Abstract
Bahaba taipingensis (Chinese bahaba) is a critically endangered fish endemic to China’s coastal waters, valued for both ecological and economic reasons and known as the “panda of the sea”. Captive breeding and stock enhancement are key conservation strategies, yet the genetic composition of released individuals directly affects program outcomes. This study combined mitochondrial and whole-genome resequencing to compare F1-generation fish with wild populations. At the mitochondrial level, 60 SNPs were detected in F1 individuals and 72 in wild populations, with haplotype analyses revealing retention of most common maternal lineages but reduced diversity. Nuclear genome analysis showed comparable genetic diversity between groups. Nucleotide diversity (π) was 0.000423 in F1 fish and 0.000401 in the wild population. However, the F1 cohort exhibited a higher inbreeding coefficient (FIS = −0.030) than the wild group (FIS = −0.118), suggesting early allele frequency shifts, thereby suggesting early genotype frequency shifts. Runs of homozygosity (ROH) analysis showed that the total number and length of ROH regions in the F1 cohort (686, 283,089.25 kb) were significantly greater than those in the wild population (171, 52,607.30 kb). Genome-wide FST between groups was 0.035, and PCA indicated genetic homogenization in F1 fish. Ne analysis showed that the wild population declined rapidly over generations and stabilized at a low level, indicating genetic diversity loss under environmental stress and highlighting the role of artificial breeding. These findings highlight the need for improved broodstock management and long-term genetic monitoring.
1. Introduction
The Bahaba taipingensis is the sole extant species of the genus Bahaba within the family Sciaenidae (order Perciformes) and is endemic to China, with its distribution largely restricted to the coastal waters of the East and South China Seas. Owing to its exceptional ecological and economic importance, as well as its extreme rarity, it has been widely referred to as the “panda of the sea” [1]. Ecologically, the Chinese bahaba is among the largest sciaenid species, occupying a high trophic level in nearshore food webs and playing a pivotal role in maintaining the structure and functioning of coastal ecosystems [2]. Economically, its swim bladder is highly prized for its perceived medicinal properties and has been traded at prices that often surpass the value of gold, making it a major target of illegal fishing [3,4]. In recent decades, accelerating industrial development has caused extensive degradation of the species’ natural habitats, while intense exploitation driven by high economic returns has further contributed to the dramatic decline of wild populations [5,6]. In recognition of its critical conservation status, the species was listed as Critically Endangered (CR) by the International Union for Conservation of Nature (IUCN) in 2006 [7] and was subsequently designated as a Class I protected aquatic wildlife species under China’s National Key Protected Wild Animals list [8]. A recent study reconstructed the population dynamics of the Chinese bahaba over the past five decades and revealed alarming trends. Since 1995, the maximum observed body length has decreased markedly—from nearly 2 m to under 1 m—and catch per unit effort (CPUE) has remained consistently near zero, indicating an extremely low level of wild abundance. Moreover, the species’ core distribution range has contracted by 69.5%, from approximately 104.4 km2 before 2000 to just 31.9 km2 in recent years [2]. Together, these findings underscore the species’ critically endangered status and highlight the urgent need for strengthened conservation interventions.
As one of the “Ten Treasures of the Ocean” selected by the World Wide Fund for Nature (WWF), the Chinese bahaba is among the most critically endangered yet least studied species on the list [9]. To mitigate the risk of extinction, captive breeding and stock enhancement have become key conservation strategies [10]. In 2005, a municipal nature reserve for the Chinese bahaba was established in Dongguan, Guangdong Province, marking the beginning of ex situ conservation efforts [11]. In 2021, the species successfully underwent artificial reproduction for the first time, representing a major milestone in conservation progress [12]. In recent years, a dedicated research program has been launched in Huidong, Guangdong, focusing on the Chinese bahaba in the Pearl River Estuary. This initiative aims to advance protective breeding and stock enhancement, providing scientific support for population recovery and habitat restoration of this endangered species [13].
Against the backdrop of ongoing conservation efforts, genetic research plays a critical role in guiding the scientific application of captive breeding and stock enhancement, ensuring that such interventions maximize genetic benefits while minimizing potential risks. Numerous studies, both domestic and international, have shown that poorly managed captive breeding and unregulated release programs can lead to local gene pool homogenization, inbreeding accumulation, and genetic drift, ultimately reducing the adaptive potential of populations and compromising the integrity of wild gene pools—often rendering enhancement programs ineffective [14,15,16]. However, genetic studies on Chinese bahaba to date have largely focused on microsatellite or SSR marker analyses within single geographic regions. While relatively high observed and expected heterozygosity values have been reported, limitations in sampling coverage and marker resolution prevent comprehensive assessment of genome-wide population structure and the genetic consequences of captive propagation [17,18]. Therefore, systematic evaluation of the genetic impact of captive interventions on the diversity and structure of Chinese bahaba populations is urgently needed not only when designing but also when implementing conservation breeding and release strategies.
A high-quality reference genome of Chinese bahaba was recently assembled [19], providing a foundation for the first integrated analysis of genetic differences between wild individuals from the Pearl River Estuary and first-generation hatchery-reared offspring (F1) using both complete mitochondrial genomes and whole-genome resequencing data. At the mitochondrial level, haplotype diversity is compared between the two groups. At the nuclear genome level, key genetic parameters including nucleotide diversity (π), observed heterozygosity (Ho), expected heterozygosity (He), inbreeding coefficient (FIS = (He − Ho)/He), and genetic differentiation index (FST) are evaluated. Runs of homozygosity (ROH) are analyzed to quantify genomic autozygosity and assess inbreeding signatures. Population structure is inferred using principal component analysis (PCA) and ADMIXTURE clustering, providing insights into potential genetic drift and structural divergence associated with early-stage domestication. Finally, effective population size (Ne) is estimated from linkage disequilibrium patterns to reconstruct recent demographic trajectories. It is hypothesized that, due to the limited broodstock base and selection pressures in captivity, the F1 cohort may already exhibit reduced genetic diversity and increased genetic homogeneity. Conversely, a lack of significant differences suggests that current breeding and release strategies effectively preserve the genetic integrity of hatchery-reared individuals.
This study not only provides a scientific basis for assessing the genetic risks associated with conservation breeding of B. taipingensis and optimizing broodstock selection strategies but also establishes a critical foundation for quantifying stock enhancement efforts, delineating management units, and conducting long-term monitoring of the species’ genetic health, while considering the impact of unequal reproductive success among broodstock on the genetic composition of the F1 cohort. Conversely, a lack of significant differences suggests that current breeding and release strategies to some degree preserve the genetic integrity of the augmented wild population. The hatchery-reared individuals provide the means for managing the imperiled wild population.
