Abstract
Non-indigenous species (NIS) introduction notoriously threatens the Mediterranean Sea. In addition, Mediterranean coastal lagoons play a crucial role as nurseries for marine species, which new NIS arrivals can threaten. Therefore, monitoring and early warning of NIS presence is essential in preserving biodiversity. An innovative technique for rapid and accurate species identification and biodiversity screening is the application of environmental DNA (eDNA) metabarcoding. In this research, different Penaeidae (Arthropoda, Crustacea, Decapoda) NIS specimens were collected from a Mediterranean coastal lagoon after an early warning about a potentially invasive NIS arising from next-generation sequencing data. DNA barcoding of the DNA extracted from tissue samples and amplified with specifically designed primer pairs led to the recognition of Penaeus aztecus in this NATURA 2000 protected ecosystem for the first time. DNA barcoding from DNA isolated from the water where the living specimens were stored further validated the possibility of identifying P. aztecus starting from eDNA. This approach demonstrated the validity of environmental DNA analysis in the early screening of potentially invasive NIS presence in Mediterranean protected areas and ecosystems. This work describes an applicative example of the efficacy in improving the biomonitoring of lagoon ecosystems using molecular tools and it represents a guideline for the validation of eDNA metabarcoding data for the presence of potentially invasive species.
1. Introduction
With a surface area of 2,500,000 km2, the Mediterranean Sea represents less than 1% of the world’s ocean area [1]. Despite this, the Mediterranean Sea is one of the richest reservoirs in the world in terms of marine and coastal biodiversity [2]. The Mediterranean Sea hosts more than 17,000 marine species, 20–30% of which are endemic, making it the sea with the highest rate of endemism [1,2,3,4,5].
The impact of non-indigenous species (NIS) on native species is notoriously increasing, leading the Mediterranean scientific community to highlight the importance of early warnings and monitoring programs on NIS presence and distribution [6,7,8,9,10].
The main recognised source of NIS arrivals in the Mediterranean Sea is represented by the Suez Canal, which allowed the so-called Lessepsian migration of Indo-Pacific origin species [11,12]. Other recognised pathways for NIS arrivals in the Mediterranean Sea include aquaculture, ornamental aquarium-pet trade, ballast waters, oil rigs, and climate change [9,11,13,14,15].
The regulation (EU) N° 1143/2014 of the European Parliament and the Council of 22 October 2014 on the prevention and management of the introduction and spread of NIS states that early warnings and monitoring programs are key requirements for the conservation and monitoring of ecosystems [16].
Although a specific gap concerning the NIS barcode availability for the Mediterranean Sea still needs to be filled, DNA-based approaches are taking place quickly for species identification and biomonitoring of ecosystems [17,18,19,20,21,22]. In particular, DNA barcoding refers to single-species identification using a short DNA fragment. In the metabarcoding technique, DNA is extracted from a pool of organisms/species, amplified, and then high-throughput sequenced [18,19,23,24].
The advent of environmental DNA (eDNA) metabarcoding is further facilitating the data collection for species assessments and ecosystem biomonitoring. Environmental DNA (eDNA) is DNA from cells, faeces, mucous, gametes, degrading cells or tissues, and fragments from predation, retrieved in environmental samples like water or sediment [17,18,19,20,22,23,24].
This approach has not been applied widely for NIS identification, although some results demonstrate its validity in early warnings in marine and freshwater environments [25,26].
In this work, we demonstrated that eDNA metabarcoding data can represent an early alert of potentially invasive NIS presence. We provide a practical guideline for DNA-based monitoring of NIS using species-specific primer pairs. Specifically, a previous eDNA metabarcoding assessment performed in a protected Mediterranean lagoon, using Cytochrome c Oxidase subunit I (COI) as the gene marker, unveiled an Operational taxonomic unit (OTU) annotated as Penaeus genus. These results raised awareness since different Penaeus NIS were discovered in the Mediterranean Sea [21]. Consequently, the sampling effort in the lagoon increased and we collected several Penaeus specimens. DNA barcoding from tissue and water samples was used for species identification through species-specific primer pairs. The results confirmed the presence of the NIS shrimp Penaeus aztecus in the lagoon.
2. Materials and Methods
2.1. Study Area and Sampling Protocol
The Aquatina di Frigole lagoon is the study area. It is a coastal lagoon included within the NATURA 2000 site “Aquatina di Frigole” (IT50003) in southeastern Italy. The lagoon represents about 3% of the NATURA 2000 site. The surface area is about 42 ha and the length is about 2 km parallel to the dune cordon. The maximum registered depth is about 1.5 m and the annual maximum registered tidal excursion is about 34 cm.
The lagoon directly connects with the Adriatic Sea through a 400 m long and 15 m wide channel on the southern boundary. On the northern boundary, a lateral ramification of the Giammatteo canal represents the freshwater input.
