Comparison of Gut Bacterial Communities in the Freshwater Mussel Sinanodonta woodiana at Different Life Stages
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Genomic DNA Extraction, PCR Amplification, and Sequencing
2.3. Data Processing
2.4. Statistical Analysis
3. Results
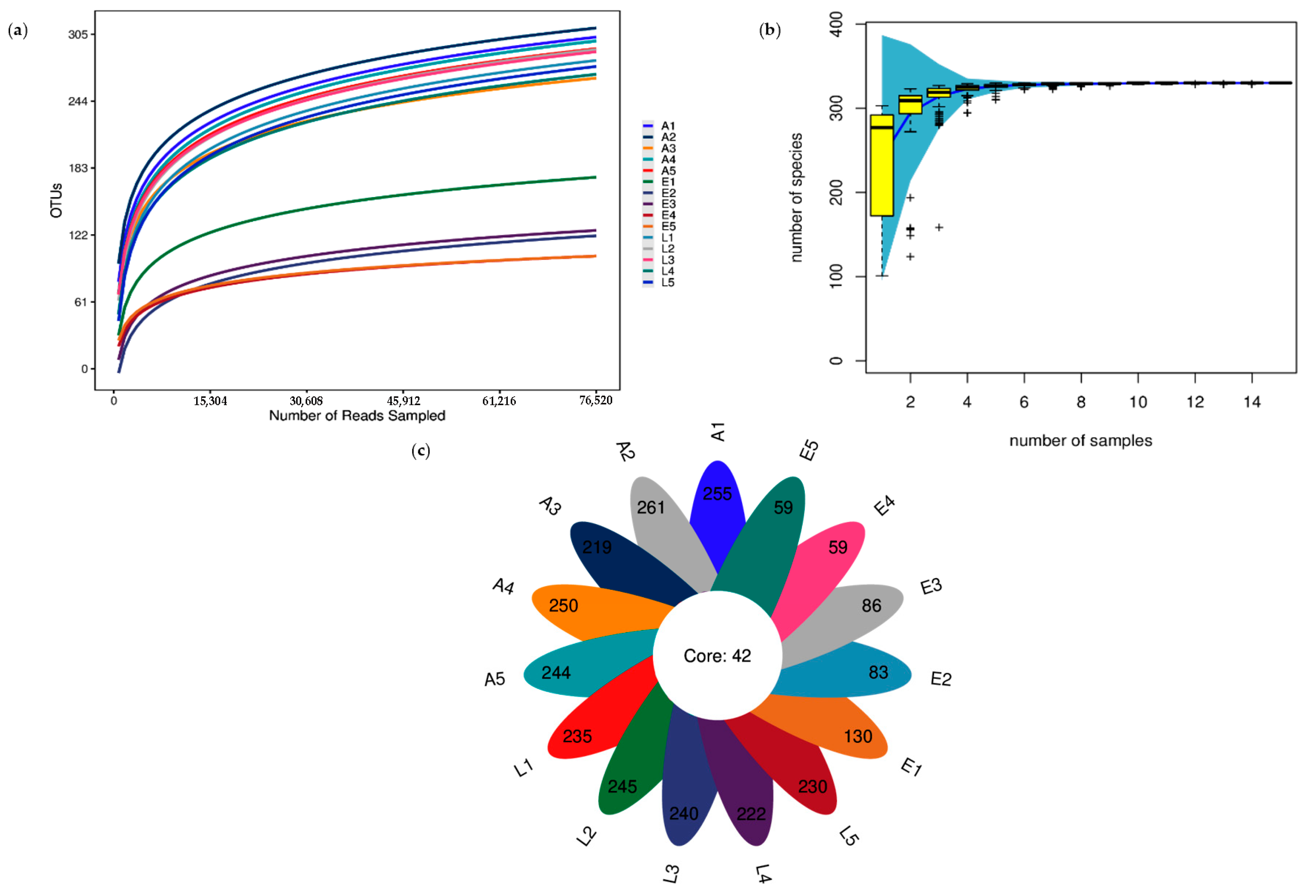
3.1. OTU Analysis and Alpha Diversity
3.2. Beta Diversity
3.3. Taxonomic Composition
3.4. Functional Prediction
4. Discussion
4.1. Diversity of Gut Bacterial Communities
4.2. Composition of Gut Bacterial Communities
4.3. Physiological Functions of Gut Bacterial Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Douda, K.; Zieritz, A.; Vodáková, B.; Urbanska, M.; Bolotov, I.N.; Marková, J.; Froufe, E.; Bogan, A.E.; Lopes-Lima, M. Review of the globally invasive freshwater mussels in the genus Sinanodonta modell, 1945. Hydrobiologia 2025, 852, 1243–1273. [Google Scholar] [CrossRef]
- Chen, X.B.; Jiang, T.; Xue, J.R.; Gu, M.Y.; Wang, M.Y.; Liu, K. Chromosome-level genome assembly of the freshwater bivalve Anodonta woodiana. Sci. Data 2025, 12, 731–740. [Google Scholar] [CrossRef]
- Zhou, C.Q.; Huang, J.C.; Liu, F.; He, S.B.; Zhou, W.L. Removal of selenium containing algae by the bivalve Sinanodonta woodiana and the potential risk to human health. Environ. Pollut. 2018, 242, 73–81. [Google Scholar] [CrossRef]
- Ningrum, R.A.; Adam, M.A.; Diniarti, N.; Anjani, Y.D.; Maulana, I. Strategies for effective plankton management in Kijing Taiwanese (Anodonta woodiana, Lea): A study on stocking density and aquaculture techniques. Indones. J. Limnol. 2024, 5, 1–10. [Google Scholar] [CrossRef]
- Yang, J.; Harino, H.; Liu, H.B.; Miyazaki, N. Monitoring the organotin contamination in the Taihu Lake of China by bivalve mussel Anodonta woodiana. Bull. Environ. Contam. Toxicol. 2008, 81, 164–168. [Google Scholar] [CrossRef]
- Konieczny, P.; Tomaszewska-Gras, J.; Andrzejewski, W.; Mikolajczak, B.; Urbanska, M.; Mazurkiewicz, J.; Stangierski, J. DSC and electrophoretic studies on protein denaturation of Anodonta woodiana (Lea, 1834). J. Therm. Anal. Calorim. 2016, 126, 69–75. [Google Scholar] [CrossRef]
- Rahayu, S.Y.S.; Solihin, D.D.; Manalu, W.; Rahayu, S. Nucleus pearl coating process of freshwater mussel Anodonta woodiana (Unionidae). Hayati J. Biosci. 2013, 20, 24–30. [Google Scholar] [CrossRef]
- Liu, J.; Gu, B.; Bian, J.; Hu, S.G.; Cheng, X.; Ke, Q.K.; Yan, H.F. Antitumor activities of liposome-incorporated aqueous extracts of Anodonta woodiana (Lea, 1834). Eur. Food Res. Technol. 2008, 227, 919–924. [Google Scholar] [CrossRef]
- Chen, X.B.; Liu, H.B.; Su, Y.P.; Yang, J. Morphological development and growth of the freshwater mussel Anodonta woodiana from early juvenile to adult. Invertebr. Reprod. Dev. 2015, 59, 131–140. [Google Scholar] [CrossRef]