2. Materials and Methods
2.1. Ethical Statement
The care and use of experimental animals complied with China animal welfare laws, guidelines and policies, as approved by South China Normal University (permit reference number SCNU-SLS-2023-025, approved on May 25 2021). The Dongguan Bahaba taipingensis Nature Reserve was officially established on 9 May 2005 under the approval of the Dongguan Municipal Government (Document No. [2005]67). In addition, the collection and artificial propagation of wild fish were conducted under the official permit issued by the Department of Agriculture and Rural Affairs of Guangdong Province (permit number: No. 0926, approved on 7 December 2022).
2.2. Sample Collection
A total of 43 fin tissue samples of B. taipingensis were collected for this study, including 16 F1 individuals and 27 wild individuals. Fin clips were taken non-lethally from the distal margin of the caudal fin (or from the soft-rayed dorsal or pectoral fins when necessary). Approximately 4–5 mm of tissue was sampled, yielding 30–50 mg of tissue, which was sufficient for high-quality DNA ex-traction. The fin tissue was immediately placed in 95–100% ethanol at a tissue-to-ethanol ratio of at least 1:10. Ethanol was refreshed after 12–24 h, and the samples were stored at −20 °C for short-term storage and −80 °C for long-term preservation. For each fish, duplicate tissue samples were preserved (one for DNA extraction and the other as a backup and voucher), with the remaining DNA stored after extraction. Voucher samples (backup fin tissue and photographic records) were archived in the laboratory biobank and are available upon request.
The 27 wild individuals were obtained between 2010 and 2025 through rescue operations conducted in specific areas of the Pearl River Estuary; these fish were all immature at the time of rescue, fully recovered after rehabilitation, and subsequently released back into their habitats. None of these wild individuals were used as broodstock. The 16 F1 individuals were offspring produced in April 2022 through artificial breeding at the B. taipingensis Rescue Center (113°38′18″ E, 22°48′36″ N) (Figure 1). Their broodstock consisted of healthy, sexually mature fish rescued from the Pearl River Estuary between 2010 and 2022 and maintained at the center as part of the breeding population for stock enhancement. Artificial breeding was conducted using three groups of broodstock, each consisting of one male and three females (three males and nine females in total). Artificial breeding was conducted under controlled water temperature (28 ± 1 °C) and water quality conditions (dissolved oxygen ≥ 6 mg/L, pH 7.5–8.0, salinity 0‰). Hormonal induction of broodstock was achieved using a combination of three hormones: luteinizing hormone-releasing hormone analog (LRH-A), dopamine antagonist (such as domperidone), and carp pituitary extract. The three hormones were freshly prepared before injection; LRH-A was dissolved in 0.9% NaCl saline to a final concentration of 1 mg/mL, domperidone was dissolved in a small volume of 95% ethanol and diluted with 0.9% NaCl to a concentration of 10 mg/mL, and carp pituitary extract was dissolved in 0.9% NaCl to a final concentration of 2 mg/mL. These hormones were mixed in the same syringe according to the calculated doses for each individual, with the total injection volume adjusted to 0.5–1.5 mL per kg of body weight. Specific doses were as follows: LRH-A—0.5 mg/kg for females and 0.25 mg/kg for males; domperidone—10 mg/kg for both sexes; carp pituitary extract—0.1 mg/kg for both sexes. Broodstock were handled individually in separate tanks, ensuring accurate dosage based on body weight. After injection, the broodstock were maintained in appropriate environmental conditions, with monitoring of ovulation starting approximately 12 h post-injection. Once natural ovulation occurred, eggs were manually stripped and fertilized by the dry method, mixing the sperm and eggs. Fertilized eggs were incubated in aerated, temperature-controlled flow-through hatching tanks at 28 ± 1 °C, with water flow and oxygen levels carefully controlled, ensuring adequate aeration. The incubation period for the eggs was approximately 24–48 h, until the larvae hatched. The larvae were initially reared separately by family group to maintain pedigree integrity, then later combined for communal rearing. High-quality artificial feed was provided in batches to ensure optimal growth conditions. Finally, fin tissues were collected from 16 juveniles with uniform body size (approximately 10–15 cm in length and 20–40 g in weight), which were randomly selected from the mixed population as F1 samples for genetic analysis.
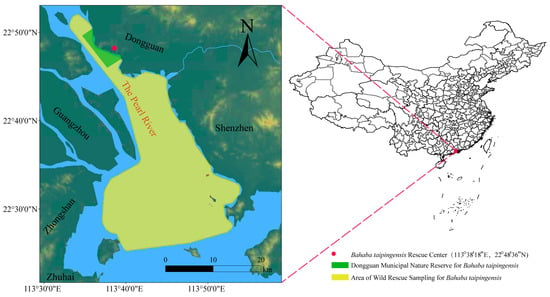
Figure 1.
Geographic locations of B. taipingensis wild sampling and conservation sites in the Pearl River Estuary. The yellow polygon marks the area of wild rescue sampling, and the green polygon indicates the Dongguan Municipal Nature Reserve for B. taipingensis. The red dot represents the B. taipingensis Rescue Center (113°38′18″ E, 22°48′36″ N), where broodstock maintenance and F1 breeding were conducted. Major geographic landmarks are shown in black and the main river channel (The Pearl River) is highlighted in brown.
2.3. DNA Extraction and Library Construction
Approximately 1.5 μg of genomic DNA from each individual was used for library preparation. Libraries were constructed using the ND627 Library Prep Kit (Novogene, Nanjing, China) following the manufacturer’s instructions. Each sample was assigned a unique index barcode to enable individual identification. Genomic DNA was sheared by sonication to an average fragment size of approximately 350 bp, followed by end-repair, A-tailing, and adaptor ligation. The ligated products were then PCR-amplified and purified using the AMPure XP system (Beckman Coulter, Brea, CA, USA).
Library quality and fragment size distribution were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and the final library concentrations were quantified by real-time quantitative PCR. Qualified libraries were sequenced at the Analytical Testing Center of the Institute of Hydrobiology, Chinese Academy of Sciences, using the DNBSEQ-T7 platform (BGI, Beijing, China), generating 150 bp paired-end reads with an average insert size of ~350 bp.
2.4. Mitochondrial Genome Assembly and Annotation
Mitochondrial genome sequences were extracted from Illumina sequencing data and assembled using the GetOrganelle v1.7.7.1 [20] and NOVOPlasty v4.3.5 [21] software. The mitochondrial genome of B. taipingensis (NCBI reference sequence: NC_018347.1) was used as a reference for annotation. Manual correction of potential annotation errors was performed using Geneious Prime v2022.2.2 [22].
2.5. Mitochondrial Data Analysis
Mitochondrial sequences were first aligned using Clustal Omega v1.2.2 [23] to ensure consistent sequence lengths and positional homology. The alignment results were visualized and manually inspected in Geneious Prime. To avoid potential biases caused by high variability, the D-loop region was excluded [24]. Haplotype types and their distribution within the F1 cohort and the wild population were identified using DnaSP v6.12.03 [25]. A median-joining haplotype network was then constructed in PopART v1.7 [26] to visualize the maternal genetic relationships between the two groups.
