DNA Barcoding Applications in Ants (Hymenoptera: Formicidae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Statistical Analysis
2.3. Barcoding Analysis
3. Results
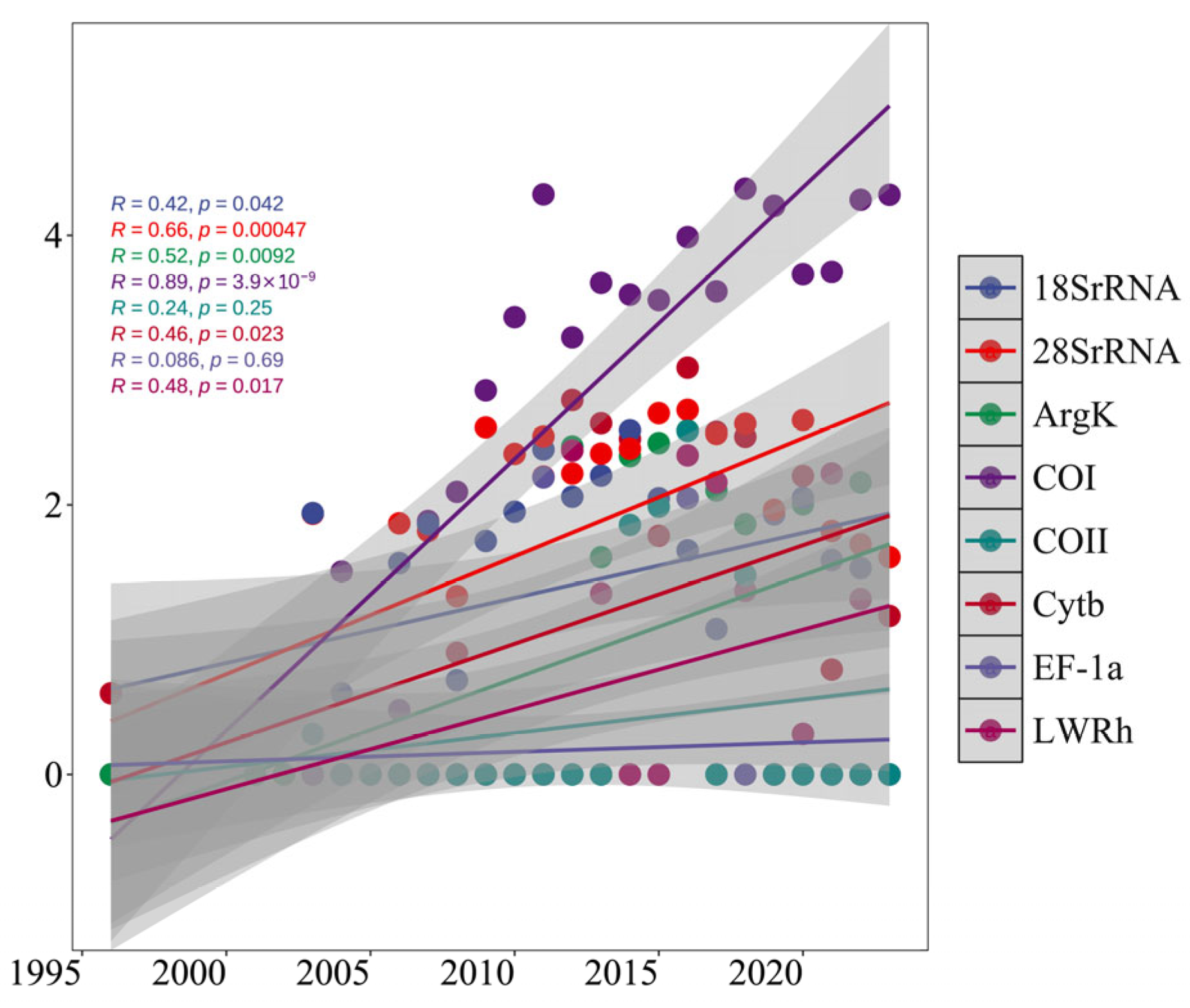
3.1. Temporal Trend of Sequence Development
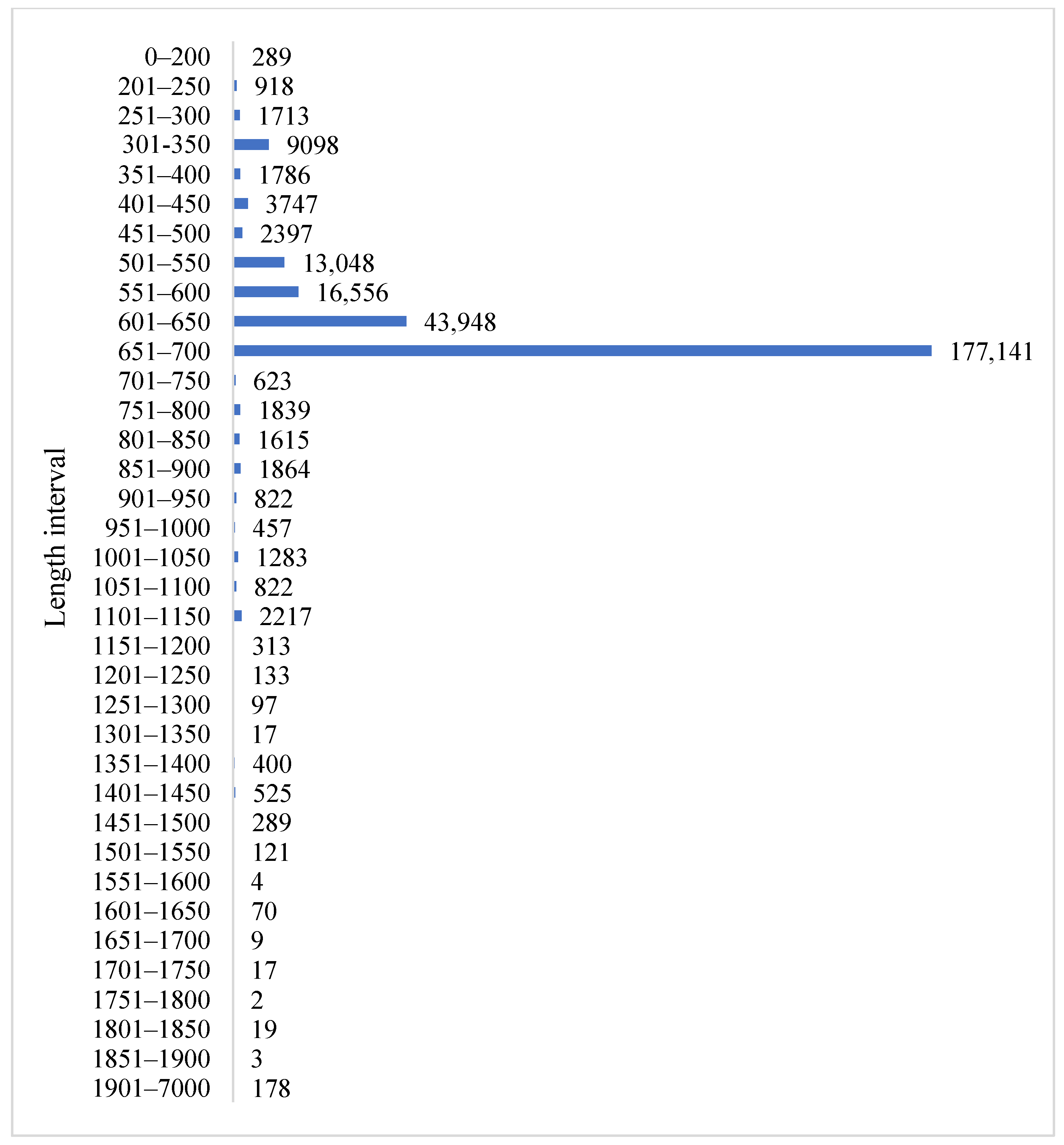
3.2. Sequence Length Distribution
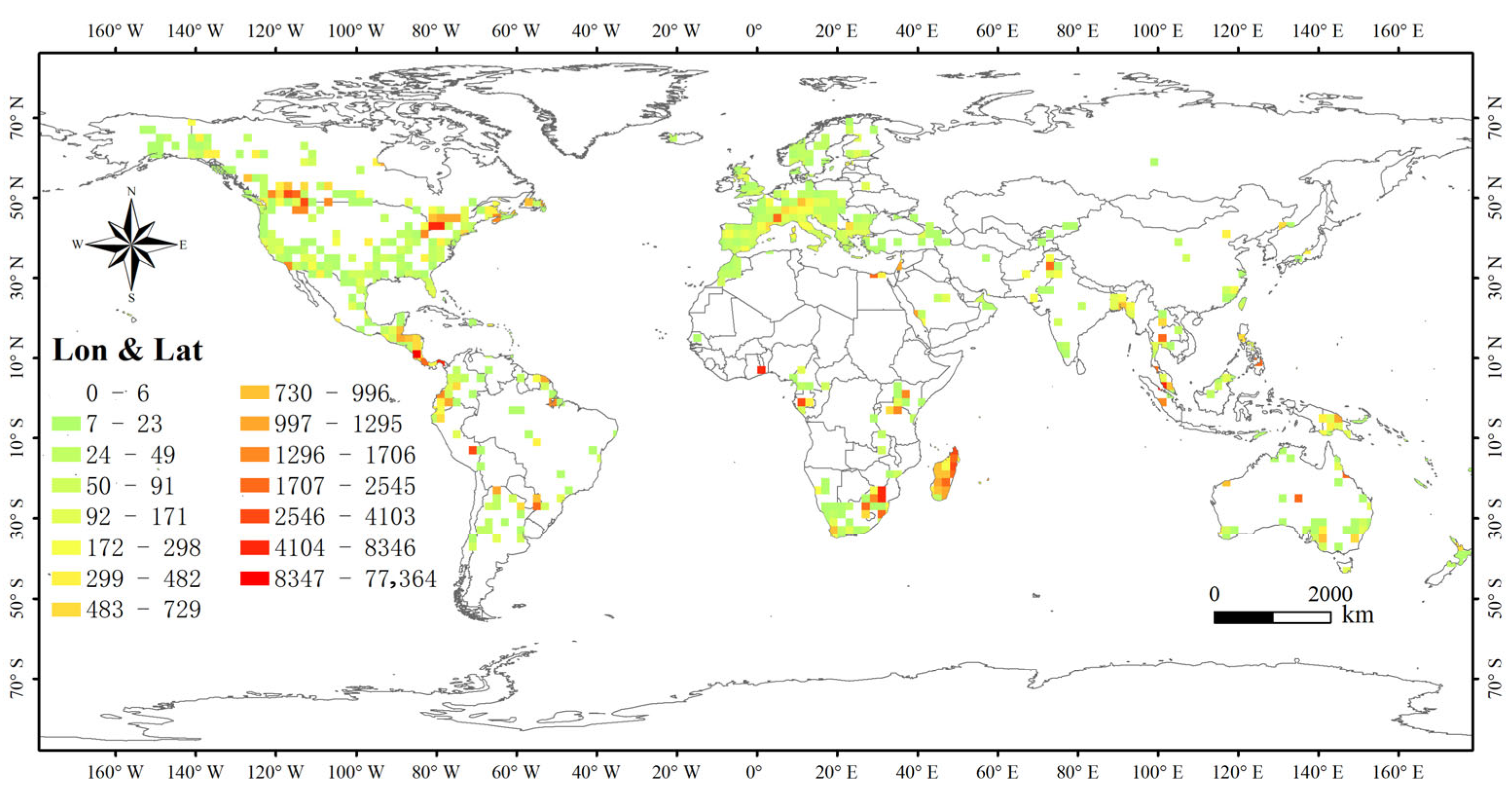
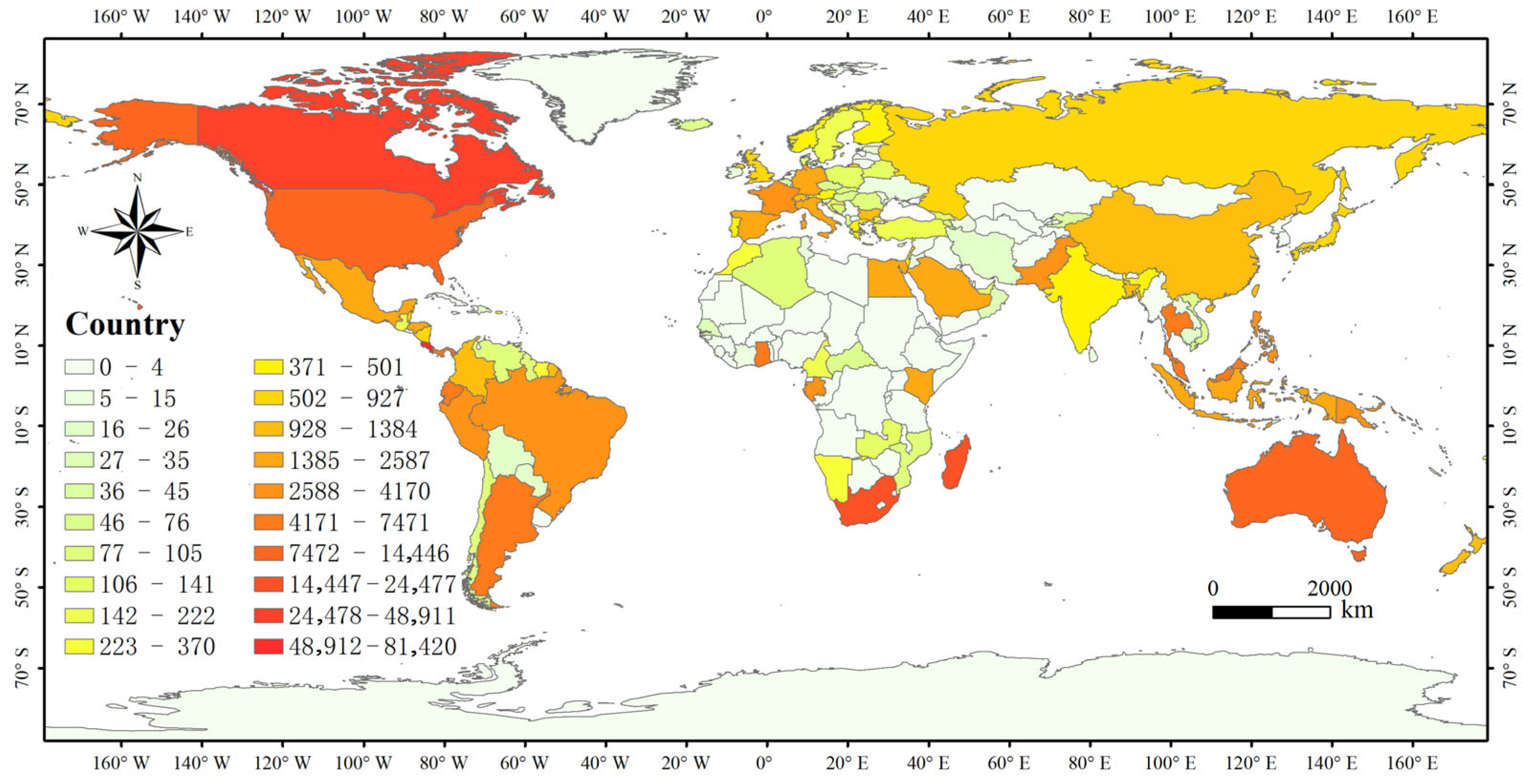
3.3. Spatial Distribution Pattern
3.4. Species Composition
