: Lacertidae) of East Asia. Mol. Phylogenetics Evol. 2002, 22, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Chen, Q. On the taxonomic status of Platyplacous sylvaticus based on 12S rRNA gene. Acta Zootaxonomica Sin. 2006, 31, 475–479. [Google Scholar]
- Chen, Q.-L.; Tang, X.-S.; Yao, W.-J.; Lu, S.-Q. Bioinformatics analysis the complete sequences of cytochrome b of Takydromus sylvaticus and modeling the tertiary structure of encoded protein. Int. J. Biol. Sci. 2009, 5, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yu, X.; Bi, W.; Li, Z.; Zhou, Y.; Ma, R.; Feng, F.; Huang, C.; Gu, J.; Wu, W.; et al. Mitogenome diversity and phylogeny of Felidae species. Diversity 2025, 17, 634. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Pett, W. Animal mitochondrial DNA as we do not know it: Mt-genome organization and evolution in nonbilaterian lineages. Genome Biol. Evol. 2016, 8, 2896–2913. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, J.; Luo, H.-Y.; Chen, X.; Zhong, J.; Ji, X. Climate-driven mitochondrial selection in lacertid lizards. Ecol. Evol. 2024, 14, e11176. [Google Scholar] [CrossRef]
- Zhan, L.; Chen, Y.; He, J.; Guo, Z.; Wu, L.; Storey, K.B.; Zhang, J.; Yu, D. The phylogenetic relationships of major lizard families using mitochondrial genomes and selection pressure analyses in Anguimorpha. Int. J. Mol. Sci. 2024, 25, 8464. [Google Scholar] [CrossRef]
- Cai, Y.; Yan, J.; Xu, X.-F.; Lin, Z.-H.; Ji, X. Mitochondrial DNA phylogeography reveals a west–east division of the northern grass lizard (Takydromus septentrionalis) endemic to China. J. Zool. Syst. Evol. Res. 2012, 50, 137–144. [Google Scholar] [CrossRef]
- Sheremetyeva, I.N.; Popova, A.A.; Gorobeyko, U.V.; Maslova, I.V. Genetic variability of the mtDNA cytochrome b gene of the Amur grass lizard Takydromus amurensis Peters, 1881 in the south of the Russian Far East. Russ. J. Genet. 2025, 61, 966–973. [Google Scholar] [CrossRef]
- Liu, J.; Yu, J.; Zhou, M.; Yang, J. Complete mitochondrial genome of Japalura flaviceps: Deep insights into the phylogeny and gene rearrangements of Agamidae species. Int. J. Biol. Macromol. 2019, 125, 423–431. [Google Scholar] [CrossRef]
- Lan, G.; Yu, J.; Liu, J.; Zhang, Y.; Ma, R.; Zhou, Y.; Zhu, B.; Wei, W.; Liu, J.; Qi, G. Complete mitochondrial genome and phylogenetic analysis of Tarsiger indicus (Aves: Passeriformes: Muscicapidae). Genes 2024, 15, 90. [Google Scholar] [CrossRef] [PubMed]
- Kumazawa, Y. Mitochondrial genomes from major lizard families suggest their phylogenetic relationships and ancient radiations. Gene 2007, 388, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.-N.; Ji, X. The complete mitochondrial genome of Takydromus wolteri (Squamata: Lacertidae). Mitochondrial DNA 2013, 24, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Chen, J.; Huang, S. Mitochondrial genome of the Chung-an ground lizard Takydromus sylvaticus (Reptilia: Lacertidae). Mitochondrial DNA 2014, 25, 319–320. [Google Scholar] [CrossRef]
- Qin, P.; Zeng, D.; Hou, L.; Yang, X.; Qin, X. Complete mitochondrial genome of Takydromus sexlineatus (Squamata, Lacertidae). Mitochondrial DNA 2015, 26, 465–466. [Google Scholar] [CrossRef]
- Ma, W.; Liu, H.; Zhao, W.; Liu, P. The complete mitochondrial genome of Takydromus amurensis (Squamata: Lacertidae). Mitochondrial DNA Part B 2016, 1, 214–215. [Google Scholar] [CrossRef]
- Sun, Y.; Kurisaki, M.; Hashiguchi, Y.; Kumazawa, Y. Variation and evolution of polyadenylation profiles in sauropsid mitochondrial mRNAs as deduced from the high-throughput RNA sequencing. BMC Genom. 2017, 18, 665. [Google Scholar] [CrossRef]