Phylogenetic analyses (excluding the D-loop region) were conducted using Bayesian inference (BI) methods in MrBayes v3.2.6 [27]. Firstly, all mitochondrial sequences were aligned in MEGA X [28] using the MUSCLE algorithm [29]. The aligned sequences were exported in NEXUS format and subjected to Bayesian Inference phylogenetic analysis in MrBayes v3.2.6. For BI analysis, the General Time Reversible (GTR) model with a gamma distribution of site rate variation (invgamma) was applied. Markov Chain Monte Carlo (MCMC) sampling was run for 1,100,000 generations with sampling every 200 generations. The first 10,000 trees were discarded as burn-in, and the remaining trees were used to calculate posterior probabilities and construct a consensus phylogenetic tree.
2.6. Whole-Genome Resequencing and SNP Detection
Raw sequencing reads in FASTQ format were processed using FASTP v0.24.0 [30] for quality control and adapter trimming. Specifically, five bases were trimmed from both ends of each read, a sliding window of five bases was applied with a minimum average Phred quality score threshold of 20, and reads shorter than 50 bp were removed. Cleaned reads were then aligned to the high-quality B. taipingensis reference genome using BWA-MEM v0.7.18 [31]. The resulting alignments were sorted and converted using SAMtools v1.21 [32].
SNP detection was performed using GATK4 v4.6.1.0 [33]. After read alignment, duplicate reads were marked and removed using the MarkDuplicates module to minimize PCR artifacts. Individual GVCF files were then generated for each sample using the HaplotypeCaller module. These were subsequently merged with CombineGVCFs by population group and jointly genotyped to produce a population-level VCF file. SNPs were filtered using VariantFiltration with the following criteria: QD < 2.0, MQ < 40.0, FS > 60.0, SOR > 3.0, MQRankSum < −12.5, and ReadPosRank-Sum < −8.0. High-quality SNPs that passed these filters in both the wild and F1 groups were retained for downstream analyses.
2.7. Population Genetic Diversity
To minimize false-positive variants caused by misalignment, all InDel sites and the surrounding ±5 bp regions were excluded using the vcfutils.pl script from SAMtools v1.21. Subsequent quality filtering was performed with BCFtools v1.21 [34], retaining only high-quality bi-allelic SNPs that met the criteria: QUAL > 30, DP > 5, MQ > 40, QD > 2, and F_MISSING ≤ 0.05. This final dataset was defined as the full genome-wide SNP set. To identify outlier variants, the OutFLANK v0.2 R package [35] was used to apply a robust method for detecting loci under selection. The analysis was performed using the full genome-wide SNP set, with parameters set as follows: LeftTrim-Fraction = 0.05, RightTrimFraction = 0.02, Hmin = 0.05, and a q-value threshold of 0.10. Loci flagged as outliers were removed and the remaining variants were considered as the hypothetical neutral SNP set, which was used for downstream population genetic analyses.
Based on the filtered dataset, the neutral SNP set was defined from the combined dataset of the two groups after removing outliers. SNP distribution across the 24 chromosomes was visualized to provide an overview of genome-wide variation patterns.
VCFtools v0.1.16 [36] was employed to compute π, Ho, and He for both the F1 cohort and the wild population, enabling assessment of within-population genetic diversity based on the neutral SNP set. Average π values between the two groups were compared, and sliding window analysis (window size: 100 kb; step size: 10 kb) was applied across the 24 chromosomes to visualize local variation in nucleotide diversity. For heterozygosity metrics, intra-population comparisons between Ho and He were conducted using the Mann–Whitney U test to evaluate deviations from Hardy–Weinberg equilibrium. The same test was applied to compare Ho and FIS between two groups, to assess differences in overall heterozygosity levels. ROHs were identified using PLINK v1.90b6.21 [36] with the --homozyg function, based on the neutral SNP set to preserve segment continuity. The following parameters were applied: --homozyg-density 50, --homozyg-gap 1000, --homozyg-kb 500, --homozyg-snp 50, --homozyg-window-het 1, --homozyg-window-snp 50, and --homozyg-window-threshold 0.05.
2.8. Population Differentiation and Structure Analysis
To evaluate the extent of genetic differentiation between the two groups, FST values and FST_NoCorr values were calculated using OutFLANK v0.2 R package based on the neutral SNP set. The frequency distribution of FST_NoCorr values was calculated using the seaborn v0.11.2 package in Python and visualized with the matplotlib v3.5.2 package [37], with an overlaid kernel density estimation (KDE) curve to depict the overall genetic differentiation across the genome. Manhattan plots were also generated to display FST values across 24 chromosomes, with a red horizontal line indicating the 99th percentile threshold, identifying regions with higher FST values.
To ensure independence among SNPs used for structure analyses, linkage disequilibrium (LD) pruning was conducted using PLINK v1.90b6.21 [38], with a sliding window of 50 SNPs, a step size of 10 SNPs, and an R2 threshold of 0.8. The pruned neutral SNP panel (excluding FST outliers) was then used for downstream structure inference. Population structure was inferred using ADMIXTURE v1.3.0 [39] by testing K values from 1 to 10, and the model with the lowest cross-validation (CV) error was selected as the optimal clustering solution. Prior to principal component analysis (PCA), a quantile–quantile (Q–Q) plot was generated by comparing the distribution of the first two principal components (PC1 and PC2) with the theoretical quantiles of a normal distribution, to evaluate potential systematic deviations or outliers and assess the suitability of the data for clustering. PCA was then performed using PLINK v1.90b6.21, and the resulting eigenvectors were subjected to K-means clustering. A 95% confidence ellipse was overlaid on the PCA scatter plot to visualize and delineate the boundaries of the inferred genetic clusters.
Ne was estimated using GONE (release updated on 20 July 2025) [40] based on the pruned neutral SNP panel, which infers recent effective population size trajectories from patterns of linkage disequilibrium. SNPs passing filters were converted to PED/MAP format as the GONE input. Analyses were run under default parameters with 40 independent replicates per dataset, and Ne estimates across replicates were summarized as the geometric mean. Ne trajectories were visualized across generations on both logarithmic and linear time scales, with particular focus on the most recent 1–100 generations.
3. Result
3.1. Mitochondrial Genetic Assessment
By stringently filtering the raw sequencing reads, approximately 427.4 Gb of high-quality clean data were obtained at an average sequencing depth of 14× across the nuclear genome (Table S1).