The NATURA 2000 site hosts the Research Centre for Fisheries and Aquaculture of Aquatina di Frigole, University of Salento, which routinely monitors the biodiversity and abiotic gradients of the Aquatina di Frigole lagoon in seven surveyed sampling points [22].
On the 3 June 2022, a monthly routine sampling was held in the lagoon. In correspondence with the fourth sampling station, four shrimps were collected using a hand net (mesh size 0.5 cm). The shrimps were placed in a 10 L bucket filled with water from the same sampling station and stored for about an hour, until the end of the sampling. All the collected samples were transported to the Research Centre for Fisheries and Aquaculture, located adjacent to the lagoon.
Once in the research centre, one litre of water was isolated from the bucket, filtered, and processed for eDNA applications.
Because of the absence of adult traits and mature gonads, the morphological identification of the collected shrimps was doubtful. The identification was approached by applying the taxonomic keys provided by Pérez Farfante (1988) [27].
2.2. Penaeus Aztecus Species-Specific Primer Pairs Design
For the molecular confirmation of the Penaeus aztecus species, we designed species-specific primer pairs. For this purpose, we downloaded from the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/ accessed on 3 March 2024) three COI DNA barcodes per each of three species: Penaeus aztecus, Penaeus vannamei, and Penaeus kerathurus.
We used the open-source software MEGA v.11 (https://www.megasoftware.net accessed on 4 March 2024) [28] to multi-align the sequences using the Clustal omega (ClustalW) algorithm. Following the multi-alignment, we identified regions of approximately 20 base pairs (bp) uniquely conserved for Penaeus aztecus along the entire COI shared sequence. These uniquely conserved regions guarantee species-specificity during the amplification phase. The potential primers identified were divided into forward (FWD) and reverse (REV), named and numbered.
The melting temperature (Tm), the probability of secondary structure generation, and the possibility of self-annealing into primer dimers were carefully evaluated to guarantee the primers’ functionality.
The primer pairs (FWD and REV) that presented the most suitable characteristics for the experimental application were tabulated and subsequently synthesised with the support of BMR Genomics (Padova, Italy).
2.3. DNA Barcoding Approach
About 800 mg of fresh tissue was isolated from the abdomen of one specimen, carefully avoiding contamination from the intestinal tract. The isolated tissue was rinsed with ultrapure sterile water and dried at 37 °C for 24 h.
The resulting 220 mg of dry tissue was divided into two equal sub-samples and processed independently. Each sub-sample was homogenised in 0.7 mL of TNES solution (Tris 50 mM, NaCl 400 mM, EDTA 100 mM, 0.5% SDS); 5 µL of Proteinase K was added and the solution was left in a bath at 65 °C for 3 h, vortexing every 30 min.
The DNeasy PowerSoil kit (QIAGEN, Hilden, Germany) was applied for DNA extraction starting from step 5 of the manufacturer’s protocol. The DNA was stored at –20 °C immediately after extraction.
A portion of the COI marker gene was amplified from both DNA samples using the species-specific primers PaztF4 (5′-GTCACAGCTCACGCTTTC-3′) and PaztR4 (5′-AGGGTCAAAGAAGGATGTATTG-3′). The obtained amplicon is about 450 bp in length.
The PCR reactions were performed in a volume of 50 μL composed of 5 μL 10X reaction buffer; 1 μL MgCl2 (50 mM); 1 μL dNTP mix (10 mM); 1 μL forward primer (10 μM); 1 μL reverse primer (10 μM); 10 ng DNA; 0.2 μL Platinum Taq (5 U/μL; Life Technologies, Carlsbad, CA, USA); and sterile water to reach a volume of 50 μL.
The amplification programs included the following phases: initial denaturation at 95 °C for 5 min, followed by thirty cycles of denaturation (95 °C for 30 s), annealing (55 °C for 30 s), extension (72 °C for 1 min), and a final extension at 72 °C for 10 min.
The amplicons were purified using the PCR Purification kit following the manufacturer’s protocol. The purified amplicons were Sanger-sequenced by BMR Genomics (Padova, Italy). The sequencings were encoded as DNA_Pazt_Rev4_1; and DNA_Pazt_Rev4_2.
2.4. Mapping of Previous Observations in the Mediterranean Sea
Scientific literature about P. aztecus occurrences in the Mediterranean Sea was obtained from multiple databases related to cross-disciplinary research, such as Web of Science (http://www.webofknowledge.com/ accessed on 8 January 2024), Science Direct (http://www.sciencedirect.com accessed on 10 January 2024), and Google Scholar (http://scholar.google.com accessed on 12 January 2024). Scientific literature published up to December 2023 was selected. Different combinations of the keywords “Penaeus aztecus”, “Farfantepenaeus aztecus”, “brown shrimp” and “Mediterranean Sea” were used.
Scientific literature data were mapped at the Mediterranean Sea scale using QGIS v3.32.0 (https://qgis.org accessed on 28 February 2024). Data published in peer-reviewed scientific journals were preferred, as this is believed to ensure data reliability owing to the peer-review process before publication.