3.5. Collaboration Network
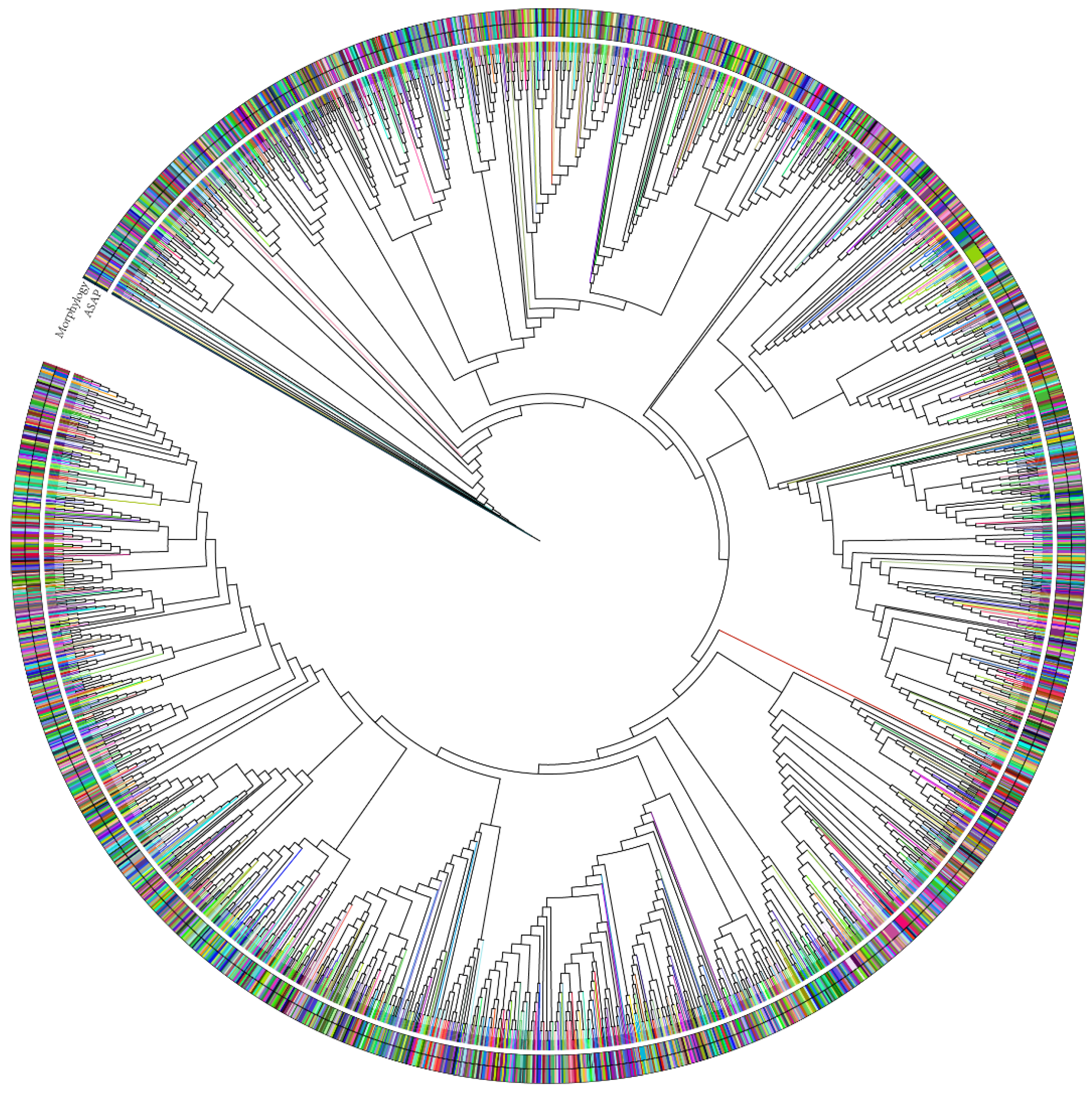
3.6. Barcoding Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DNA | Deoxyribonucleic Acid |
| NCBI | National Center for Biotechnology Information |
| BOLD | Barcode of Life Data System |
| ML | Maximum Likelihood |
| K2P | Kimura 2-parameter |
| MOTU | Molecular Operational Taxonomic Unit |
| PCR | Polymerase Chain Reaction |
| ASAP | Assemble Species by Automatic Partitioning |
| iTOL | Interactive Tree of Life |
Appendix A
- ###1. R package bold (version 1.3.0)|download the molecular data of Formicidae from BOLD
- library(bold)
- formicidae = bold_seq(taxon = ‘Formicidae’)
- formicidae_data = bold_specimens(taxon = ‘Formicidae’)
- write.csv(formicidae, “formicidae_sequences.csv”, row.names = FALSE)
- write.csv(formicidae_data, “formicidae_specimens.csv”, row.names = FALSE)
- ###2. R package refdb (version 0.1.1)|download the molecular data of Formicidae from NCBI.