Mitochondrial genomes from the F1 cohort and the wild population were then assembled and aligned to assess maternal genetic differences. SNPs were detected across most regions of the mitochondrial genome, with a pronounced clustering of variation in the D-loop region. Nevertheless, the total number of SNPs in the F1 cohort was slightly lower than in the wild population, suggesting a relatively limited maternal lineage diversity in the hatchery stock. In terms of SNP counts across functional regions of the mitochondrial genome (Figure 2a,b), a total of 72 SNPs were identified in the wild population and 60 in the F1 cohort, indicating a relatively small difference. Most major coding regions in the F1 mitochondrial genome showed detectable SNP variation, with comparable counts to those in the wild population. For instance, six and five SNPs were detected in the ND2 region, four and three in ND4, eight and six in ND5, and four and three in COX3 for wild and F1 cohort, respectively. These results suggest that the F1 cohort retains many haplotypes found in the wild population. Notably, some SNPs in certain regions such as ATP8 were exclusively detected in the wild population, indicating the absence of variation at some sites in the F1 cohort. Additionally, both groups exhibited limited but consistent variation in tRNA regions, such as tRNA-Asn, tRNA-Lys, tRNA-Glu, and tRNA-Thr, reflecting a degree of conservation and lineage representation.
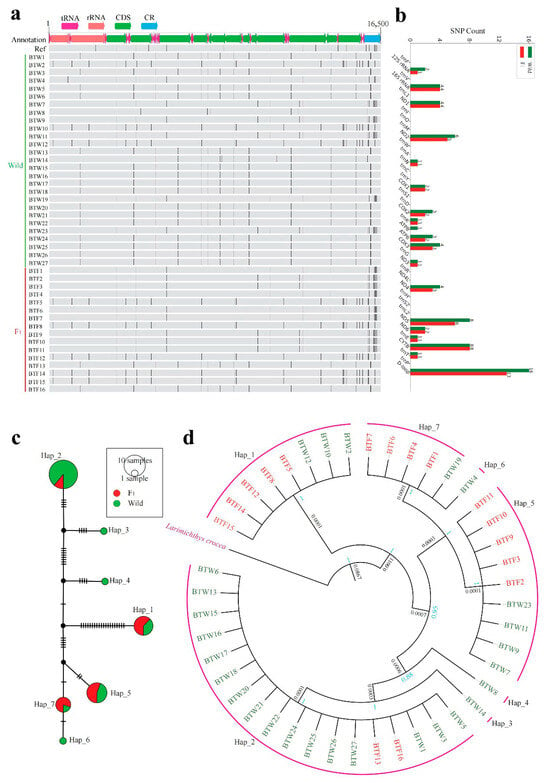
Figure 2.
Mitochondrial genome variation and maternal lineage structure of the wild population and the F1 cohort. (a) SNP distribution across the mitochondrial genome. Each black vertical line represents a SNP. Wild individuals (BTW1–27) are shown in green, F1 individuals (BTF1–16) in red. Genomic features include tRNA, rRNA, CDS, and the control region (CR). (b) Comparison of SNP counts across mitochondrial functional regions between the wild population and the F1 cohort. (c) Median-joining haplotype network based on mitochondrial sequences (excluding the D-loop). Circle size reflects haplotype frequency; colors denote group origin (green = wild, red = F1). (d) Phylogenetic tree inferred from mitochondrial sequences (excluding the D-loop) using Bayesian inference. Larimichthys crocea served as the outgroup. Node values indicate Bayesian posterior probabilities (blue); the black number represents the genetic distance at the basal divergence between the outgroup and the ingroup.
Haplotype network analysis (Figure 2c) identified seven maternal haplotypes, with the F1 cohort primarily grouped into Hap_1, Hap_2, Hap_5, and Hap_7. Among these, Hap_1, Hap_2, and Hap_5 were also the most frequent haplotypes in the wild population, suggesting that the F1 cohort successfully encompassed the most common maternal lineages. The phylogenetic tree (Figure 2d) further supported this pattern, showing that F1 cohort did not exhibit extreme homogeneity but rather formed several lineage branches, with some individuals clustering alongside wild counterparts—indicating a degree of genetic continuity.
3.2. Population Genetic Diversity Analysis
To compare the nuclear genomic diversity between the F1 cohort and the wild populations, several genetic parameters were calculated based on high-quality SNPs obtained after stringent filtering, including π, Ho, He, FIS, and ROH (Figure 3 and Figure 4, Table 1).
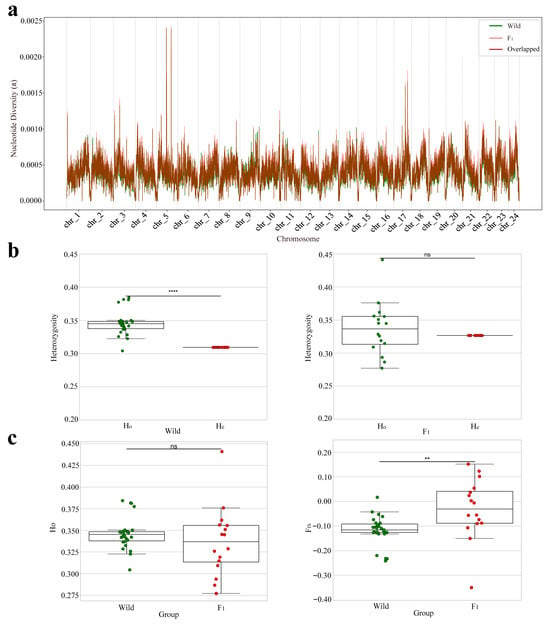
Figure 3.
Nuclear genomic diversity and population genetic structure in the wild population and the F1 cohort. (a) Sliding window analysis of nucleotide diversity (π) along each chromosome (window size: 100 kb; step size: 10 kb). The wild population and the F1 cohort are shown in green and red, respectively, with overlapping regions indicated in dark brown. (b) Comparison of Ho and He within each group. Statistical significance is indicated as follows: ns = not significant (p > 0.05); ** = p < 0.01; **** = p < 0.0001. (c) Between-group comparisons of Ho and FIS. Significance levels follow the same notation as in panel (b).
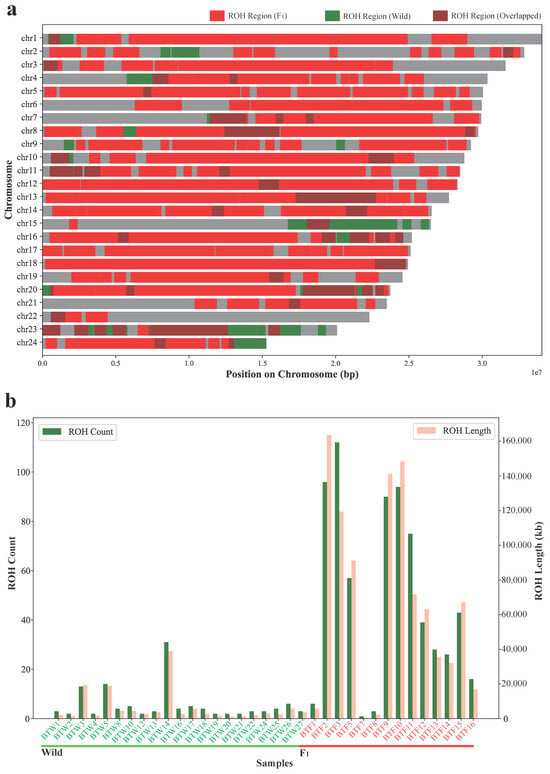
Figure 4.