2.5. Environmental DNA (eDNA) Barcoding for NIS Biomonitoring
One litre of the water in which the living shrimps were stored and transported to the Research Centre for Fisheries and Aquaculture of Aquatina di Frigole was filtered through a 0.45 μm cellulose filter of 47 mm diameter (Mixed Cellulose Ester filters, Advantec®, Taipei, Taiwan).
The DNA was extracted from the filter using the DNeasy PowerWater kit (QIAGEN, Hilden, Germany) following the manufacturer’s protocol. The extracted DNA was immediately stored at –20 °C.
A portion of the COI marker gene was amplified using the species-specific primers PaztF3 (5′-TACAGCTCTTAGACTTATCATT-3′), PaztR2 (5′-GTGTTGATATAAGACAGGATCT-3′), PaztF4, and PaztR4. The obtained amplicon for the first primer set is about 560 bp long, and about 450 bp for the second primer set.
Both PCR reactions were performed in a volume of 50 μL composed of 5 μL 10X reaction buffer; 1 μL MgCl2 (50 mM); 1 μL dNTP mix (10 mM); 1 μL forward primer (10 μM); 1 μL reverse primer (10 μM); 10 ng DNA; 0.2 μL Platinum Taq (5 U/μL; Life Technologies, Carlsbad, CA, USA); and sterile water to reach a volume of 50 μL.
The first amplification program included the following phases: initial denaturation at 95 °C for 5 min, followed by thirty cycles of denaturation (95 °C for 30 s), annealing (50 °C for 30 s), extension (72 °C for 1 min), and a final extension at 72 °C for 10 min.
The second amplification program included the following phases: initial denaturation at 95 °C for 5 min, followed by thirty cycles of denaturation (95 °C for 30 s), annealing (55 °C for 30 s), extension (72 °C for 1 min), and a final extension at 72 °C for 10 min.
The amplicons were purified using the PCR Purification kit following the manufacturer’s protocol. The purified amplicons were Sanger-sequenced by BMR Genomics (Padova, Italy). The sequencings were encoded as eDNA_Pazt_For4; eDNA_Pazt_For3; eDNA_Pazt_Rev4; and eDNA_Pazt_Rev2.
2.6. Maximum Likelihood Species Delimitation Analysis
Chromatograms were examined for both DNA and eDNA barcoding approaches. Sequences whose chromatograms displayed baseline noise were not included in further analysis. Chromatograms were analysed using the software MEGA v.11. The initial and final regions of the retained sequences were discarded.
GenBank was queried for downloading 5 COI sequences per each of three species: Penaeus aztecus, Penaeus vannamei, and Penaeus kerathurus. The search engine was queried as (“Species name” [Organism] OR Species name [All Fields]) AND COI [All Fields]. The first five results for each species were downloaded for analysis.
The sequences were multi-aligned using the ClustalW algorithm using the MEGA v.11 alignment options. The multi-alignment was used to construct the maximum likelihood tree in MEGA v.11. For the tree construction, the test of phylogeny was set on bootstrap with 1000 bootstrap replications. The nucleotide substitution model was set on Jukes–Cantor with no outgroup. The statistical method was set on maximum likelihood.
The resulting tree was used to infer putative species boundaries by applying a Bayesian implementation of the Poisson tree processes (bPTP) model for species delimitation [29]. For the bPTP model, 100,000 Markov chain Monte Carlo (MCMC) generations with 123 seed and 0.1 burn-in were applied.
The final rendering of the maximum likelihood phylogenetic tree with species delimitation was obtained through the interactive tree of life (iTOL) v.5 tool [30].
3. Results
3.1. Early Warning and Taxonomic Validation
The first early warning about a living population of non-indigenous penaeids in the lagoon dates back to October 2020 from an eDNA metabarcoding survey [21]. In this survey, the COI marker gene was used to assess the biodiversity of Aquatina di Frigole lagoon [21]. As reported in the Supplementary Table S2 [21], a COI OTU was annotated as Penaeus genus.
In June 2022, four uncommon juvenile shrimps were collected during biological samplings held in the Aquatina di Frigole lagoon (Figure 1). Because of the absence of adult traits and mature gonads, the morphological identification was doubtful. The morphological keys suggested the identification of juvenile specimens of Penaeus aztecus [27].

Figure 1.
One specimen of the shrimps collected in June 2022 in the coastal lagoon of Aquatina, located in the Aquatina di Frigole NATURA 2000 site (IT9150003): (a) dorsal view; (b) lateral view; (c) detail of the cephalothorax; (d) detail of the last abdominal segment.
DNA barcoding was performed to overcome the taxonomic annotation issue. Through BLASTn in NCBI, the results of the Sanger sequencing confirmed the annotation of the collected shrimps as Penaeus aztecus (Table 1).

Table 1.
Synthetic table of the BLASTn results for the Sanger sequences of the DNA and eDNA amplicons.