- Douda, K.; Vrtílek, M.; Slavík, O.; Reichard, M. The role of host specificity in explaining the invasion success of the freshwater mussel Anodonta woodiana in Europe. Biol. Invasions 2012, 14, 127–137. [Google Scholar] [CrossRef]
- Su, Y.P.; Chen, X.B.; Liu, H.B.; Wu, X.G.; Yang, J. Characteristics of dietary algae selection for Anodonta woodiana at different growth stages from the aquaculture pond. Acta Hydrobiol. Sin. 2016, 40, 634–640. [Google Scholar]
- Christian, A.D.; Smith, B.N.; Berg, D.J.; Smoot, J.C.; Findlay, R.H. Trophic position and potential food sources of 2 species of unionid bivalves (Mollusca: Unionidae) in 2 small Ohio streams. J. N. Am. Benthol. Soc. 2004, 23, 101–113. [Google Scholar]
- Liu, X.J.; Cao, Y.L.; Ouyang, S.; Wu, X.P. Comparative analysis of gut microbiota diversity in endangered, economical, and common freshwater mussels using 16S rRNA gene sequencing. Ecol. Evol. 2020, 10, 12015–12023. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.P.; Learman, D.R.; Annis, M.L.; Woolnough, D.A. Freshwater mussels and host fish gut microbe community composition shifts after agricultural contaminant exposure. J. Appl. Microbiol. 2022, 133, 3645–3658. [Google Scholar] [CrossRef]
- Li, S.; Young, T.; Archer, S.; Lee, K.; Alfaro, A.C. Gut microbiome resilience of green-lipped mussels, Perna canaliculus, to starvation. Int. Microbiol. 2024, 27, 571–580. [Google Scholar] [CrossRef]
- Chen, X.; Sun, C.F.; Dong, J.J.; Li, W.H.; Tian, Y.Y.; Hu, J.; Ye, X. Comparative analysis of the gut microbiota of mandarin fish (Siniperca chuatsi) feeding on compound diets and live baits. Front. Genet. 2022, 13, 797420. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, X.Y.; Liu, M.Y.; Liu, W.X.; Xu, J.Z.; Li, Y.W. Comparative evaluation of 16S rRNA primer pairs in identifying nitrifying guilds in soils under long-term organic fertilization and water management. Front. Microbiol. 2024, 15, 1424795. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. Uchime improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Prodan, A.; Tremaroli, V.; Brolin, H.; Zwinderman, A.H.; Nieuwdorp, M.; Levin, E. Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing. PLoS ONE 2020, 15, e0227434. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The silva ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Li, X.Q.; Wu, P.; Zeng, X.J.; Lang, Q.L.; Lin, Y.; Huang, H.; Qian, P.X. Protocol for correlation analysis of the murine gut microbiome and meta-metabolome using 16S rDNA sequencing and UPLC-MS. Star Protoc. 2022, 3, 101494. [Google Scholar] [CrossRef]