Comparison of the ROH patterns between the wild population and the F1 cohort. (a) Genomic distribution of ROH regions across 24 chromosomes. ROH regions specific to wild individuals (BTW1–27) are shown in red, those specific to F1 individuals (BTF1–16) in green, and overlapped ROH regions shared by both groups are marked in brown. Each chromosome is shown as a horizontal bar scaled to its actual length. (b) ROH statistics for each individual, including the number of ROH segments (green bars, left y-axis) and total ROH length (pink bars, right y-axis). Wild individuals are labeled in green (BTW1–27) and F1 individuals in red (BTF1–16).

Table 1.
Summary of genome-wide genetic diversity and differentiation metrics in the wild population and the F1 cohort.
In terms of SNP abundance, the F1 cohort harbored a total of 2.255 × 106 of high-quality SNPs, slightly fewer than the 2.565 × 106 identified in the wild population (Table S2). To reduce the potential confounding effects of loci under selection, we further applied OutFLANK to detect and remove outlier SNPs, resulting in a final neutral SNP set of 824,587 variants that was used for downstream population genetic analyses (Figure S1).
Nucleotide diversity analysis (Figure 3a, Table 1 and Table S3) revealed that the average π value in the F1 cohort was 0.000423, slightly higher than that of the wild population (0.000401). The chromosomal patterns of π were highly concordant between the two groups, with only minor local deviations, suggesting that the F1 individuals retained a relatively high level of genome-wide genetic diversity.
Heterozygosity analysis (Figure 3b, Table 1 and Table S4) showed that the wild population exhibited significantly higher Ho (0.3460) than He (0.3094) (****, p < 0.0001), indicating a heterozygote excess (negative FIS). In contrast, the F1 cohort showed no significant difference between Ho (0.3366) and He (0.3267) (ns, p > 0.05), implying that it was approximately in Hardy–Weinberg equilibrium. Between-population comparisons of heterozygosity (Figure 3c) indicated that the F1 cohort had a slightly lower Ho than the wild population, though the difference was not statistically significant (ns, p > 0.05). However, the average FIS in the F1 cohort was −0.030, which—although negative—was significantly higher than that of the wild population (−0.118; **, p < 0.01), indicating a weaker heterozygote excess in F1 and a broader inter-individual spread, which may reflect allele frequency shifts caused by the limited number of broodstock or unequal parental contributions.
The ROH analysis (Figure 4, Table 1 and Table S5) revealed significant differences between the wild population and the F1 cohort in terms of both the total count and length of ROH regions. The wild population exhibited a relatively low total of 117 ROH regions, with a combined length of 52,607.30 kb. In contrast, the F1 cohort showed a significantly higher total of 686 ROH regions, spanning 283,089.25 kb. These results indicate a marked increase in homozygosity in the F1 cohort, which could be a result of inbreeding or limited genetic variation due to a restricted broodstock. The distribution of ROH across chromosomes (Figure 4a) showed that while the majority of ROH regions overlapped between both populations, the F1 cohort had a higher density of ROH regions, especially in specific chromosomal regions. The differences in ROH length and count between the two groups (Figure 4b) further support the idea that the F1 cohort has experienced greater levels of homozygosity.
Overall, the F1 cohort retained comparable genetic diversity to the wild population in terms of nucleotide diversity and heterozygosity. However, the F1 cohort exhibited a weaker heterozygote excess, as indicated by a higher FIS value. The ROH analysis showed a significant increase in homozygosity in the F1 cohort, suggesting a narrowing of the genetic base, likely due to inbreeding or a limited broodstock.
3.3. Population Structure Analysis
To explore whether there is underlying genetic differentiation structure between the wild population and the F1 cohort, an FST analysis was performed. The genome-wide average FST between the two groups was 0.035 (Table 1), within the range of low genetic differentiation. A total of 8232 neutral SNPs were identified as exceeding the top 1% FST threshold (FST > 0.2602). These highly differentiated loci were distributed across multiple chromosomal regions (Figure 5a).
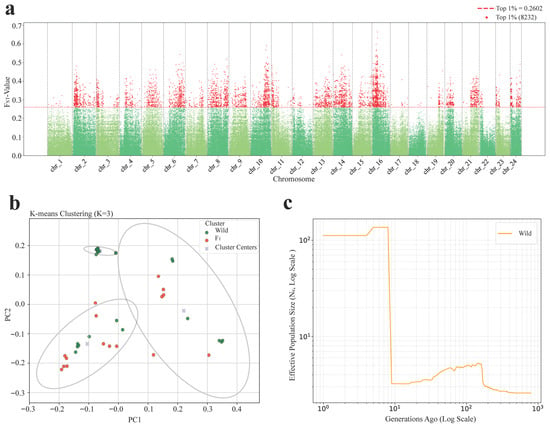
Figure 5.
Genome-wide population differentiation and structure of the wild population and the F1 cohort. (a) Manhattan plot of genetic differentiation (FST) between the wild population and the F1 cohort. Red dots represent loci in the top 1% of FST values and the red dashed line indicates the threshold (FST = 0.2602). (b) Principal component analysis (PCA) with K-means clustering (K = 3). Points represent individuals, with ellipses indicating 95% confidence intervals. (c) Ne trajectories of the wild (orange) and F1 (blue) groups, inferred from linkage disequilibrium. Left: long-term dynamics on a log scale.
Principal component analysis (PCA), combined with K-means clustering with the optimal cluster number K = 3 (Figure S3), further supported the population structure differences observed (Figure 5b). The first two principal components (PC1 and PC2) clearly separated individuals into three clusters. While most F1 individuals clustered tightly together, a few were scattered into other clusters and showed partial overlap with wild individuals, indicating a degree of genetic continuity between the two groups. Meanwhile, wild individuals displayed a more dispersed distribution, suggesting higher structural heterogeneity.
The Ne analysis for the wild population (Figure 5c and Figure S5) provided insight into the genetic dynamics. The wild population showed a relatively high Ne in the early phase, followed by a sharp decline and stabilization at a small value. This small stable estimate may partly reflect historical bottlenecks in the natural population but is also likely influenced by the limitations of the GONE under small sample sizes [40].
These results, together with the FST and population structure analyses, demonstrate that although the F1 cohort retains traces of ancestral lineages from wild individuals, its overall genetic composition has become more homogeneous and exhibits genetic convergence.