These results confirmed the taxonomic annotation of the collected shrimps as juvenile Penaeus aztecus and the presence of a population of this NIS in the Aquatina di Frigole lagoon for the first time.
3.2. Validation of the Presence of P. aztecus DNA in Water
The water in which the shrimps were stored was used for DNA extraction and amplification to validate the P. aztecus DNA release in water. Sanger sequencing of COI amplicons and BLASTn in NCBI confirmed the annotation of the barcodes belonging to Penaeus. aztecus (Table 1).
The results from this experiment confirmed the validity of eDNA extracted from water samples to monitor the presence and distribution of Penaeus aztecus in Mediterranean coastal waters. Also, these results demonstrate the validity of this approach in monitoring potentially invasive NIS through eDNA isolated from environmental matrices.
3.3. Maximum Likelihood Species Delimitation
The sequences obtained through DNA and eDNA barcoding and the reference sequences obtained from GenBank (File S1 were used to create a maximum likelihood phylogenetic tree. The tree was then used for the bPTP species delimitation analysis.
Applying the bPTP model to infer putative species boundaries resulted in good MCMC chains convergence (Figure S1). This validates the reliability of the Bayesian support values reported in the highest Bayesian-supported solution tree (Figure S2 and Table S1).
Through iTOL v.5, the maximum likelihood phylogenetic tree displaying species boundaries was built (Figure 2). The sequences obtained in our experiments clustered with Penaeus aztecus COI sequences.
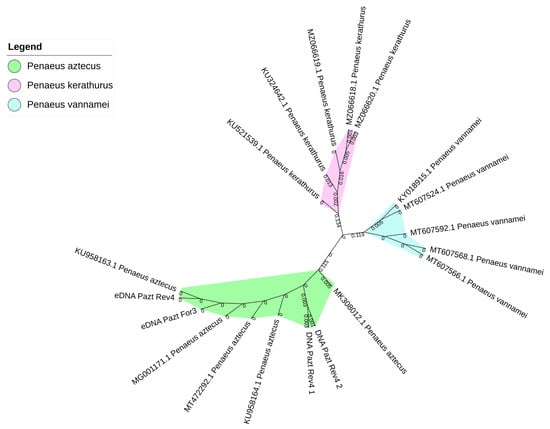
Figure 2.
Unrooted maximum likelihood phylogenetic tree (Jukes–Cantor model with 1000 bootstrap replications) of the ClustalW multiple alignments of Penaeus aztecus, P. vannamei, and P. kerathurus COI sequences for bPTP species delimitation analysis. The reported codes refer to GenBank accession numbers.
3.4. Distribution of P. aztecus in the Mediterranean Sea
Our results add a new coastal lagoon ecosystem within the known distribution range of the NIS Penaeus aztecus in the Mediterranean Sea. The first observation in the Mediterranean Sea dates back to 2009 in the Gulf of Antalya [29], probably due to the unintentional transport of larvae via ballast waters. In the following years, Penaeus aztecus presence was reported in different localities in the Mediterranean Sea (Table 2 and Figure 3), demonstrating its potential invasiveness.

Table 2.
First records of Penaeus aztecus in the Mediterranean Sea, ordered by date.
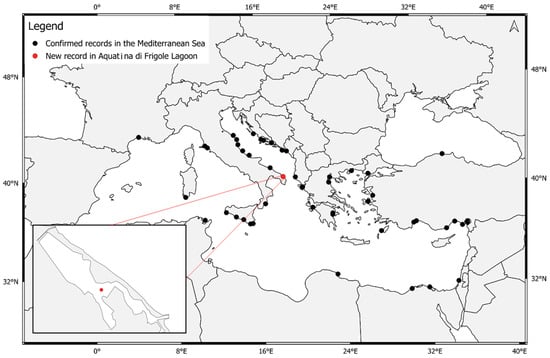
Figure 3.
First confirmed records of Penaeus aztecus, showing its distribution in the Mediterranean Sea. The focus is on the Aquatina di Frigole coastal lagoon, in the present study. The map was generated using QGIS v3.32.0 (https://qgis.org accessed on 28 February 2024).
4. Discussion
The Directive 2008/56/EC of the European Parliament and of the Council of 17 June 2008 established a framework for community action in marine environmental policy (Marine Strategy Framework Directive). To help EU countries achieve good environmental status (GES), the directive set out 11 illustrative qualitative descriptors. Specifically, descriptor 2 states “non-indigenous species do not adversely alter ecosystems”.
Reaching the descriptor 2 objective requires much information about NIS in the Mediterranean Sea. Gathering information about NIS presence and distribution requires constant early warnings and monitoring programs.
In this work, results from an eDNA metabarcoding survey [21] raised awareness concerning the presence of non-indigenous penaeids in a NATURA 2000 protected Mediterranean coastal lagoon. The OTU annotations were limited to the genera, but the unusual retrieving of the genus Penaeus in a shallow Mediterranean coastal lagoon represented an alert about the presence of a new NIS in the area. This led to the need for biomonitoring actions to shed light on the presence of potentially invasive NIS in the lagoon. The following samplings allowed the collection of some uncommon juvenile shrimps.