- Hu, J.; Peng, L.; Tang, X.; Huang, S. The complete mitochondrial genome of Takydromus septentrionalis (Reptilia: Lacertidae). Mitochondrial DNA Part B 2019, 4, 2193–2194. [Google Scholar] [CrossRef]
- Wu, L.-X.; Luo, K.-N.; Ding, G.-H. Complete mitochondrial genome of Takydromus kuehnei (Squamata: Takydromus) and its phylogenetic analysis. Mitochondrial DNA Part B 2022, 7, 764–765. [Google Scholar] [CrossRef]
- Kirchhof, S.; Lyra, M.L.; Rodríguez, A.; Ineich, I.; Müller, J.; Rödel, M.-O.; Trape, J.-F.; Vences, M.; Boissinot, S. Mitogenome analyses elucidate the evolutionary relationships of a probable Eocene wet tropics relic in the xerophilic lizard genus Acanthodactylus. Sci. Rep. 2021, 11, 4858. [Google Scholar] [CrossRef]
- Liu, J.; Yu, J.; Yu, X.; Bi, W.; Yang, H.; Xue, F.; Zhang, G.; Zhang, J.; Yi, D.; Ma, R.; et al. Complete mitogenomes of ticks Ixodes acutitarsus and Ixodes ovatus parasitizing giant panda: Deep insights into the comparative mitogenomic and phylogenetic relationship of Ixodidae species. Genes 2022, 13, 2049. [Google Scholar] [CrossRef]
- Zhu, T.; Sato, Y.; Sado, T.; Miya, M.; Iwasaki, W. MitoFish, MitoAnnotator, and MiFish Pipeline: Updates in 10 Years. Mol. Biol. Evol. 2023, 40, msad035. [Google Scholar] [CrossRef] [PubMed]
- Donath, A.; Jühling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Poczai, P.; Hyvönen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An online program for the versatile plotting of organelle genomes. Front. Genet. 2020, 11, 576124. [Google Scholar] [CrossRef]
- Xiang, C.; Gao, F.; Jakovlić, I.; Lei, H.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef]
- Du, L.; Zhang, C.; Liu, Q.; Zhang, X.; Yue, B. Krait: An ultrafast tool for genome-wide survey of microsatellites and primer design. Bioinformatics 2018, 34, 681–683. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Ranwez, V.; Douzery, E.J.P.; Cambon, C.; Chantret, N.; Delsuc, F. MACSE v2: Toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol. Biol. Evol. 2018, 35, 2582–2584. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Braband, A.; Schierwater, B.; Stadler, P.F. Genetic aspects of mitochondrial genome evolution. Mol. Phylogenetics Evol. 2013, 69, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Kumazawa, Y.; Miura, S.; Yamada, C.; Hashiguchi, Y. Gene rearrangements in gekkonid mitochondrial genomes with shuffling, loss, and reassignment of tRNA genes. BMC Genom. 2014, 15, 930. [Google Scholar] [CrossRef]
- Lü, Z.; Zhu, K.; Jiang, H.; Lu, X.; Liu, B.; Ye, Y.; Jiang, L.; Liu, L.; Gong, L. Complete mitochondrial genome of Ophichthus brevicaudatus reveals novel gene order and phylogenetic relationships of Anguilliformes. Int. J. Biol. Macromol. 2019, 135, 609–618. [Google Scholar] [CrossRef]
- Podnar, M.; Pinsker, W.; Mayer, W. Complete mitochondrial genomes of three lizard species and the systematic position of the Lacertidae (Squamata). J. Zool. Syst. Evol. Res. 2009, 47, 35–41. [Google Scholar] [CrossRef]
- Murtskhvaladze, M.; Tarkhnishvili, D.; Anderson, C.L.; Kotorashvili, A. Phylogeny of caucasian rock lizards (Darevskia) and other true lizards based on mitogenome analysis: Optimisation of the algorithms and gene selection. PLoS ONE 2020, 15, e0233680. [Google Scholar] [CrossRef]
- Omote, K.; Nishida, C.; Dick, M.H.; Masuda, R. Limited phylogenetic distribution of a long tandem-repeat cluster in the mitochondrial control region in Bubo (Aves, Strigidae) and cluster variation in Blakiston’s fish owl (Bubo blakistoni). Mol. Phylogenetics Evol. 2013, 66, 889–897. [Google Scholar] [CrossRef]
