Cordyceps biarmica sp. nov., an Entomopathogenic Fungus from Boreal Forests of North European Russia
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Fungal Isolation
2.2. DNA Extraction, PCR and Sequencing
2.3. Phylogenetic Studies
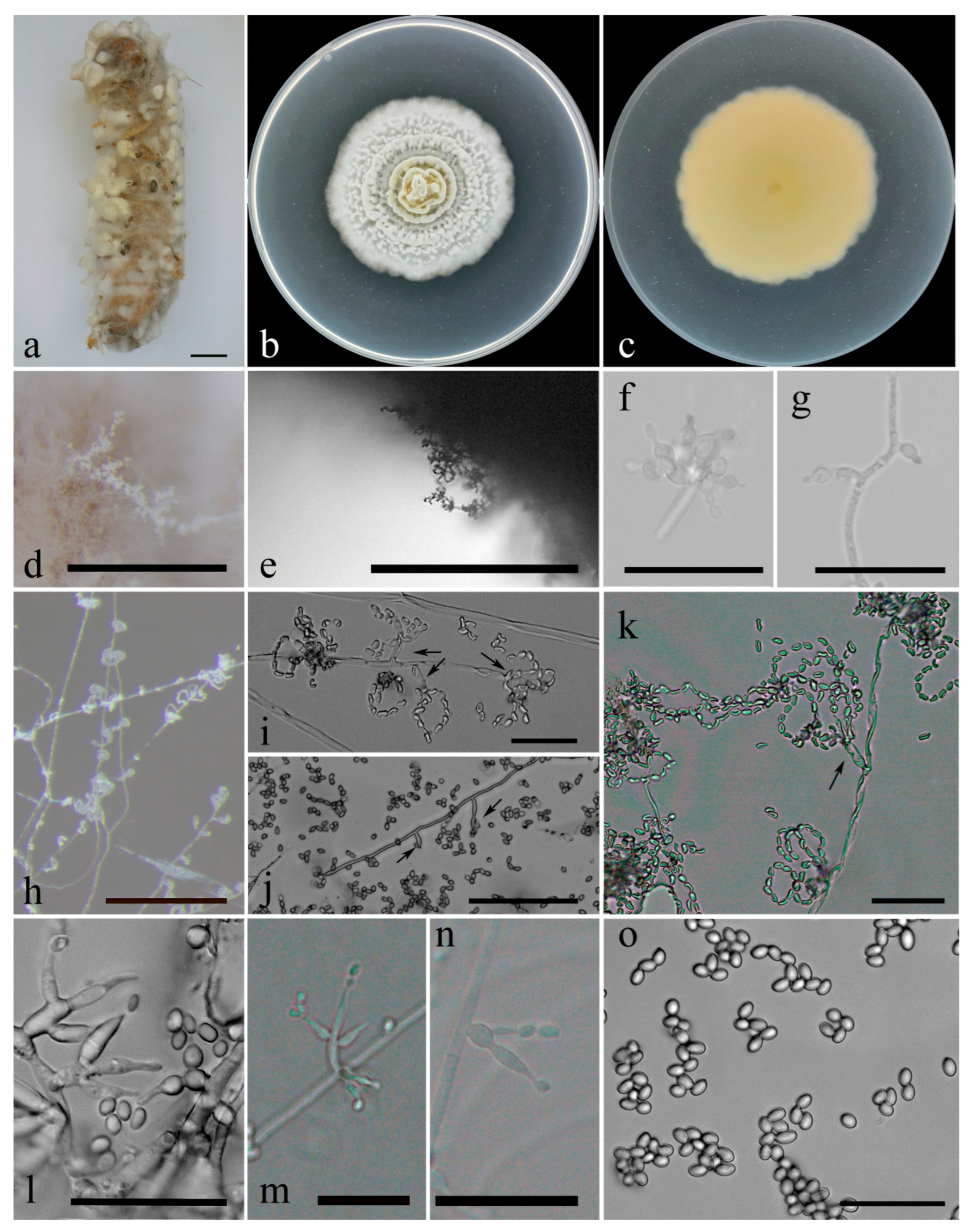
2.4. Morphology
2.5. Artificial Infection
3. Results
3.1. Phylogeny
3.2. Taxonomy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, G.-H.; Hywel-Jones, N.L.; Sung, J.-M.; Luangsa-Ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef] [PubMed]
- Kepler, R.M.; Luangsa-Ard, J.J.; Hywel-Jones, N.L.; Quandt, C.A.; Sung, G.-H.; Rehner, S.A.; Aime, M.C.; Henkel, T.W.; Sanjuan, T.; Zare, R.; et al. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 2017, 8, 335–353. [Google Scholar] [CrossRef]
- Xiao, Y.-P.; Wang, Y.B.; Hyde, K.D.; Eleni, G.; Sun, J.-Z.; Yang, Y.; Meng, J.; Yu, H.; Wen, T.-C. Polycephalomycetaceae, a new family of clavicipitoid fungi segregates from Ophiocordycipitaceae. Fungal Divers. 2023, 120, 1–76. [Google Scholar] [CrossRef]
- Koval, E.Z. Klavitsipitalnye Griby SSSR [Clavicipitalean Fungi of the USSR]; Naukova Dumka: Kiev, Russia, 1984; 288p. [Google Scholar]
- Kobayasi, Y. The genus Cordyceps and its allies. Sci. Rep. Tokyo Bunrika Daigaku Sect. B 1941, 84, 53–260. [Google Scholar]
- Kobayasi, Y. Keys to the taxa of the genera Cordyceps and Torrubiella. Trans. Mycol. Soc. Jpn. 1982, 23, 329–364. [Google Scholar]
- Koval, E.Z. Opredelitel Entomofilnych Gribov SSSR [Key to the Entomophilous Fungi of the USSR]; Naukova Dumka: Kiev, Russia, 1974; 260p. [Google Scholar]
- Mains, E.B. North American entomogenous species of Cordyceps. Mycologia 1958, 50, 169–222. [Google Scholar] [CrossRef]
- Borisov, B.A. Pathogens of fungal diseases of invertebrates in the “Kaluzhskiye Zaseki” Nature Reserve: Study results from the southern tract and adjacent areas. In Trudy Gosudarstvennogo prirodnogo zapovednika «Kaluzhskie zaseki» [State Nature Reserve “Kaluzhskiye Zaseki”]; Alekseev, S.K., Ed.; Ejdos: Kaluga, Russia, 2012; Volume 2, pp. 29–73. [Google Scholar]
- Borisov, B.A.; Alexandrova, A.V. Zooparazitic Cordycipitoid Fungi of the Moscow Region. In Proceedings of the Sovremennaya Mikologiya v Rossii, Materialy Tretego Sjezda Mikologov Rossii [Modern Mycology in Russia, Third Congress of Russian Mycologists], Moscow, Russia, 10–12 October 2012; Volume 3. [Google Scholar]