Molecular tools can face some difficulties, e.g., the need for short DNA fragments, the requirement of specific template concentrations, and the correct binding efficiency of the primers during reactions. However, the application of DNA-based tools, exploiting both DNA isolated from tissue and eDNA isolated from water, allowed the first identification of Penaeus aztecus in the Aquatina di Frigole lagoon.
The results from a molecular survey conducted in October 2020 [21] and the identification of juvenile Penaeus aztecus in 2022 suggest the presence of a stable and reproducing population in the protected Aquatina di Frigole lagoon.
The application of DNA barcoding on DNA samples isolated from water further confirmed the efficacy of eDNA-based approaches for the early warnings and monitoring programs that are essential for gathering information about NIS in the Mediterranean Sea. These are fast, reliable, cost-effective approaches requiring low personnel effort.
The application of the bPTP model for maximum likelihood species delimitation analysis and the obtained results confirmed the reliability of COI as a marker gene for species identification, not only in tissue samples but also in environmental samples. Although a barcode gap still exists for Mediterranean-confirmed NIS [20], these findings highlight that eDNA metabarcoding surveys are effective in raising awareness and extensively monitoring the presence and distribution of NIS in the Mediterranean Sea.
Moreover, coastal lagoons are transitional water bodies between marine and freshwater ecosystems that represent important nurseries for many species reproducing in these areas [56,57,58,59]. The finding of juvenile Penaeus aztecus specimens in a NATURA 2000 protected coastal lagoon demonstrates the importance of monitoring these ecosystems, in which potentially invasive NIS may find the ideal conditions for reproduction.
In addition, the design and validation of species-specific primer pairs for Penaeus aztecus allowed the identification of this NIS, providing a useful guideline for monitoring the presence and distribution of P. aztecus and other NIS in the Mediterranean Sea. For such reasons, we not only encourage collaboration among researchers in this sector, but we also stress the importance of training and involvement of researchers from countries that are potential sources of new and hard-to-detect NIS.
In conclusion, it is essential to underline that molecular techniques represent a great opportunity to improve studies about the occurrence and distribution of NIS. Hence, a specific gap needs to be filled by the scientific community to make molecular identification efficient and independent at a regional, national, and international level.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16090525/s1, File S1: The sequences obtained through DNA and eDNA barcoding and the reference sequences obtained from GenBank; Figure S1: Likelihood trace plot of MCMC iterations after thinning; Figure S2: Highest Bayesian supported solution tree annotated with Bayesian support values; Table S1: Most supported partition found by simple heuristic search.
Author Contributions
F.Z. performed experiments and analysed data; M.P. and V.S. conceptualized the research and analysed data. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported from the PRO-COAST project funded by EU HORIZON-CL6-2022-BIODIV-01 to Maurizio Pinna, from BlueDiversity project funded by Interreg Italy-Croatia 2021-2027 First Call to Maurizio Pinna. Post doc grant of F. Zangaro was supported by the National Biodiversity Future Center (NBFC) project CN_00000033 funded by MUR of Italy (PNRR, Mission 4 Component 2 Investment 1.4). The authors acknowledge Mariangela Eleonora Pinna for English language revision.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Coll, M.; Piroddi, C.; Albouy, C.; Ben Rais Lasram, F.; Cheung, W.W.L.; Christensen, V.; Karpouzi, V.S.; Guilhaumon, F.; Mouillot, D.; Paleczny, M.; et al. The Mediterranean Sea under Siege: Spatial Overlap between Marine Biodiversity, Cumulative Threats and Marine Reserves. Glob. Ecol. Biogeogr. 2012, 21, 465–480. [Google Scholar] [CrossRef]