- install.packages(“refdb”)###The package calls rentrez to retrieve information from the NCBI, and also uses the bioseq package processing sequence.
- install.packages(“rentrez”)
- rm(list = ls())
- library(refdb)
- library(dplyr)
- library(stringr)
- library(rentrez)
- ###Download NCBI Nucleotide data
- ###Search and download data from NCBI Nucleotide database. In addition, the NCBI Taxonomy database will be called to obtain the classification.
- Formicidae_ncbi <- refdb_import_NCBI(query = “Formicidae COI”, ###Search sequence.
- full = TRUE, ###Whether to get complete information. If FALSE, only columns of important information are obtained.
- max_seq_length = 100,00,
- seq_bin = 200, ###Number of articles downloaded at one time. Does not affect the final result.
- verbose = TRUE ### Whether to display download prompt information
- )
- write.csv(Formicidae_ncbi, ’Formicidae_COI.csv’)
- ###Environmental preparation
- library(tidyr)
- library(readr)
- library(magrittr)
- ###Extract columns ID, Sequence, Categories, Genus, Family, Super Family, Order, Gene, Length, Latitude and Longitude from Solenopsis_ncbi data frame.
- ###Rename the data frame formed by the extracted target column as Solenopsis_seq_1, and then output the bit csv table.
- Formicidae_seq_1 <- Formicidae_ncbi % > % dplyr::select(id,sequence,species,genus,family,superfamily,order,gene,length,latitude,longitude)
- write.csv(Formicidae_seq_1, file = “Formicidae.csv”)###Save some column output in Solenopsis_ncbi data frame.
- ###In the target column to be extracted, ID and sequence are used as fasta information to be output. The first two columns are the necessary fasta id and fasta sequences, respectively. Here we combine the following columns as the description part of fasta. Description is listed as an optional part.
- ### Prepare the sequence to be written in data frame format. The first column is id, the second column is Sequence, and the third column is description. Rename it as Solenopsis_seq_2.
- ###Here, the contents of two columns, Categories and length, are merged as the description part.
- Formicidae_seq_2_COI <- Formicidae_ncbi % > % dplyr::select(id,sequence,species,genus,family,superfamily,order,gene,length,latitude,longitude) % > % tidyr::unite(col = “description”,species,length,genus,family,superfamily,order,gene,latitude,longitude,sep = “ “,remove = TRUE, na.rm = FALSE)### Unite the columns of a merged data frame.
- write.csv(Formicidae_seq_2_COI, file = “Formicidae_seq_2_COI.csv”)###Save some column output in Solenopsis_ncbi data frame.
- ###Define fasta sequence to write function: write_bio_fasta()
- write_bio_fasta <- function(data, file, width = 60, append = FALSE){
- ### check data
- stopifnot(is.data.frame(data)) ###Must be in data frame format.
- n_col <- ncol(data)
- if(n_col! = 2 & n_col! = 3){
- stop(“‘data’: the input must be a data frame with 2 or 3 columns!”)
- }
- ### prepare the list for ‘write_lines()’
- fa <- list() # Define in advance
- ###width = 60 # Width of the output sequence
- for (i in seq(nrow(data))){
- n = length(fa) + 1 # Add to fa list the nth element, that is, the number of lines in the fasta sequence file. if(n_col == 2){
- id <- data[i,1] % > % stringr::str_replace(pattern = “^”, “>”) %>% as.character()
- fa[[n]] <- id
- } else {
- id <- data[i,1] % > % stringr::str_replace(pattern = “^”, “>”) %>% as.character()
- description <- data[i,3] % > % as.character()
- fa[[n]] <- paste(id,description,sep = “ “)
- }seq <- data[i,2] % > % as.character() ###The seq column in the result of refdb is not a simple vector. Turn it around.
- kz <- ceiling(stringr::str_length(seq)/width)
- if(is.finite(kz)){
- for (x in seq(ceiling(stringr::str_length(seq)/width))) {
- fa[[n + 1]] <- stringr::str_sub(seq,start = ((x − 1)*width + 1), end = x*width)
- n = length(fa)
- }
- }
- }
- ### Write the sequence and set append and num_threads.
- readr::write_lines(fa,file = file, sep = “\n”, append = FALSE) ###List or vector is required. If it is a list, write the list content, excluding name.
- }
- ###Based on the write_bio_fasta function defined above, Solenopsis_seq_2 is output as a file in fas format.