4. Discussion
4.1. Decline in Maternal Lineage Diversity
In the F1 cohort of B. taipingensis, 60 mitochondrial SNP sites were detected—compared to 72 in the wild—and a marked reduction in haplotype number was observed, with only four major lineages represented. Although these haplotypes include the most common wild maternal lineages (e.g., Hap_2, Hap_5 and Hap_7), indicating that the breeding strategy has preserved key variants, the overall loss of lineage coverage signifies a contraction of maternal diversity. Similar reductions in mitochondrial haplotype richness have been documented in other endangered fishes. In a restocking study of Brycon opalinus in Brazil, Hilsdorf et al. (2002) reported that hatchery-reared cohorts only partially encompassed the wild haplotypic spectrum, attributable to limited broodstock collection areas [41]. Zhang et al. (2017) found that cultured Larimichthys polyactis retained only seven COI haplotypes versus 27 in wild populations [16]. In Larimichthys crocea, Yuan et al. (2021) identified 199 combined COI + Cyt b haplotypes in wild fish (haplotype diversity = 0.983) but only 30 in cultured stocks (haplotype diversity = 0.704), underscoring substantial lineage loss during broodstock assembly [42]. Collectively, these studies underscore the point that genetic homogenization may emerge before significant declines in diversity, highlighting the need for ongoing genetic monitoring of cultured broodstock and outplanted stock. Allendorf et al. (2010) emphasize that the depletion of mitochondrial diversity can limit a population’s resilience to environmental stressors, a critical consideration in intensive restocking programs [43].
4.2. Nuclear Genomic Diversity and Genetic Homogenization in the F1 Generation
Genome-wide analyses indicate that the F1 cohort maintains a level of nuclear genomic diversity comparable to that of the wild population. The average π value for the F1 cohort was 0.000423, slightly higher than that of the wild population (0.000401), with highly similar distribution patterns of the average π values across chromosomal intervals. The mean Ho was nearly equal to the mean He, suggesting that no significant genetic erosion occurred after one generation of captive breeding. Further analysis at the individual level revealed that, in the wild population, Ho was significantly higher than He, suggesting the presence of underlying population structure. In contrast, no significant difference between Ho and He was observed in the F1 cohort, indicating that it was approximately in Hardy–Weinberg equilibrium. When comparing between groups, the mean Ho of the F1 cohort was slightly lower than that of the wild population but not significantly so, whereas the FIS was markedly elevated in the F1 cohort. This suggests that, despite the overall retention of genetic diversity, there may already be signs of genotype frequency shifts or genetic homogenization arising from captive breeding. The analysis of ROH further supports this observation. The F1 cohort showed a significant increase in both the total number and length of ROH regions, with 686 regions spanning 283,089.25 kb, compared to 117 regions and 52,607.30 kb in the wild population. This higher homozygosity suggests that the F1 cohort may have experienced increased homozygosity in certain regions, which could be related to the limited number of broodstock or restricted genetic variation. Moreover, the higher density of ROH in specific chromosomal regions indicates that long-term artificial breeding may lead to a gradual reduction in genetic diversity. Similar findings have been reported in Piaractus mesopotamicus and Labeo rohita, where cultured stocks exhibited Ho and He values equivalent to those of the wild population, although long-term mismanagement of broodstock can still lead to elevated FIS and reduced effective population size [44,45].
This potential genetic convergence is further supported by population structure analyses. The genome-wide FST average between the F1 cohort and the wild population is 0.035, indicating a certain degree of genetic similarity between the two groups. An OutFLANK scan identified 8232 highly differentiated neutral loci (FST > 0.2602, in the top 1%), scattered across multiple chromosomes, primarily reflecting the effects of genetic drift or other non-selective factors, which may be associated with the limited number of broodstock and restricted genetic variation in artificial breeding. The PCA and K-means clustering results showed that individuals of the F1 cohort were overall more tightly clustered in genetic structure, whereas the wild population displayed a more dispersed distribution, reflecting greater genetic heterogeneity. This pattern is consistent with the increased FIS and the higher number of ROH regions observed in the F1 cohort. Such a pattern is closely related to the limited number of broodstock and relatively fixed mating schemes in artificial breeding, which compress the genetic differences among offspring and lead to a more homogeneous population structure. At the same time, a few F1 individuals overlapped with wild individuals, indicating that the artificially bred cohort has retained a certain degree of genetic continuity with the wild population. However, long-term reliance on a limited number of broodstock in artificial breeding may exacerbate genetic convergence and increase the risk of inbreeding. The Ne analysis shows that the wild population experienced a rapid decline in Ne after several generations, this may be associated with environmental pressures, habitat changes, and genetic drift, as well as the influence of the population’s historical dynamics. Additionally, the limitation of the current sample size may also contribute to these observations. Overall, the wild population is facing multiple threats, including environmental pressures, habitat loss, overfishing, and genetic drift, which have led to a decline in population size and erosion of genetic diversity. Against this backdrop, artificial breeding and stock enhancement have become essential measures for maintaining and restoring wild populations. However, the limited number of broodstock and restricted mating schemes in artificial breeding can easily result in genetic convergence and inbreeding, thereby reducing the adaptive potential of the population.
Such structural homogenization is common in cultured populations. Brown et al. (2024) and Geletu et al. (2023) both demonstrated that closed breeding systems with limited broodstock promote genetic homogenization [46,47]. Zhang et al. (2022) used microsatellites in Procypris rabaudi to show that hatchery cohorts’ Ho/He declined from 0.79/0.86 to 0.75/0.77, with FST = 0.05–0.06 and tight clustering in structure analyses [48]. Guo et al. (2022) similarly found significant structuring in Pelteobagrus fulvidraco, with cultured stocks forming distinct clusters and exhibiting higher FST than some wild groups [49]. Ukenye and Megbowon (2023) [50] reported that wild Oreochromis niloticus populations had higher effective numbers of alleles, Shannon diversity, Ho, and He compared to cultured populations. They also found that cultured fish clustered distinctly in PCA, suggesting that suboptimal broodstock management contributed to reduced diversity and genetic homogenization [50]. Finally, Yuan et al. (2024) showed in Larimichthys crocea that cultured individuals exhibit a ~20% drop in Ho/He, ~25% reduction in π, LD decay extending from ~5 kb to ~30 kb, and ROH lengths increasing from 0.5 Mb to 1.1 Mb, with all cultured fish clustering in structure analyses apart from wild mixtures [15]. Collectively, these studies underscore that genetic homogenization may emerge even before significant declines in diversity, highlighting the need for ongoing genetic monitoring of cultured broodstock.
4.3. Recommendations for Genetic Management and Optimization
Although the F1 cohort has not yet shown significant declines in diversity metrics, its incipient genetic homogenization and emergence of highly differentiated genomic regions represent an early warning of potential genetic erosion. To safeguard long-term diversity and adaptive potential, we recommend that future broodstock management should (1) expand broodstock size and optimize mating designs—for example, through multi-parent crosses—to avoid over-reliance on a few families and reduce inbreeding accumulation and genetic homogenization; (2) introduce new broodstock every year to enhance genetic diversity; (3) balance family contributions to prevent over-representation from certain parents [46,47]; and (4) establish a standardized molecular monitoring framework—using ddRAD-seq or whole-genome resequencing—to track changes in population structure across generations and build a long-term genetic baseline database [14,48]. By integrating these measures with continuous genetic assessments, breeding programs can counteract early signs of genetic homogenization, maintain high levels of genomic diversity, and enhance the resilience and restoration potential of cultured cohorts.