- Bianchi, C.N.; Morri, C. Marine Biodiversity of the Mediterranean Sea: Situation, Problems and Prospects for Future Research. Mar. Pollut. Bull. 2000, 40, 367–376. [Google Scholar] [CrossRef]
- Coll, M.; Piroddi, C.; Steenbeek, J.; Kaschner, K.; Ben Rais Lasram, F.; Aguzzi, J.; Ballesteros, E.; Bianchi, C.N.; Corbera, J.; Dailianis, T.; et al. The Biodiversity of the Mediterranean Sea: Estimates, Patterns, and Threats. PLoS ONE 2010, 5, e11842. [Google Scholar] [CrossRef]
- Marrocco, V.; Zangaro, F.; Sicuro, A.; Pinna, M. A Scaling down Mapping of Pinna nobilis (Linnaeus, 1758) through the Combination of Scientific Literature, NATURA 2000, Grey Literature and Citizen Science Data. Nat. Conserv. 2019, 33, 43–53. [Google Scholar] [CrossRef]
- Zangaro, F.; Schifano, V.; Specchia, V.; Tzafesta, E.; Pinna, M. A New Extralimital Sighting of Monachus monachus (Hermann, 1779) in the Aquatina di Frigole NATURA 2000 Site (IT9150003) Beach (Salento Peninsula, Apulia Region, Italy) after Two Decades: Strategies for Conservation Are Needed. Biodivers. Data J. 2020, 8, e53950. [Google Scholar] [CrossRef]
- Katsanevakis, S.; Tempera, F.; Teixeira, H. Mapping the Impact of Alien Species on Marine Ecosystems: The Mediterranean Sea Case Study. Divers. Distrib. 2016, 22, 694–707. [Google Scholar] [CrossRef]
- Darling, J.A.; Galil, B.S.; Carvalho, G.R.; Rius, M.; Viard, F.; Piraino, S. Recommendations for Developing and Applying Genetic Tools to Assess and Manage Biological Invasions in Marine Ecosystems. Mar. Policy 2017, 85, 54–64. [Google Scholar] [CrossRef]
- Marrocco, V.; Sicuro, A.; Zangaro, F.; Pinna, M. First Record of the Protected Species Pinna nobilis (Linnaeus, 1758) in the Aquatina Lagoon (NATURA 2000 Site IT9150003, South-East Italian Coastline). Nat. Conserv. 2018, 28, 51–59. [Google Scholar] [CrossRef]
- Bariche, M.; Al-Mabruk, S.A.A.; Ateş, M.A.; Büyük, A.; Crocetta, F.; Dritsas, M.; Edde, D.; Fortič, A.; Gavriil, E.; Gerovasileiou, V.; et al. New Alien Mediterranean Biodiversity Records (March 2020). Mediterr. Mar. Sci. 2020, 21, 129–145. [Google Scholar] [CrossRef]
- Tsiamis, K.; Azzurro, E.; Bariche, M.; Çinar, M.E.; Crocetta, F.; De Clerck, O.; Galil, B.; Gómez, F.; Hoffman, R.; Jensen, K.R.; et al. Prioritizing Marine Invasive Alien Species in the European Union through Horizon Scanning. Aquat. Conserv. 2020, 30, 794–845. [Google Scholar] [CrossRef]
- Galil, B.S.; Marchini, A.; Occhipinti-Ambrogi, A.; Minchin, D.; Narščius, A.; Ojaveer, H.; Olenin, S. International Arrivals: Widespread Bioinvasions in European Seas. Ethol. Ecol. Evol. 2014, 26, 152–171. [Google Scholar] [CrossRef] [PubMed]
- Galil, B.; Marchini, A.; Occhipinti-Ambrogi, A.; Ojaveer, H. The Enlargement of the Suez Canal—Erythraean Introductions and Management Challenges. Manag. Biol. Invasions 2017, 8, 141–152. [Google Scholar] [CrossRef]
- Atalah, J.; Davidson, I.C.; Thoene, M.; Georgiades, E.; Hutson, K.S. Evaluating Importation of Aquatic Ornamental Species for Biosecurity Purposes. Front. Ecol. Evol. 2022, 9, 804160. [Google Scholar] [CrossRef]
- Galanidi, M.; Zenetos, A. Data-Driven Recommendations for Establishing Threshold Values for the NIS Trend Indicator in the Mediterranean Sea. Diversity 2022, 14, 57. [Google Scholar] [CrossRef]
- Galanidi, M.; Aissi, M.; Ali, M.; Bakalem, A.; Bariche, M.; Bartolo, A.G.; Bazairi, H.; Beqiraj, S.; Bilecenoglu, M.; Bitar, G.; et al. Validated Inventories of Non-Indigenous Species (NIS) for the Mediterranean Sea as Tools for Regional Policy and Patterns of NIS Spread. Diversity 2023, 15, 962. [Google Scholar] [CrossRef]