- write_bio_fasta(data = Formicidae_seq_2_COI, file = “Formicidae_COI.fas”,width = 70)
References
- Henke, W.; Tattersall, I.; Hardt, T. Handbook of Paleoanthropology; Reference; Springer: Berlin/Heidelberg, Germany, 2007; ISBN 978-3-540-32474-4. [Google Scholar]
- Wilson, E.O. Taxonomy as a Fundamental Discipline. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 2004, 359, 739. [Google Scholar] [CrossRef]
- Hong, D.Y. World Peonies and Herbaceous Peonies Research: With Discussion on the Fundamental Principles of Taxonomy; Science Press: Beijing, China, 2024. [Google Scholar]
- Löbl, I. The Silent Extinction of Species and Taxonomists—An Appeal to Science Policymakers and Legislators. Diversity 2023, 15, 1053. [Google Scholar] [CrossRef]
- Deyuan, H. A Brief Discussion on Methodology in Taxonomy. Biodivers. Sci. 2025, 33, 24541. [Google Scholar] [CrossRef]
- Godfray, H.C.J. Challenges for Taxonomy. Nature 2002, 417, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. London Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Karbstein, K.; Kösters, L.; Hodač, L.; Hofmann, M.; Hörandl, E.; Tomasello, S.; Wagner, N.D.; Emerson, B.C.; Albach, D.C.; Scheu, S.; et al. Species Delimitation 4.0: Integrative Taxonomy Meets Artificial Intelligence. Trends Ecol. Evol. 2024, 39, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.L.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic Species as a Window on Diversity and Conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Wang, H.-H.; He, L.-L.; Hua, B.-Z. Postembryonic Development and Morphological Variations of Larval Instars in the Scorpionfly Panorpa liui Hua (Mecoptera: Panorpidae). Arthropod Struct. Dev. 2025, 85, 101414. [Google Scholar] [CrossRef]
- Punzalan, D.; Hosken, D.J. Sexual Dimorphism: Why the Sexes Are (and Are Not) Different. Curr. Biol. 2010, 20, R972–R973. [Google Scholar] [CrossRef]
- Vinton, A.C.; Gascoigne, S.J.L.; Sepil, I.; Salguero-Gómez, R. Plasticity’s Role in Adaptive Evolution Depends on Environmental Change Components. Trends Ecol. Evol. 2022, 37, 1067–1078. [Google Scholar] [CrossRef]
- Adams, M.; Raadik, T.A.; Burridge, C.P.; Georges, A. Global Biodiversity Assessment and Hyper-Cryptic Species Complexes: More Than One Species of Elephant in the Room? Syst. Biol. 2014, 63, 518–533. [Google Scholar] [CrossRef]
- de León, G.P.-P.; Nadler, S.A. What We Don’t Recognize Can Hurt Us: A Plea for Awareness About Cryptic Species. J. Parasitol. 2010, 96, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Jinbo, U.; Kato, T.; Ito, M. Current Progress in DNA Barcoding and Future Implications for Entomology. Entomol. Sci. 2011, 14, 107–124. [Google Scholar] [CrossRef]
- Watanabe, S.; Masamura, N.; Satoh, S.; Hirao, T. Investigating the Efficiency of DNA Barcoding in Insect Classification: A Review Study. Entomol. Appl. Sci. Lett. 2024, 11, 15–23. [Google Scholar] [CrossRef]
- Narváez-Vásquez, A.; Gaviria, J.; Vergara-Navarro, E.V.; Rivera-Pedroza, L.; Löhr, B. Ant (Hymenoptera: Formicidae) Species Diversity in Secondary Forest and Three Agricultural Land Uses of the Colombian Pacific Coast. Rev. Chil. De Entomol. 2021, 47, 441–458. [Google Scholar] [CrossRef]
- Arredondo, B.S.; Guerrero, R.J. The Ant Genus Nesomyrmex Wheeler (Formicidae, Myrmicinae) from the Threatened Colombian Tropical Dry Forest: Three New Species, a New Synonymy, and New Distributional Data. Zookeys 2025, 1232, 131–172. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, Z.; Xu, Z.; Fu, Q.; Fu, L. Two New Ant Species of the Genus Leptogenys (Hymenoptera, Formicidae) from Hainan, China, with a Key to the Known Chinese Species. Zookeys 2024, 1195, 199–217. [Google Scholar] [CrossRef]