- Amato, K.R.; Yeoman, C.J.; Kent, A.; Righini, N.; Carbonero, F.; Estrada, A.; Gaskins, H.R.; Stumpf, R.M.; Yildirim, S.; Torralba, M.; et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013, 7, 1344–1353. [Google Scholar] [CrossRef]
- Thormar, E.A.; Hansen, S.B.; Jorgensen, L.V.; Limborg, M.T. Sampling fish gut microbiota—A genome-resolved metagenomic approach. Ecol. Evol. 2024, 14, e70302. [Google Scholar] [CrossRef] [PubMed]
- Maughan, H.; Wang, P.W.; Caballero, J.D.; Fung, P.; Gong, Y.C.; Donaldson, S.L.; Yuan, L.J.; Keshavjee, S.; Zhang, Y.; Yau, Y.C.W.; et al. Analysis of the cystic fibrosis lung microbiota via serial Illumina sequencing of bacterial 16S rRNA hypervariable regions. PLoS ONE 2012, 7, e45791. [Google Scholar] [CrossRef]
- Xu, H.X.; Ren, J.D.; Xu, X.J.; Lou, B.; Zhang, D.M. The composition and diversity of intestinal microbes at different growth stages of giant freshwater prawns (Macrobrachium rosenbergii). Fishes 2023, 8, 473. [Google Scholar] [CrossRef]
- Weingarten, E.A.; Atkinson, C.L.; Jackson, C.R. The gut microbiome of freshwater Unionidae mussels is determined by host species and is selectively retained from filtered seston. PLoS ONE 2019, 14, e0224796. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.D.; Kong, Q.H.; Liu, S.Z.; Tan, Z.K.; Shang, P.; Wang, H.H. Dynamic distribution of intestinal microbes in Schizothorax o’connori at different growth stages. Fish. Sci. 2020, 86, 87–95. [Google Scholar] [CrossRef]
- Yang, J.C.; Lin, Y.Y.; Wei, Z.H.; Wu, Z.B.; Zhang, Q.Q.; Hao, J.W.; Wang, S.Y.; Li, A.H. Edwardsiella ictaluri almost completely occupies the gut microbiota of fish suffering from enteric septicemia of catfish (Esc). Fishes 2023, 8, 30. [Google Scholar] [CrossRef]
- Zhang, Z.; Ran, C.; Ding, Q.W.; Liu, H.L.; Xie, M.X.; Yang, Y.L.; Xie, Y.D.; Gao, C.C.; Zhang, H.L.; Zhou, Z.G. Ability of prebiotic polysaccharides to activate a HIF1α-antimicrobial peptide axis determines liver injury risk in zebrafish. Commun. Biol. 2019, 2, 274. [Google Scholar] [CrossRef]
- Wang, A.R.; Zhang, Z.; Ding, Q.W.; Yang, Y.L.; Bindelle, J.; Ran, C.; Zhou, Z.G. Intestinal Cetobacterium and acetate modify glucose homeostasis via parasympathetic activation in zebrafish. Gut Microbes 2021, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Liu, S.B.; Teame, T.; Luo, J.; Liu, Y.; Zhou, Q.W.; Ding, Q.W.; Yao, Y.Y.; Yang, Y.L.; Ran, C.; et al. Effect of Bacillus velezensis T23 solid-state fermentation product on growth, gut and liver health, and gut microbiota of common carp (Cyprinus carpio). Aquaculture 2025, 596, 741733. [Google Scholar] [CrossRef]
- Sugita, H.; Miyajima, C.; Deguchi, Y. The vitamin B12-producing ability of the intestinal microflora of freshwater fish. Aquaculture 1991, 92, 267–276. [Google Scholar] [CrossRef]