- Komissarov, A.; Korchagin, V.; Kliver, S.; Dobrynin, P.; Semyenova, S.; Vergun, A.; O’Brien, S.; Ryskov, A. The complete mitochondrial genome of the parthenogenetic Caucasian rock lizard Darevskia unisexualis (Squamata: Lacertidae) contains long tandem repeat formed by 59 bp monomer. Mitochondrial DNA Part B 2016, 1, 875–877. [Google Scholar] [CrossRef]
- Huang, A.; Liu, S.; Li, H.; Luo, H.; Ni, Q.; Yao, Y.; Xu, H.; Zeng, B.; Li, Y.; Wei, Z.; et al. The revised complete mitogenome sequence of the tree frog Polypedates megacephalus (Anura, Rhacophoridae) by next-generation sequencing and phylogenetic analysis. PeerJ 2019, 7, e7415. [Google Scholar] [CrossRef]
- Tensen, L.; Jansen van Vuuren, B.; Groom, R.; du Plessis, C.; Fischer, K. Little variation, lots of repetition: Mitogenomic diversity in African wild dogs. Mitochondrial DNA Part A 2025, 35, 197–206. [Google Scholar] [CrossRef]
- Juen, L.; Koroiva, R.; Geraldo de Carvalho, F.; Mendoza-Penagos, C.C.; Brito, J.D.; Calvão, L.B.; Ferreira, V.R.; Ribeiro-dos-Santos, Â.; Silva, C.S.; Guerreiro, S.; et al. The first mitochondrial genome of an Odonata endemic to South America, Chalcopteryx rutilans (Rambur, 1842) (Odonata: Polythoridae), and its implications for the phylogeny of the Zygoptera. Diversity 2023, 15, 908. [Google Scholar] [CrossRef]
- Zhou, Q.; Xiang, H.-M.; Zhang, M.-Y.; Liu, Y.; Gu, Z.-R.; Lan, X.-Y.; Wang, J.-X.; Jiang, W.-S. Two complete mitochondrial genomes of Leptobrachium (Anura: Megophryidae: Leptobrachiinae): Characteristics, population divergences, and phylogenetic implications. Genes 2023, 14, 768. [Google Scholar] [CrossRef]
- Hu, Y.-J.; Jia, F.-F.; Hu, L.; Wu, C.; Tian, T.; Li, T.-J.; Chen, B. Comparative mitogenome research revealed the phylogenetics and evolution of the superfamily Tenebrionoidea (Coleoptera: Polyphage). Ecol. Evol. 2024, 14, e11520. [Google Scholar] [CrossRef]
| Species | GenBank Accession No. | Locality | Mitogenome Size (bp) | References |
|---|---|---|---|---|
| Takydromus amurensis Peters, 1881 | KU641018/NC_030209 | Changbai Mountai, Jilin, China | 17,333 | [29] |
| PQ177901 | South Korea | 17,334 | Direct submission | |
| PQ177902 | South Korea | 17,327 | Direct submission | |
| Takydromus intermedius Stejneger, 1924 | OQ632596/NC_077637 | Bashu, Chongqing, China | 17,713 | [19] |
| PX101964 | Dujiangyan, Sichuan, China | 18,770 | This study | |
| Takydromus kuehnei Van Denbugeh, 1909 | MZ435950 | Quzhou, Zhejiang, China | 17,224 | [32] |
| Takydromus septentrionalis Günther, 1864 | MK630237 | Huangshan, Anhui, China | 18,304 | [31] |
| Takydromus sexlineatus Daudin, 1802 | KF425529/NC_022703 | Guangxi, China | 18,943 | [28] |
| Takydromus sylvaticus (Pope, 1928) | JX290083 | Chung-an, Fujian, China | 17,838 | [27] |
| OK513036/NC_067055 | Suichang, Zhejiang, China | 17,518 | Direct submission | |
| Takydromus tachydromoides (Schlegel, 1838) | AB080237 | Nagoya University campus, Japan | 18,245 | [25] |
| LC101816 | Unknown | 17,923 | [30] | |
| Takydromus wolteri Fischer, 1885 | JX181764/NC_018777 | China | 18,236 | [26] |
| PP972212 | South Korea | 18,237 | Direct submission | |
| PP972213 | South Korea | 18,242 | Direct submission | |
| Gallotia atlantica (Peters & Doria, 1882) | MW496111/NC_059771 | 15,552 | [33] |
| Species | Accession ID | ATP6 | ATP8 | COX1 | COX2 | COX3 | CYTB | NAD1 |
|---|---|---|---|---|---|---|---|---|
| T. amurensis | KU641018 | ATG/TA- 1 | ATG/TAA | ATG/AGG | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA |
| PQ177901 | ATG/TA- 1 | ATG/TAA | ATG/AGG | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | |
| PQ177902 | ATG/TA- 1 | ATG/TAA | ATG/AGG | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | |
| T. intermedius | OQ632596 | ATG/TA- 1 | ATG/TAA | ATG/AGG 2 | ATG/T-- | ATG/T-- | ATG/TAG | ATG/TAA |
| PX101964 | ATG/TA- | ATG/TAA | ATG/AGG | ATG/T-- | ATG/T-- | ATG/TAG | ATG/TAA | |