- Borisov, B.A. Zooparazitic Cordycipitoid Fungi (Ascomycota: Hypocreales) of the Sochi National Park and Adjacent Territory. In Proceedings of the Bioraznoobrazie i Ekologiya Gribov i Gribopodobnyh Organizmov Severnoj Evrazii, Materialy Vserossijskoj Konferencii s Mezhdunarodnym Uchastiem [Biodiversity and Ecology of Fungi and Fungus-like Organisms of Northern Eurasia, Proceedings of the All-Russian Conference with International Participation], Ekaterinburg, Russia, 20–24 April 2015. [Google Scholar]
- Prokhorov, V.P.; Borisov, B.A.; Voronina, E.Y.; Alexandrova, A.V. New data on Sordariomycetes and Dothideomycetes at the territory of Moscow State University Zvenigorod biological station and Sima open pit wildlife refuge. Mikol. Fitopatol. 2015, 49, 359–365. [Google Scholar]
- Borisov, B.A. Detection of tropical arthropod-pathogenic fungi Conoideocrella luteorostrata and Cordyceps thaxteri in the Moscow Region—New examples of the biota’s response to climate warming? In Proceedings of the Sovremennaya Mikologiya v Rossii, Materialy Chetvyortogo Sjezda Mikologov Rossii [Modern Mycology in Russia, Proceedings of the Fourth Congress of Russian mycologists], Moscow, Russia, 12–14 April 2017; Volume 6. [Google Scholar]
- Shrestha, B.; Tanaka, E.; Han, J.G.; Oh, J.; Han, S.K.; Lee, K.H.; Sung, G.H. A brief chronicle of the genus Cordyceps Fr., the oldest valid genus in Cordycipitaceae (Hypocreales, Ascomycota). Mycobiology 2014, 42, 93–99. [Google Scholar] [CrossRef]
- Borisov, B.A.; Alexandrova, A.V.; Antonov, E.A. An unusual discovery on the Kola Peninsula: The Cordyceps militaris fungi as an aphid parasite. In Proceedings of the Sovremennaya Mikologiya v Rossii, Materialy Mezhdunarodnogo Mikologicheskogo Foruma [Modern Mycology in Russia, Proceedings of the International Mycological Forum], Moscow, Russia, 22–23 May 2024; Volume 10. [Google Scholar]
- Zha, L.-S.; Kryukov, V.Y.; Ding, J.-H.; Jeewon, R.; Chomnunti, P. Novel taxa and species diversity of Cordyceps sensu lato (Hypocreales, Ascomycota) developing on wireworms (Elateroidea and Tenebrionoidea, Coleoptera). MycoKeys 2021, 78, 79–117. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Rehner, S.A.; Samuels, G.J. Molecular systematics of the Hypocreales: A teleomorph gene phylogeny and the status of the anamorphs. Can. J. Bot. 1995, 73, 816–823. [Google Scholar] [CrossRef]