- Fisher, B.L.; Smith, M.A. A Revision of Malagasy Species of Anochetus Mayr and Odontomachus Latreille (Hymenoptera: Formicidae). PLoS ONE 2008, 3, e1787. [Google Scholar] [CrossRef]
- Siddiqui, J.A.; Chen, Z.; Li, Q.; Deng, J.; Lin, X.; Huang, X. DNA Barcoding of Aphid-Associated Ants (Hymenoptera, Formicidae) in a Subtropical Area of Southern China. Zookeys 2019, 879, 117–136. [Google Scholar] [CrossRef]
- Chesnokova, S.V.; Vaulin, O.V.; Zhigulskaya, Z.A.; Novgorodova, T.A. Formica gagatoides Ruzsky, 1904, and Siberian F. kozlovi Dlussky, 1965 (Hymenoptera: Formicidae); Two or One Species? Diversity 2023, 15, 686. [Google Scholar] [CrossRef]
- Cardenas, C.R.; Luo, A.R.; Jones, T.H.; Schultz, T.R.; Adams, R.M.M. Using an Integrative Taxonomic Approach to Delimit a Sibling Species, Mycetomoellerius mikromelanos Sp. Nov. (Formicidae: Attini: Attina). PeerJ 2021, 9, e11622. [Google Scholar] [CrossRef] [PubMed]
- Klimeš, P.; Drescher, J.; Buchori, D.; Hidayat, P.; Nazarreta, R.; Potocký, P.; Rimandai, M.; Scheu, S.; Matos-Maraví, P. Uncovering Cryptic Diversity in the Enigmatic Ant Genus Overbeckia and Insights into the Phylogeny of Camponotini (Hymenoptera: Formicidae: Formicinae). Invertebr. Syst. 2022, 36, 557–579. [Google Scholar] [CrossRef]
- Ouellette, G.D.; Fisher, B.L.; Girman, D.J. Molecular Systematics of Basal Subfamilies of Ants Using 28S rRNA (Hymenoptera: Formicidae). Mol. Phylogenetics Evol. 2006, 40, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Keck, F.; Altermatt, F. Management of DNA Reference Libraries for Barcoding and Metabarcoding Studies with the R Package Refdb. Mol. Ecol. Resour. 2023, 23, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble Species by Automatic Partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef]
- Zhou, T.; Xu, K.; Zhao, F.; Liu, W.; Li, L.; Hua, Z.; Zhou, X. Itol.Toolkit Accelerates Working with iTOL (Interactive Tree of Life) by an Automated Generation of Annotation Files. Bioinformatics 2023, 39, btad339. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Zhang, D.-X.; Hewitt, G.M. Assessment of the Universality and Utility of a Set of Conserved Mitochondrial COI Primers in Insects. Insect Mol. Biol. 1997, 6, 143–150. [Google Scholar] [CrossRef]
- Timm, V.F.; Gonçalves, L.T.; Valente, V.L.D.S.; Deprá, M. The Efficiency of the COI Gene as a DNA Barcode and an Overview of Orthoptera (Caelifera and Ensifera) Sequences in the BOLD System. Can. J. Zool. 2022, 100, 710–718. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; De Waard, J.R. Barcoding Animal Life: Cytochrome c Oxidase Subunit 1 Divergences among Closely Related Species. Proc. R. Soc. London Ser. B Biol. Sci. 2003, 270. [Google Scholar] [CrossRef] [PubMed]
- Twyford, A.D.; Beasley, J.; Barnes, I.; Allen, H.; Azzopardi, F.; Bell, D.; Blaxter, M.L.; Broad, G.; Campos-Dominguez, L.; Choonea, D.; et al. A DNA Barcoding Framework for Taxonomic Verification in the Darwin Tree of Life Project. Wellcome Open Res. 2024, 9, 339. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Zhang, H.; Bu, W. Exploring Large-Scale Patterns of Genetic Variation in the COI Gene among Insecta: Implications for DNA Barcoding and Threshold-Based Species Delimitation Studies. Insects 2022, 13, 425. [Google Scholar] [CrossRef] [PubMed]
- Rubinoff, D.; Cameron, S.; Will, K. A Genomic Perspective on the Shortcomings of Mitochondrial DNA for “Barcoding” Identification. J. Hered. 2006, 97, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Deagle, B.E.; Jarman, S.N.; Coissac, E.; Pompanon, F.; Taberlet, P. DNA Metabarcoding and the Cytochrome c Oxidase Subunit I Marker: Not a Perfect Match. Biol. Lett. 2014, 10, 20140562. [Google Scholar] [CrossRef]