| T. kuehnei | MZ435950 | GTG/TA- 1 | ATG/TAA | ATG/AGG | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA |
| T. septentrionalis | MK630237 | ATG/TA- 1 | ATG/TAA | ATG/AGG | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA |
| T. sexlineatus | KF425529 | ATG/TA- | ATG/TAA | GTG/AGG | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAG |
| T. sylvaticus | JX290083 | ATG/TA- 1 | ATG/TAA | ATG/AGG | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAG |
| OK513036 | ATG/TA- 1 | ATG/TAA | ATG/AGG | ATG/T-- | ATG/T-- 4 | ATG/TAA | ATG/TAG | |
| T. tachydromoides | AB080237 | ATG/TA- | ATG/TAA | GTG/AGG | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA |
| LC101816 | ATG/TA- | ATG/TAA | GTG/AGG | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | |
| T. wolteri | JX181764 | ATG/TA- 1 | ATG/TAA | ATG/AGG | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA |
| PP972212 | ATG/TA- 1 | ATG/TAA | ATG/AGG | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | |
| PP972213 | ATG/TA- 1 | ATG/TAA | ATG/AGG | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | |
| Species | Accession ID | NAD2 | NAD3 | NAD4 | NAD4L | NAD5 | NAD6 | |
| T. amurensis | KU641018 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | ATG/AGA | |
| PQ177901 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | ATG/AGA | ||
| PQ177902 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | ATG/AGA | ||
| T. intermedius | OQ632596 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | ATG/AGA | |
| PX101964 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | ATG/AGA | ||
| T. kuehnei | MZ435950 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | ATG/AGG | |
| T. septentrionalis | MK630237 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | ATG/AGA | |
| T. sexlineatus | KF425529 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | ATG/AGA | |
| T. sylvaticus | JX290083 | ATG/T-- | ATG/T-- 3 | ATG/T-- | ATG/TAA | ATA/TAA | ATG/AGA | |
| OK513036 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | GTG/TAA | ATG/AGA | ||
| T. tachydromoides | AB080237 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATA/TAA | ATG/AGG | |
| LC101816 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATA/TAA | ATG/AGG | ||
| T. wolteri | JX181764 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | ATG/AGA | |
| PP972212 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | ATG/AGA | ||
| PP972213 | ATG/T-- | ATG/T-- | ATG/T-- | ATG/TAA | ATG/TAA | ATG/AGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Chen, M.; Shang, B.; Yi, D.; Wu, W.; Liu, J. Decoding the Mitogenome of Takydromus intermedius: Insights into the Comparative Mitogenomics and Phylogenetic Relationships of Takydromus Lizards. Diversity 2025, 17, 813. https://doi.org/10.3390/d17120813
Yu J, Chen M, Shang B, Yi D, Wu W, Liu J. Decoding the Mitogenome of Takydromus intermedius: Insights into the Comparative Mitogenomics and Phylogenetic Relationships of Takydromus Lizards. Diversity. 2025; 17(12):813. https://doi.org/10.3390/d17120813
Chicago/Turabian StyleYu, Jiaojiao, Minghua Chen, Buqie Shang, Dejiao Yi, Wei Wu, and Jiabin Liu. 2025. "Decoding the Mitogenome of Takydromus intermedius: Insights into the Comparative Mitogenomics and Phylogenetic Relationships of Takydromus Lizards" Diversity 17, no. 12: 813. https://doi.org/10.3390/d17120813
APA StyleYu, J., Chen, M., Shang, B., Yi, D., Wu, W., & Liu, J. (2025). Decoding the Mitogenome of Takydromus intermedius: Insights into the Comparative Mitogenomics and Phylogenetic Relationships of Takydromus Lizards. Diversity, 17(12), 813. https://doi.org/10.3390/d17120813