- Tiralongo, F.; Lillo, A.O.; Tibullo, D.; Tondo, E.; Martire, C.L.; D’Agnese, R.; Macali, A.; Mancini, E.; Giovos, I.; Coco, S.; et al. Monitoring Uncommon and Non-Indigenous Fishes in Italian Waters: One Year of Results for the AlienFish Project. Reg. Stud. Mar. Sci. 2019, 28, 100606. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Hajibabaei, M.; Rieseberg, L.H. Environmental DNA. Mol. Ecol. 2012, 21, 1789–1793. [Google Scholar] [CrossRef]
- Leese, F.; Altermatt, F.; Bouchez, A.; Ekrem, T.; Hering, D.; Meissner, K.; Mergen, P.; Pawlowski, J.; Piggott, J.; Rimet, F.; et al. DNAqua-Net: Developing New Genetic Tools for Bioassessment and Monitoring of Aquatic Ecosystems in Europe. Res. Ideas Outcomes 2016, 2, e11321. [Google Scholar] [CrossRef]
- Pawlowski, J.; Kelly-Quinn, M.; Altermatt, F.; Apothéloz-Perret-Gentil, L.; Beja, P.; Boggero, A.; Borja, A.; Bouchez, A.; Cordier, T.; Domaizon, I.; et al. The Future of Biotic Indices in the Ecogenomic Era: Integrating (e)DNA Metabarcoding in Biological Assessment of Aquatic Ecosystems. Sci. Total Environ. 2018, 637, 1295–1310. [Google Scholar] [CrossRef]
- Zangaro, F.; Saccomanno, B.; Tzafesta, E.; Bozzeda, F.; Specchia, V.; Pinna, M. Current Limitations and Future Prospects of Detection and Biomonitoring of NIS in the Mediterranean Sea through Environmental DNA. NeoBiota 2021, 70, 151–165. [Google Scholar] [CrossRef]
- Specchia, V.; Saccomanno, B.; Zangaro, F.; Tzafesta, E.; Pinna, M. Exploring the Biodiversity of a European NATURA 2000 Mediterranean Lagoon through eDNA Metabarcoding. Diversity 2022, 14, 991. [Google Scholar] [CrossRef]
- Specchia, V.; Zangaro, F.; Tzafesta, E.; Saccomanno, B.; Vadrucci, M.R.; Pinna, M. Environmental DNA Detects Biodiversity and Ecological Features of Phytoplankton Communities in Mediterranean Transitional Waters. Sci. Rep. 2023, 13, 15192. [Google Scholar] [CrossRef]
- Ji, Y.; Ashton, L.; Pedley, S.M.; Edwards, D.P.; Tang, Y.; Nakamura, A.; Kitching, R.; Dolman, P.M.; Woodcock, P.; Edwards, F.A.; et al. Reliable, Verifiable and Efficient Monitoring of Biodiversity via Metabarcoding. Ecol. Lett. 2013, 16, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N.; et al. Environmental DNA Metabarcoding: Transforming How We Survey Animal and Plant Communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef]
- Holman, L.E.; de Bruyn, M.; Creer, S.; Carvalho, G.; Robidart, J.; Rius, M. Detection of Introduced and Resident Marine Species Using Environmental DNA Metabarcoding of Sediment and Water. Sci. Rep. 2019, 9, 11559. [Google Scholar] [CrossRef]
- Jeunen, G.J.; Lipinskaya, T.; Gajduchenko, H.; Golovenchik, V.; Moroz, M.; Rizevsky, V.; Semenchenko, V.; Gemmell, N.J. Environmental DNA (eDNA) Metabarcoding Surveys Show Evidence of Non-Indigenous Freshwater Species Invasion to New Parts of Eastern Europe. Metabarcoding Metagenom 2022, 6, e68575. [Google Scholar] [CrossRef]
- Farfante, I.P. Illustrated key to penaeoid shrimps of commerce in the Americas. NOAA/Natl. Mar. Fish. Serv. 1988. Available online: https://repository.library.noaa.gov/view/noaa/5793 (accessed on 23 August 2024).
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A General Species Delimitation Method with Applications to Phylogenetic Placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Deval, M.C.; Kaya, Y.; Güven, O.; Gökoğlu, M.; Froglia, C. An unexpected find of the western Atlantic shrimp, Farfantepenaeus aztecus (Ives, 1891) (Decapoda, Penaeidae) in Antalya Bay, eastern Mediterranean Sea. Crustaceana 2010, 83, 1531–1537. [Google Scholar] [CrossRef]
- Nikolopoulou, I.; Baxevanis, A.D.; Kampouris, T.E.; Abatzopoulos, T.J. Farfantepenaeus aztecus (Ives, 1891) (Crustacea: Decapoda: Penaeidae) in Aegean: First Record in Greece by Morphological and Genetic Features. J. Biol. Res. 2013, 20, 367–375. [Google Scholar]
- Sadek, S.; El-Soud, W.; Galil, B. The Brown Shrimp Penaeus aztecus Ives, 1891 (Crustacea, Decapoda, Penaeidae) in the Nile Delta, Egypt: An Exploitable Resource for Fishery and Mariculture? Bioinvasions Rec. 2018, 7, 51–54. [Google Scholar] [CrossRef]