5. Conclusions
This study provides the first comprehensive comparison of mitochondrial and nuclear genomic diversity between wild and F1 captive-bred populations of B. taipingensis. The results show that while the F1 cohort retains comparable levels of nuclear genomic diversity to wild populations, mitochondrial analyses revealed a contraction in maternal haplotype diversity, with most common wild haplotypes preserved but overall lineage representation reduced. Additionally, early signs of genetic homogenization and genotype frequency shifts were observed in the F1 cohort, as reflected by elevated FIS values, a significant increase in ROH regions, and clustering patterns seen in PCA analyses. Genome-wide FST analysis identified thousands of highly differentiated neutral loci, suggesting that the divergence at these loci may be associated with genetic drift or other non-selective factors. Ne analysis further revealed that the wild population experienced a rapid decline after several generations and then stabilized at a lower level, reflecting the gradual loss of genetic diversity under environmental pressures, habitat changes, and genetic drift, thereby highlighting the importance of artificial breeding. These findings highlight potential genetic risks in the early stages of stock enhancement and underscore the need for optimized broodstock management, particularly by maintaining a broad parental genetic pool and implementing rigorous genetic monitoring. Establishing long-term molecular surveillance will be essential for preserving genetic diversity, mitigating inbreeding, and supporting the sustainable recovery of this critically endangered species.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d17100676/s1, Figure S1: The neutral SNP count across the 24 chromosomes of the F1 cohort; Figure S2: Genome-wide distribution of raw FST estimates (FST_NoCorr) between the wild population and the F1 cohort based on OutFLANK analysis; Figure S3: Cross-validation error plot from ADMIXTURE analysis to determine the optimal number of ancestral clusters (K); Figure S4: Quantile–Quantile (Q–Q) plots of principal components for normality assessment prior to clustering; Figure S5: Effective population size (Ne) of the wild Bahaba taipingensis population inferred over the past 100 generations; Table S1: Statistical Table of Effective Data Output, Mapping Results and Coverage; Table S2: SNP counts per chromosome in the wild population and the F1 cohort; Table S3: Mean π-value per chromosome in the wild population and the F1 cohort; Table S4: Comparison of Ho, He, and FIS between F1 and Wild Samples; Table S5: Comparison of ROH Counts and Lengths Between Wild and F1 Groups.
Author Contributions
Y.H., methodology, software, formal analysis, validation, visualization, writing—original draft preparation; Q.C., investigation, visualization, data curation; J.C., investigation, data curation, resources; W.C., visualization, data curation; J.W., (Jujing Wang), visualization, writing—original draft preparation; H.L. (Haimei Lin), investigation, data curation; G.C., investigation, data curation; J.X., visualization; H.L. (Hungdu Lin), visualization, writing—original draft preparation; W.F., visualization, project administration, supervision; J.W. (Junjie Wang), supervision, funding acquisition, project administration, conceptualization. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by a grant (#441901202109379) from Monitoring of Marine Biological Resources and Environment in Dongguan Bahaba taipingensis Nature Reserve. It was also supported by the Project of Financial Funds of Ministry of Agriculture and Rural Affairs: Investigation of Fishery Resources and Habitat in the Pearl River Basin [ZJZX-06], and by the China–ASEAN Maritime Cooperation Fund [CAMC-2018F].
Data Availability Statement
The sequencing data generated in this study have been deposited in the National Genomics Data Center (NGDC) under BioProject accession number PRJCA045418 (https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA045418, accessed on 4 September 2025)). The scripts used for data processing and intermediate files are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gu, Y.G.; Huang, H.H.; Liang, Y.; Fang, Y.; Dai, M.; Ou, Y.J.; Wang, L.G.; Wang, X.N. Micro-CT and SEM investigation of sound absorption structure and chambers in the otoliths of Giant Panda fish species-Chinese Bahaba (Bahaba taipingensis). Micron 2022, 161, 103342. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, Y.; Li, M.; Zhang, X. Decadal population depletion, size class reduction, and range contraction of the giant yellow croaker in China: Implications for conservation and management. Ocean Coast. Manag. 2025, 265, 107659. [Google Scholar] [CrossRef]
- Boilevin, V.; Crosta, A.; Hennige, S.J. Addressing illegal transnational trade of totoaba and its role in the possible extinction of the vaquita. J. Int. Wildl. Law Policy 2023, 26, 104–134. [Google Scholar] [CrossRef]
- Cruz-López, H.; Rodríguez-Morales, S.; Enríquez-Paredes, L.M.; Villarreal-Gómez, L.J.; True, C.; Olivera-Castillo, L.; Fernández-Velasco, D.A.; López, L.M. Swim bladder of farmed Totoaba macdonaldi: A source of value-added collagen. Mar. Drugs 2023, 21, 173. [Google Scholar] [CrossRef]
- Rodenbiker, J. Shark fin city: Transitional marine wildlife economies in global Hong Kong. Urban Geogr. 2025, 46, 155–179. [Google Scholar] [CrossRef]
- Ntho, A.; Ny, W. How ‘cocaine of the seas’ is wreaking ecological mayhem. Nature 2024, 634, 10. [Google Scholar]
- Wang, Y.; Hu, M.; Sadovy, Y.; Cheung, S.G.; Shin, P.K. Threatened fishes of the world: Bahaba taipingensis Herre, 1932 (Sciaenidae). Environ. Biol. Fishes 2009, 85, 335–336. [Google Scholar] [CrossRef]
- National Forestry and Grassland Administration; Ministry of Agriculture and Rural Affairs of China. List of National Key Protected Wild Animals; Announcement No. 3; National Forestry and Grassland Administration: Beijing, China, 2021.
- WWF Hong Kong. Oceans 10: Sitemap. Available online: www.wwf.org.hk (accessed on 4 September 2025).