- Castlebury, L.A.; Rossman, A.Y.; Sung, G.-H.; Hyten, A.S.; Spatafora, J.W. Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol. Res. 2004, 108, 864–872. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorph. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Gillespie, D. Preparative and analytical purification of DNA from agarose. Proc. Natl. Acad. Sci. USA 1979, 76, 615–619. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MegaX: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Samson, R.A.; Hoekstra, E.S.; Frisvad, J.C. Introduction to Food- and Airborne Fungi, 3rd ed.; Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands, 2004; p. 389. [Google Scholar]
- Liang, Z.Q. Two new species of Paecilomyces from insects. Acta Microbiol. Sin. 1981, 21, 31–34. [Google Scholar]
- Liang, Z.Q.; Han, Y.F.; Chu, H.L.; Liu, A.Y. Studies on the genus Paecilomyces in China. I. Fungal Divers. 2005, 20, 83–101. [Google Scholar]
- Shimazu, M. Paecilomyces cateniannulatus Liang, a commonly found, but an unrecorded entomogenous fungus in Japan. Appl. Entomol. Zool. 2001, 36, 283–288. [Google Scholar] [CrossRef]
- Wei, D.P.; Gentekaki, E.; Wanasinghe, D.N.; Tang, S.M.; Hyde, K.D. Diversity, molecular dating and ancestral characters state reconstruction of entomopathogenic fungi in Hypocreales. Mycosphere 2022, 13, 281–351. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kazartsev, I.; Gomzhina, M.; Levchenko, M.; Lednev, G. Cordyceps biarmica sp. nov., an Entomopathogenic Fungus from Boreal Forests of North European Russia. Diversity 2025, 17, 762. https://doi.org/10.3390/d17110762
Kazartsev I, Gomzhina M, Levchenko M, Lednev G. Cordyceps biarmica sp. nov., an Entomopathogenic Fungus from Boreal Forests of North European Russia. Diversity. 2025; 17(11):762. https://doi.org/10.3390/d17110762
Chicago/Turabian StyleKazartsev, Igor, Maria Gomzhina, Maxim Levchenko, and Georgy Lednev. 2025. "Cordyceps biarmica sp. nov., an Entomopathogenic Fungus from Boreal Forests of North European Russia" Diversity 17, no. 11: 762. https://doi.org/10.3390/d17110762
APA StyleKazartsev, I., Gomzhina, M., Levchenko, M., & Lednev, G. (2025). Cordyceps biarmica sp. nov., an Entomopathogenic Fungus from Boreal Forests of North European Russia. Diversity, 17(11), 762. https://doi.org/10.3390/d17110762