- Rasoamanana, N.; Fisher, B.L. A Taxonomic Revision of the Malagasy Endemic Subgenus Mayria of the Genus Camponotus (Hymenoptera, Formicidae) Based on Qualitative Morphology and Quantitative Morphometric Analyses. Zookeys 2022, 1081, 137–231. [Google Scholar] [CrossRef]
- Varela-Hernández, F.; Riquelme, F.; Guerrero, R.J. New Genus and Species of Ponerine Ant (Hymenoptera: Formicidae) from Mexican Amber. Hist. Biol. 2024, 37, 1134–1140. [Google Scholar] [CrossRef]
- Dos Santos-Neto, E.A.; Chaul, J.C.M.; Delabie, J.H.C. New Species and New Records of Strumigenys Smith, 1860 (Formicidae: Myrmicinae) from the Neotropical Schulzi Species Group. Taxonomy 2024, 4, 633–648. [Google Scholar] [CrossRef]
- Andersen, A.N.; Brassard, F.; Hoffmann, B.D. Unrecognised Ant Megadiversity in the Australian Monsoonal Tropics: The Melophorus hirsutipes Heterick, Castelanelli & Shattuck Species Group. Diversity 2023, 15, 892. [Google Scholar] [CrossRef]
- Csősz, S.; Alicata, A.; Báthori, F.; Galkowski, C.; Schifani, E.; Yusupov, Z.; Herczeg, G.; Prebus, M.M. Integrative Taxonomy Reveals Inflated Biodiversity in the European Temnothorax unifasciatus Complex (Hymenoptera: Formicidae). Zool. Scr. 2025, 54, 33–49. [Google Scholar] [CrossRef]
- Hazarika, H.N.; Khanikor, B. Integration of Morphological and Molecular Taxonomic Characters for Identification of Odontoponera denticulata (Hymenoptera: Formicidae: Ponerinae) with the Description of the Antennal Sensilla. Zool. Anz. 2021, 293, 89–100. [Google Scholar] [CrossRef]
- Kass, J.M.; Guénard, B.; Dudley, K.L.; Jenkins, C.N.; Azuma, F.; Fisher, B.L.; Parr, C.L.; Gibb, H.; Longino, J.T.; Ward, P.S.; et al. The Global Distribution of Known and Undiscovered Ant Biodiversity. Sci. Adv. 2022, 8, eabp9908. [Google Scholar] [CrossRef] [PubMed]
- Titley, M.A.; Snaddon, J.L.; Turner, E.C. Scientific Research on Animal Biodiversity Is Systematically Biased towards Vertebrates and Temperate Regions. PLoS ONE 2017, 12, e0189577. [Google Scholar] [CrossRef] [PubMed]
- Stocks, G.; Seales, L.; Paniagua, F.; Maehr, E.; Bruna, E.M. The Geographical and Institutional Distribution of Ecological Research in the Tropics. Biotropica 2008, 40, 397–404. [Google Scholar] [CrossRef]
- Zamani, A.; Pos, D.D.; Fric, Z.F.; Orfinger, A.B.; Scherz, M.D.; Bartoňová, A.S.; Gante, H.F. The Future of Zoological Taxonomy Is Integrative, Not Minimalist. Syst. Biodivers. 2022, 20, 1–14. [Google Scholar] [CrossRef]
- Ward, P.S. The Phylogeny and Evolution of Ants. Annu. Rev. Ecol. Evol. Syst. 2014, 45, 23–43. [Google Scholar] [CrossRef]
- Gui, Q.; Liu, C.; Du, D. Globalization of Science and International Scientific Collaboration: A Network Perspective. Geoforum 2019, 105, 1–12. [Google Scholar] [CrossRef]
- Hennemann, S.; Rybski, D.; Liefner, I. The Myth of Global Science Collaboration—Collaboration Patterns in Epistemic Communities. J. Informetr. 2012, 6, 217–225. [Google Scholar] [CrossRef]
- Virola-V, B.S.; Abrego, J.; Castillo, D.; Bonilla, E.; Gálvez, D. Who Is Working on Ant Physiology? There Is Room to Improve International Collaborations. Myrmecol. News 2022, 32, 115–125. [Google Scholar] [CrossRef]
- Zachos, F.E. (New) Species Concepts, Species Delimitation and the Inherent Limitations of Taxonomy. J. Genet. 2018, 97, 811–815. [Google Scholar] [CrossRef] [PubMed]
- Dayrat, B. Towards Integrative Taxonomy. Biol. J. Linn. Soc. 2005, 85, 407–415. [Google Scholar] [CrossRef]
- Mera-Rodríguez, D.; Fernández-Marín, H.; Rabeling, C. Phylogenomic Approach to Integrative Taxonomy Resolves a Century-old Taxonomic Puzzle and the Evolutionary History of the Acromyrmex octospinosus Species Complex. Syst. Entomol. 2025, 50, 469–494. [Google Scholar] [CrossRef]