- Larsen, A.M.; Mohammed, H.H.; Arias, C.R. Characterization of the gut microbiota of three commercially valuable warmwater fish species. J. Appl. Microbiol. 2014, 116, 1396–1404. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Liu, F.L.; Li, B.; Duan, L.J.; Yu, J.M.; Huang, Z.Y.; Huang, S.Y.; Liu, H.W.; Liu, Q.F.; Liu, S.J. Multi-omics reveals the molecular mechanisms of rapid growth in distant hybrid fish. Aquaculture 2025, 596, 741783. [Google Scholar] [CrossRef]
- Li, F.; Huang, Y.L.; Hu, W.J.; Li, Z.; Wang, Q.Z.; Huang, S.S.; Jiang, F.J.; Pan, X.L. Roseomonas coralli sp. nov., a heavy metal resistant bacterium isolated from coral. Int. J. Syst. Evol. Microbiol. 2021, 71, 004624. [Google Scholar] [CrossRef]
- Liu, Y.; Hao, A.; Iseri, Y.; Li, C.; Zhang, Z.; Kuba, T. Possible survival mechanisms of Sinanodonta woodiana exposed to microcystin. J. JSCE 2015, 3, 259–267. [Google Scholar] [CrossRef]
- Barone, G.D.; Cernava, T.; Ullmann, J.; Liu, J.; Lio, E.; Germann, A.T.; Nakielski, A.; Russo, D.A.; Chavkin, T.; Knufmann, K.; et al. Recent developments in the production and utilization of photosynthetic microorganisms for food applications. Heliyon 2023, 9, e14708. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The firmicutes/bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Spence, C.; Wells, W.G.; Smith, C.J. Characterization of the primary starch utilization operon in the obligate anaerobe Bacteroides fragilis: Regulation by carbon source and oxygen. J. Bacteriol. 2006, 188, 4663–4672. [Google Scholar] [CrossRef]
- Zheng, X.C.; Xu, X.D.; Liu, M.Y.; Yang, J.; Yuan, M.; Sun, C.X.; Zhou, Q.L.; Chen, J.M.; Liu, B. Bile acid and short chain fatty acid metabolism of gut microbiota mediate high-fat diet induced intestinal barrier damage in Macrobrachium rosenbergii. Fish Shellfish Immunol. 2024, 146, 109376. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, Y.; Ye, T.; Liu, F.; Lou, B. Thyroid-active agents triiodothyronine, thyroxine and propylthiouracil differentially affect growth, intestinal short chain fatty acids and microbiota in little yellow croaker Larimichthys polyactis. Fishes 2025, 10, 69. [Google Scholar] [CrossRef]
- Sethuraman, A.; Stancheva, R.; Sanders, C.; Caceres, L.; Castro, D.; Hausknecht-Buss, H.; Henry, S.; Johansen, H.; Kasler, A.; Lastor, S.; et al. Genome of a novel Sediminibacterium discovered in association with two species of freshwater cyanobacteria from streams in Southern California. G3-Genes Genomes Genet. 2022, 12, jkac123. [Google Scholar] [CrossRef] [PubMed]