- Tave, D. Conservation Aquaculture: An Evolution-Based Approach for the Production of Fish for Aquaculture-Assisted Fisheries Programs; Springer Nature: Cham, Switzerland, 2025. [Google Scholar]
- Zhu, H.L. Dongguan establishes its first aquatic flora and fauna nature reserve. Guangdong Sci. Technol. News 2005, 19, 22. [Google Scholar]
- Lu, W.H. Study on Rescue, Acclimatization, and Artificial Breeding Techniques of Bahaba taipingensis; South China Institute of Special Aquatic Products: Dongguan, China, 2021. [Google Scholar]
- Cai, W.J.; Li, A.P.; Wang, X.Y. Huizhou launches national key R&D project on Bahaba taipingensis. Huizhou Daily, 16 January 2025; p. 6. [Google Scholar] [CrossRef]
- Hohenlohe, P.A.; Funk, W.C.; Rajora, O.P. Population genomics for wildlife conservation and management. Mol. Ecol. 2021, 30, 62–82. [Google Scholar] [CrossRef]
- Yuan, J.; Zhuang, X.; Wu, L.; Lin, H.; Li, Y.; Wu, L.; Yao, J.; Liu, J.; Ding, S. Assessing the population genetic structure of yellow croaker in China: Insights into the ecological and genetic consequences of artificial breeding on natural populations. Aquaculture 2024, 590, 741026. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, F.; Wang, Z.; You, Q.; Lou, B.; Xu, D.; Chen, R.; Zhan, W.; Liu, F. Mitochondrial DNA variation and population genetic structure in the small yellow croaker at the coast of Yellow Sea and East China Sea. Biochem. Syst. Ecol. 2017, 71, 236–243. [Google Scholar] [CrossRef]
- Zhao, Y.; Ou, Y.; Wen, J.; Li, J.; Zhou, H. Analysis of genetic diversity of Bahaba taipingensis based on microsatellite markers. South China Fish. Sci. 2019, 15, 127–132. [Google Scholar]
- Zhao, Y.; Ou, Y.; Wen, J.; Li, J.; Zhou, H. Screening of SSR molecular markers in Bahaba taipingensis based on transcriptome sequencing technology. South China Fish. Sci. 2019, 15, 133–139. [Google Scholar]
- Cui, R.; Wu, J.; Yan, K.; Luo, S.; Hu, Y.; Feng, W.; Lu, B.; Wang, J. Phased genome assemblies reveal haplotype-specific genetic load in the critically endangered Chinese Bahaba (Teleostei, Sciaenidae). Mol. Ecol. 2024, 33, e17250. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [PubMed]
- Jaimes, J.A.; André, N.M.; Chappie, J.S.; Millet, J.K.; Whittaker, G.R. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020, 432, 3309–3325. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal Omega, accurate alignment of very large numbers of sequences. In Multiple Sequence Alignment Methods; Humana Press: Totowa, NJ, USA, 2013; pp. 105–116. [Google Scholar]
- Bronstein, O.; Kroh, A.; Haring, E. Mind the gap! The mitochondrial control region and its power as a phylogenetic marker in echinoids. BMC Evol. Biol. 2018, 18, 80. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D.; Nakagawa, S. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; O’Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra; O’Reilly Media: Sebastopol, CA, USA, 2020. [Google Scholar]
- Genovese, G.; Rockweiler, N.B.; Gorman, B.R.; Bigdeli, T.B.; Pato, M.T.; Pato, C.N.; Ichihara, K.; McCarroll, S.A. BCFtools/liftover: An accurate and comprehensive tool to convert genetic variants across genome assemblies. Bioinformatics 2024, 40, btae038. [Google Scholar] [CrossRef]
- Whitlock, M.C.; Lotterhos, K.E. Reliable detection of loci responsible for local adaptation: Inference of a null model through trimming the distribution of FST. Am. Nat. 2015, 186 (Suppl. S1), S24–S36. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Bisong, E. Matplotlib and seaborn. In Building Machine Learning and Deep Learning Models on Google Cloud Platform: A Comprehensive Guide for Beginners; Apress: Berkeley, CA, USA, 2019; pp. 151–165. [Google Scholar]
- Chen, Z.L.; Meng, J.M.; Cao, Y.; Yin, J.L.; Fang, R.Q.; Fan, S.B.; Liu, C.; Zeng, W.F.; Ding, Y.H.; Tan, D.; et al. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 2019, 10, 3404. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Novo, I.; Ordás, P.; Moraga, N.; Santiago, E.; Quesada, H.; Caballero, A. Impact of population structure in the estimation of recent historical effective population size by the software GONE. Genet. Sel. Evol. 2023, 55, 86. [Google Scholar] [CrossRef]
- Hilsdorf, A.W.S.; Azeredo-Espin, A.M.L.; Krieger, M.H.; Krieger, J.E. Mitochondrial DNA diversity in wild and cultured populations of Brycon opalinus (Cuvier, 1819) (Characiformes, Characidae, Bryconinae) from the Paraíba do Sul Basin, Brazil. Aquaculture 2002, 214, 81–91. [Google Scholar] [CrossRef]
- Yuan, J.; Lin, H.; Wu, L.; Zhuang, X.; Ma, J.; Kang, B.; Ding, S. Resource status and effect of long-term stock enhancement of large yellow croaker in China. Front. Mar. Sci. 2021, 8, 743836. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Hohenlohe, P.A.; Luikart, G. Genomics and the future of conservation genetics. Nat. Rev. Genet. 2010, 11, 697–709. [Google Scholar] [CrossRef]
- Del Pazo, F.; Sánchez, S.; Posner, V.; Sciara, A.A.; Arranz, S.E.; Villanova, G.V. Genetic diversity and structure of the commercially important native fish pacu (Piaractus mesopotamicus) from cultured and wild fish populations: Relevance for broodstock management. Aquac. Int. 2021, 29, 289–305. [Google Scholar] [CrossRef]
- Noorullah, M.; Zuberi, A.; Zaman, M.; Younas, W.; Hussain, S.; Kamran, M. Assessment of genetic diversity among wild and captive-bred Labeo rohita through microsatellite markers and mitochondrial DNA. Fish. Aquat. Sci. 2023, 26, 752–761. [Google Scholar] [CrossRef]
- Brown, K.T.; Southgate, P.C.; Loganimoce, E.M.; Kaure, T.; Stockwell, B.; Lal, M.M. Sandfish generations: Loss of genetic diversity due to hatchery practices in the sea cucumber Holothuria (Metriatyla) scabra. Aquaculture 2024, 578, 740048. [Google Scholar] [CrossRef]
- Geletu, T.T.; Zhao, J. Genetic resources of Nile tilapia (Oreochromis niloticus Linnaeus, 1758) in its native range and aquaculture. Hydrobiologia 2023, 850, 2425–2445. [Google Scholar] [CrossRef]
- Zhang, X.; Ouyang, M.; Zhang, F.; Wang, J. Study on the genetic structure of wild and hatchery populations of Procypris rabaudi Tchang, an endemic fish in the upper Yangtze River. Fish. Res. 2022, 245, 106134. [Google Scholar] [CrossRef]
- Guo, X.Z.; Chen, H.M.; Wang, A.B.; Qian, X.Q. Population genetic structure of the yellow catfish (Pelteobagrus fulvidraco) in China inferred from microsatellite analyses: Implications for fisheries management and breeding. J. World Aquac. Soc. 2022, 53, 174–191. [Google Scholar] [CrossRef]
- Ukenye, E.A.; Megbowon, I. Comparison of genetic diversity of farmed Oreochromis niloticus and wild unidentified tilapia (Wesafu) using microsatellite markers. Biodiversitas 2023, 24, 5. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).