- Wang, Y.; Nansen, C.; Zhang, Y. Integrative Insect Taxonomy Based on Morphology, Mitochondrial DNA, and Hyperspectral Reflectance Profiling: Integrative Insect Taxonomy of Bundera. Zool. J. Linn. Soc. 2016, 177, 378–394. [Google Scholar] [CrossRef]
- Schlick-Steiner, B.C.; Steiner, F.M.; Seifert, B.; Stauffer, C.; Christian, E.; Crozier, R.H. Integrative Taxonomy: A Multisource Approach to Exploring Biodiversity. Annu. Rev. Entomol. 2010, 55, 421–438. [Google Scholar] [CrossRef]
- Mayr, A.V.; Keller, A.; Peters, M.K.; Grimmer, G.; Krischke, B.; Geyer, M.; Schmitt, T.; Steffan-Dewenter, I. Cryptic Species and Hidden Ecological Interactions of Halictine Bees along an Elevational Gradient. Ecol. Evol. 2021, 11, 7700–7712. [Google Scholar] [CrossRef]
- LeMay, G.A.; Johnson, A.J.; Hulcr, J. Integrative Taxonomy Sheds Light on Species Boundaries and Relationships in the Inbred Haplodiploid Ambrosia Beetle Genus Xyleborus. Insect Syst. Evol. 2024, 56, 47–63. [Google Scholar] [CrossRef]
- Hew, Y.X.; Ya’cob, Z.; Adler, P.H.; Chen, C.D.; Lau, K.W.; Sofian-Azirun, M.; Muhammad-Rasul, A.H.; Putt, Q.Y.; Izwan-Anas, N.; Hadi, U.K.; et al. DNA Barcoding of Black Flies (Diptera: Simuliidae) in Indonesia. Parasites Vectors 2023, 16, 248. [Google Scholar] [CrossRef]
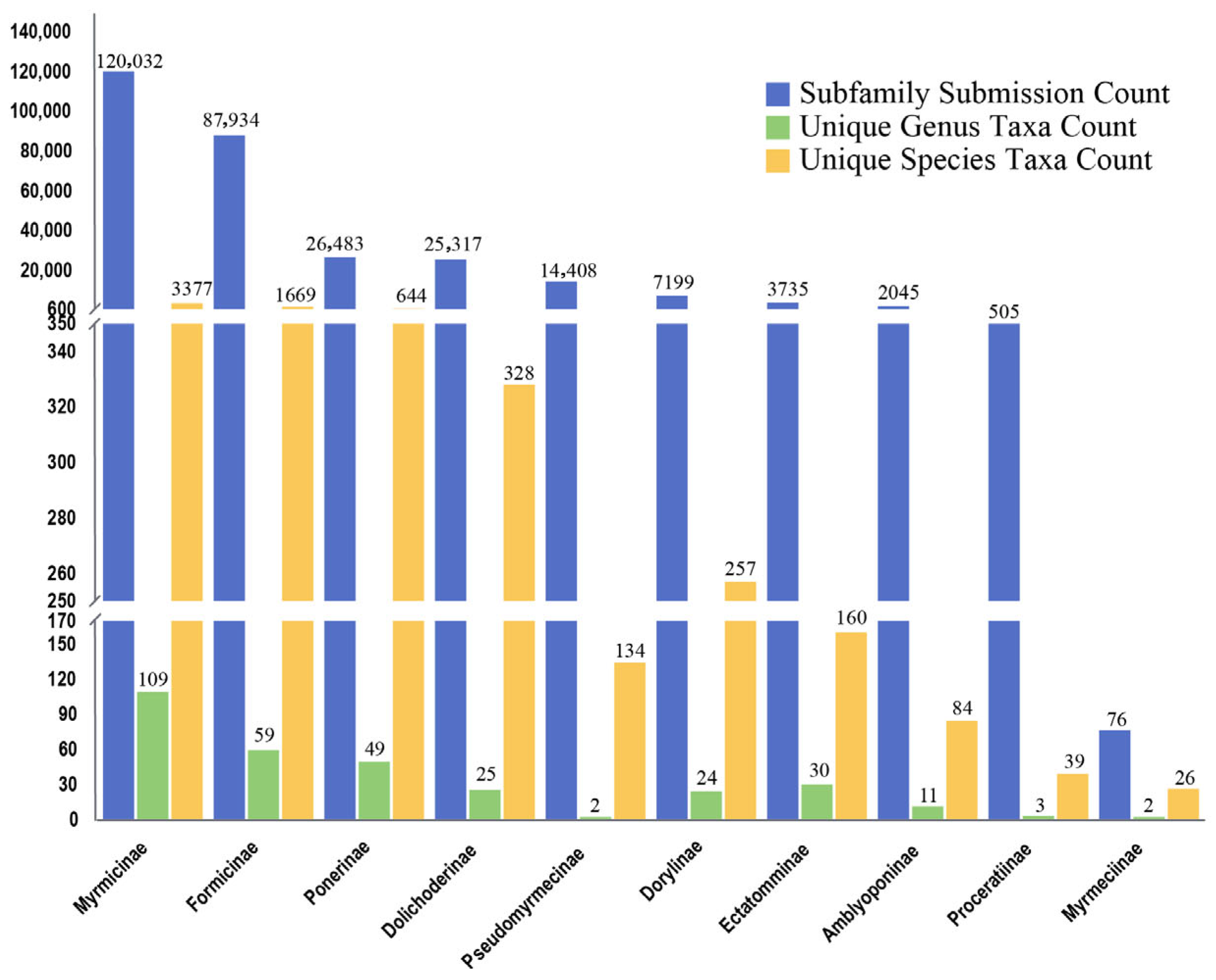
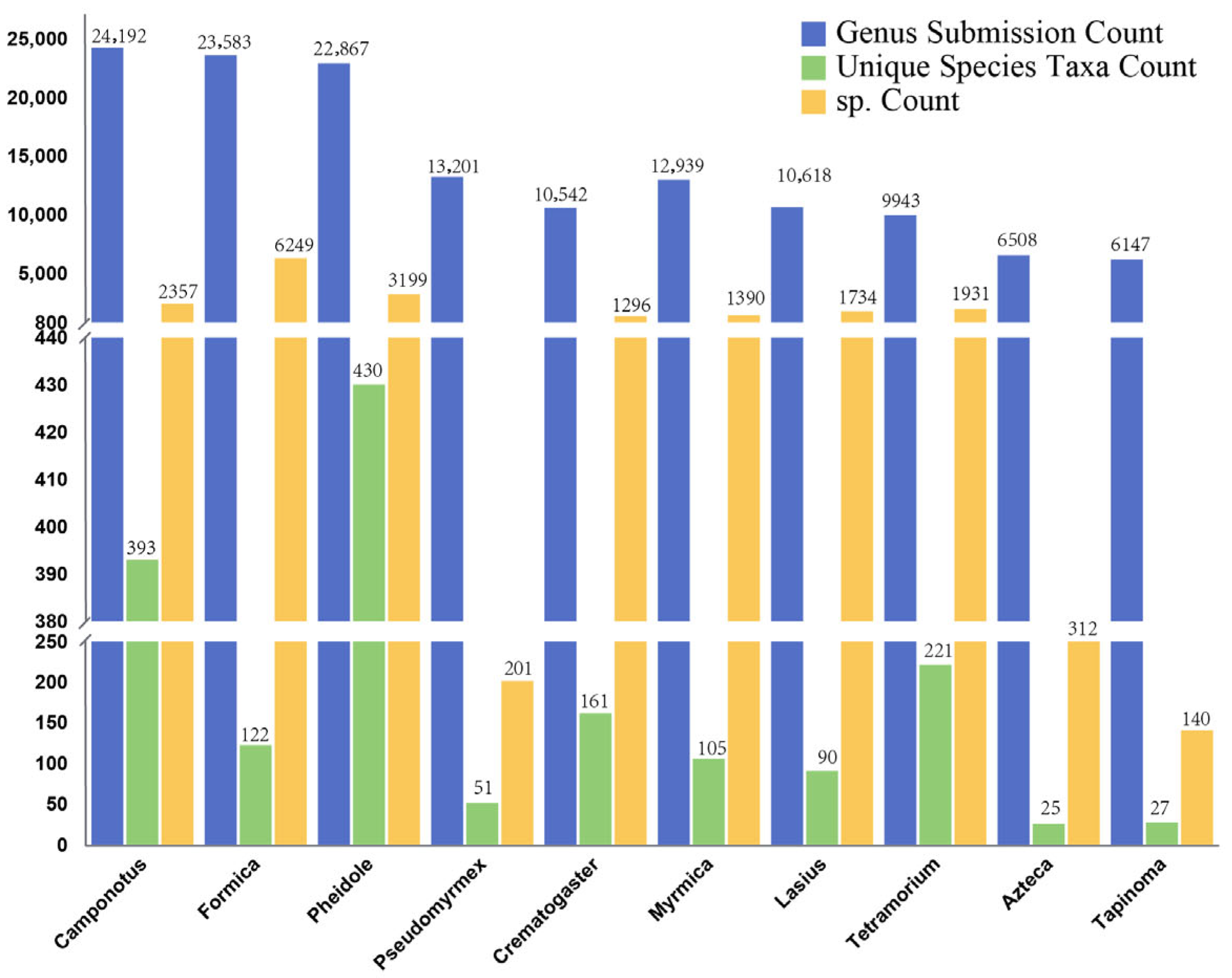

| Gene | Sequence No. | Genus No. | Species No. | sp. No. |
|---|---|---|---|---|
| COI | 337,887 | 270 | 4317 | 32,444 |
| 28S rRNA | 4560 | 304 | 1396 | 784 |
| Cytb | 3509 | 73 | 623 | 270 |
| ArgK | 1635 | 289 | 1004 | 324 |
| 18S rRNA | 1357 | 289 | 961 | 221 |
| LWRh | 1061 | 34 | 245 | 162 |
| COII | 565 | 27 | 165 | 81 |
| EF-1α | 112 | 11 | 78 | 28 |
| Subfamily | Genus |
|---|---|
| Myrmicinae | Acromyrmex, Adelomyrmex, Aphaenogaster, Apterostigma, Atta, Cardiocondyla, Carebara, Cataulacus, Cephalotes, Crematogaster, Daceton, Eurhopalothrix, Hylomyrma, Lachnomyrmex, Leptothorax, Melissotarsus, Meranoplus, Messor, Monomorium, Mycetarotes, Mycocepurus, Myrmica, Myrmicaria, Myrmicocrypta, Nesomyrmex, Octostruma, Paratrachymyrmex, Pheidole, Pogonomyrmex, Rhopalothrix, Rogeria, Royidris, Sericomyrmex, Solenopsis, Stegomyrmex, Stenamma, Strumigenys, Syllophopsis, Temnothorax, Terataner, Tetramorium, Trachymyrmex, Trichomyrmex, Wasmannia |
| Formicinae | Acropyga, Brachymyrmex, Camponotus, Cataglyphis, Formica, Lasius, Myrmecocystus, Notostigma, Nylanderia, Opisthopsis, Paraparatrechina, Plagiolepis, Polyrhachis, Proformica, Pseudolasius |
| Ponerinae | Anochetus, Dinoponera, Euponera, Hypoponera, Leptogenys, Neoponera, Odontomachus, Odontoponera, Pachycondyla, Platythyrea, Ponera |
| Dolichoderinae | Azteca, Dolichoderus, Forelius, Iridomyrmex, Leptomyrmex, Linepithema, Tapinoma, Technomyrmex |
| Dorylinae | Dorylus, Eciton, Neivamyrmex, Nomamyrmex, Ooceraea, |
| Ectatomminae | Boltonia, Ectatomma, Holcoponera, Poneracantha |
| Amblyoponinae | Amblyopone, Mystrium, Stigmatomma |
| Myrmeciinae | Myrmecia, Myrmecina |
| Prionopelta | Prionopelta |
| Proceratiinae | Proceratium |
| Pseudomyrmecinae | Pseudomyrmex |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Liu, D.; Chen, Z.; Du, C. DNA Barcoding Applications in Ants (Hymenoptera: Formicidae). Diversity 2025, 17, 783. https://doi.org/10.3390/d17110783
Wang J, Liu D, Chen Z, Du C. DNA Barcoding Applications in Ants (Hymenoptera: Formicidae). Diversity. 2025; 17(11):783. https://doi.org/10.3390/d17110783
Chicago/Turabian StyleWang, Jue, Dongqing Liu, Zhilin Chen, and Congcong Du. 2025. "DNA Barcoding Applications in Ants (Hymenoptera: Formicidae)" Diversity 17, no. 11: 783. https://doi.org/10.3390/d17110783
APA StyleWang, J., Liu, D., Chen, Z., & Du, C. (2025). DNA Barcoding Applications in Ants (Hymenoptera: Formicidae). Diversity, 17(11), 783. https://doi.org/10.3390/d17110783