- Orellana, L.H.; Ben Francis, T.; Ferraro, M.; Hehemann, J.H.; Fuchs, B.M.; Amann, R. Verrucomicrobiota are specialist consumers of sulfated methyl pentoses during diatom blooms. ISME J. 2022, 16, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Foucault, P.; Halary, S.; Duval, C.; Goto, M.; Marie, B.; Hamlaoui, S.; Jardillier, L.; Lamy, D.; Lance, E.; Raimbault, E.; et al. A summer in the greater Paris: Trophic status of peri-urban lakes shapes prokaryotic community structure and functional potential. Environ. Microbiome 2025, 20, 24. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.L.; Tourlousse, D.M.; Matsuura, N.; Ohashi, A.; Sekiguchi, Y. Draft genome sequence of Terrimicrobium sacchariphilum NM-5T, a facultative anaerobic soil bacterium of the class spartobacteria. Genome Announc. 2017, 5, 10–1128. [Google Scholar] [CrossRef]
- Piloto-Sardiñas, E.; Abuin-Denis, L.; Maitre, A.; Foucault-Simonin, A.; Corona-González, B.; Díaz-Corona, C.; Roblejo-Arias, L.; Mateos-Hernández, L.; Marrero-Perera, R.; Obregon, D.; et al. Dynamic nesting of Anaplasma marginale in the microbial communities of Rhipicephalus microplus. Ecol. Evol. 2024, 14, e11228. [Google Scholar] [CrossRef]
- Mathai, P.P.; Magnone, P.; Dunn, H.M.; Sadowsky, M.J. Water and sediment act as reservoirs for microbial taxa associated with invasive dreissenid mussels. Sci. Total Environ. 2020, 703, 134915. [Google Scholar] [CrossRef] [PubMed]
- Gómez, M.A.C.; Melo-Bolívar, J.F.; Pardo, R.Y.R.; Rodriguez, J.A.; Villamil, L.M. Unveiling the probiotic potential of the anaerobic bacterium Cetobacterium sp. nov. C33 for enhancing Nile tilapia (Oreochromis niloticus) cultures. Microorganisms 2023, 11, 2922. [Google Scholar] [CrossRef]
- Kreutzer, M.F.; Kage, H.; Nett, M. Structure and biosynthetic assembly of cupriachelin, a photoreactive siderophore from the bioplastic producer Cupriavidus necator H16. J. Am. Chem. Soc. 2012, 134, 5415–5422. [Google Scholar] [CrossRef]
- Kreutzer, M.F.; Nett, M. Genomics-driven discovery of taiwachelin, a lipopeptide siderophore from Cupriavidus taiwanensis. Org. Biomol. Chem. 2012, 10, 9338–9343. [Google Scholar] [CrossRef]
- Huang, Z.K.; Peng, Z.J.; Zhang, M.L.; Li, X.H.; Qiu, X.T. Structure, function and engineering of the nonribosomal peptide synthetase condensation domain. Int. J. Mol. Sci. 2024, 25, 11774. [Google Scholar] [CrossRef] [PubMed]
- Ashniev, G.A.; Petrov, S.N.; Iablokov, S.N.; Rodionov, D.A. Genomics-based reconstruction and predictive profiling of amino acid biosynthesis in the human gut microbiome. Microorganisms 2022, 10, 740. [Google Scholar] [CrossRef] [PubMed]


| Group | Age (Month) | Shell Length (cm) | Shell Width (cm) | Shell Height (cm) | Weight (g) |
|---|---|---|---|---|---|
| Early juveniles | 3 | 2.98 ± 2.61 | 0.81 ± 1.18 | 2.23 ± 2.17 | 2.25 ± 0.68 |
| Late juveniles | 12 | 7.59 ± 3.04 | 3.21 ± 1.97 | 5.05 ± 3.45 | 46.33 ± 6.59 |
| Adults | 36 | 15.48 ± 5.74 | 5.78 ± 4.34 | 10.98 ± 8.24 | 384.44 ± 38.98 |
| Diversity Indices | Early Juveniles | Late Juveniles | Adults |
|---|---|---|---|
| Chao1 | 142.40 ± 33.10 b | 293.94 ± 10.47 a | 308.98 ± 15.48 a |
| Shannon | 0.83 ± 0.26 c | 2.33 ± 0.26 b | 2.95 ± 0.32 a |
| Simpson | 0.30 ± 0.12 c | 0.71 ± 0.07 b | 0.86 ± 0.06 a |
| goods_coverage | 1.00 ± 0.00 a | 1.00 ± 0.00 a | 1.00 ± 0.00 a |
| Phyla Name | Early Juveniles | Late Juveniles | Adults |
|---|---|---|---|
| Fusobacteriota | 83.82 ± 8.34 a | 56.85 ± 5.15 b | 34.12 ± 7.68 c |
| Verrucomicrobiota | 9.65 ± 6.85 a | 9.77 ± 4.38 a | 15.24 ± 8.71 a |
| Pseudomonadota | 2.61 ± 1.39 c | 12.69 ± 2.87 b | 22.04 ± 10.70 a |
| Cyanobacteriota | 2.47 ± 0.70 c | 9.24 ± 2.00 b | 16.84 ± 6.24 a |
| Bacillota | 0.94 ± 1.05 *,b | 7.05 ± 1.60 a | 2.72 ± 3.00 b |
| Bacteroidota | 0.11 ± 0.09 *,b | 2.20 ± 0.84 b | 5.38 ± 3.54 a |
| Chloroflexota | 0.09 ± 0.04 *,b | 1.06 ± 0.34 ab | 2.09 ± 1.27 a |
| Genus Name | Early Juveniles | Late Juveniles | Adults |
|---|---|---|---|
| Cetobacterium | 83.81 ± 8.34 a | 56.79 ± 5.16 b | 34.09 ± 7.68 c |
| LD29 | 8.77 ± 6.83 a | 7.35 ± 4.12 a | 13.80 ± 8.25 a |
| Cyanobium_PCC-6307 | 2.35 ± 0.65 b | 5.81 ± 0.90 b | 9.84 ± 4.58 a |
| Cupriavidus | 1.08 ± 0.90 a | 0.26 ± 0.19 *,b | 0.19 ± 0.10 *,b |
| Roseomonas | 0.25 ± 0.28 *,b | 7.82 ± 2.30 a | 11.56 ± 7.36 a |
| Limnolyngbya_CHAB4449 | 0.06 ± 0.04 *,b | 1.42 ± 0.79 ab | 1.77 ± 1.70 a |
| Terrimicrobium | 0.86 ± 0.69 *,a | 1.39 ± 0.75 a | 1.29 ± 0.71 a |
| Limnothrix | 0 *,b | 1.33 ± 0.90 b | 4.13 ± 2.49 a |
| Sediminibacterium | 0 *,b | 0.61 ± 0.30 *,b | 1.54 ± 0.93 a |
| Acidibacter | 0.02 ± 0.03 *,c | 0.36 ± 0.06 *,b | 1.15 ± 0.34 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, M.; Wang, H.; Wang, M.; Bah, I.; Jiang, T.; Xue, J.; Ding, X.; Chen, X. Comparison of Gut Bacterial Communities in the Freshwater Mussel Sinanodonta woodiana at Different Life Stages. Diversity 2025, 17, 814. https://doi.org/10.3390/d17120814
Gu M, Wang H, Wang M, Bah I, Jiang T, Xue J, Ding X, Chen X. Comparison of Gut Bacterial Communities in the Freshwater Mussel Sinanodonta woodiana at Different Life Stages. Diversity. 2025; 17(12):814. https://doi.org/10.3390/d17120814
Chicago/Turabian StyleGu, Mengying, Huan Wang, Meiyi Wang, Ibrahim Bah, Tao Jiang, Junren Xue, Xinyu Ding, and Xiubao Chen. 2025. "Comparison of Gut Bacterial Communities in the Freshwater Mussel Sinanodonta woodiana at Different Life Stages" Diversity 17, no. 12: 814. https://doi.org/10.3390/d17120814
APA StyleGu, M., Wang, H., Wang, M., Bah, I., Jiang, T., Xue, J., Ding, X., & Chen, X. (2025). Comparison of Gut Bacterial Communities in the Freshwater Mussel Sinanodonta woodiana at Different Life Stages. Diversity, 17(12), 814. https://doi.org/10.3390/d17120814