- Bilecenoglu, M.; Alfaya, J.E.F.; Azzurro, E.; Baldacconi, R.; Boyaci, Y.Ö.; Circosta, V.; Compagno, L.J.V.; Coppola, F.; Deidun, A.; Durgham, H.; et al. New Mediterranean Marine Biodiversity Records (December 2013). Mediterr. Mar. Sci. 2013, 14, 463. [Google Scholar] [CrossRef]
- Kapiris, K.; Apostolidis, C.; Baldacconi, R.; Başusta, N.; Bilecenoglu, M.; Bitar, G.; Bobori, D.C.; Boyaci, Y.Ö.; Dimitriadis, C.; Djurović, M.; et al. New Mediterranean Biodiversity Records (April 2014). Mediterr. Mar. Sci. 2014, 15, 198. [Google Scholar] [CrossRef]
- Kevrekidis, K. The Occurrence of the Atlantic Penaeid Prawn Farfantepenaeus aztecus (Ives, 1891) in the Thermaikos Gulf (Aegean Sea, Eastern Mediterranean): Considerations on the Potential Establishment and Impact on the Autochthonous Melicertus kerathurus (Forskål, 1775). Crustaceana 2014, 87, 1606–1619. [Google Scholar]
- Marković, O.; Gökoğlu, M.; Petović, S.; Mandić, M. First Record of the Northern Brown Shrimp, Farfantepenaeus aztecus (Ives, 1891) (Crustacea: Decapoda: Penaeidae) in the South Adriatic Sea, Montenegro. Mediterr. Mar. Sci. 2013, 15, 165. [Google Scholar] [CrossRef][Green Version]
- Minos, G.; Kokokiris, L.; Imsiridou, A.; Karachle, P.K.; Kapiris, K. Notes on the Distribution and Biology of Northern Brown Shrimp Farfantepenaeus aztecus (Ives, 1891) in the Eastern Mediterranean. Turk. J. Zool. 2015, 39, 467–473. [Google Scholar] [CrossRef]
- Crocetta, F.; Agius, D.; Balistreri, P.; Bariche, M.; Bayhan, Y.K.; Çakir, M.; Ciriaco, S.; Corsini-Foka, M.; Deidun, A.; El Zrelli, R.; et al. New Mediterranean Biodiversity Records (October 2015). Mediterr. Mar. Sci. 2015, 16, 682. [Google Scholar] [CrossRef]
- Cruscanti, M.; Innocenti, G.; Alvarado Bremer, J.; Galil, B.S. First Report of the Brown Shrimp Penaeus aztecus Ives, 1891 (Crustacea, Decapoda, Penaeidae) in the Tyrrhenian Sea. Mar. Biodivers. Rec. 2015, 8, e81. [Google Scholar] [CrossRef]
- Kondylatos, G.; Corsini-Foka, M. First Record of Penaeus aztecus Ives, 1891 (Crustacea, Decapoda) and Melibe viridis (Kelaart, 1858)(Gastropoda, Nudibranchia) in the South-Eastern Aegean Sea (Greece). Mediterr. Mar. Sci. 2015, 16, 278–279. [Google Scholar]
- Galil, B.S.; Innocenti, G.; Douek, J.; Paz, G.; Rinkevich, B. Foul Play? On the Rapid Spread of the Brown Shrimp Penaeus aztecus Ives, 1891 (Crustacea, Decapoda, Penaeidae) in the Mediterranean, with New Records from the Gulf of Lion and the Southern Levant. Mar. Biodivers. 2017, 47, 979–985. [Google Scholar] [CrossRef]
- Scannella, D.; Falsone, F.; Geraci, M.; Froglia, C.; Fiorentino, F.; Giusto, G.; Zava, B.; Insacco, G.; Colloca, F. First Report of Northern Brown Shrimp Penaeus aztecus Ives, 1891 in Strait of Sicily. BioInvasions Rec. 2017, 6, 67–72. [Google Scholar] [CrossRef]
- Zava, B.; Insacco, G.; Galil, B. The First Record of the Brown Shrimp Penaeus aztecus Ives, 1891 in the Central Adriatic Coast of Italy. BioInvasions Rec. 2018, 7, 293–296. [Google Scholar] [CrossRef]
- Bakir, K.; Aydin, İ. New Localities in the Aegean Sea for Alien Shrimps Penaeus aztecus (Ives, 1891) and Metapenaeus affinis (H. Milne Edwards, 1837). Acta Adriat. 2016, 57, 273–280. [Google Scholar]
- Gönülal, O.; Türetken, P.S.Ç. One of the Most Invasive Alien Species, Penaeus aztecus Ives, 1891 Reached the Black Sea Coasts. BioInvasions Rec. 2019, 8, 871–875. [Google Scholar] [CrossRef]
- Ugarković, P.; Crocetta, F. The Brown Shrimp Penaeus aztecus Ives, 1891 (Crustacea: Decapoda: Penaeidae) Spreading Northern in the Adriatic Sea: A First Record from Croatia. BioInvasions Rec. 2021, 10, 636–643. [Google Scholar] [CrossRef]
- Kapiris, K.; Minos, G. Weight-Length Relationship of the Northern Brown Shrimp Penaeus aztecus Ives, 1891 (Decapoda: Penaeidae) from the Central Aegean Sea, Greece. Mediterr. Mar. Sci. 2017, 18, 562–563. [Google Scholar]
- Özcan, T.; Ateş, A.S.; Özcan, G. The Distribution of the Alien Species Penaeus aztecus Ives, 1891 (Decapoda, Penaeidae) in the Mediterranean Sea. Transylv. Rev. Syst. Ecol. Res. 2019, 21, 41–48. [Google Scholar] [CrossRef]
- Kampouris, T.E.; Tiralongo, F.; Golemaj, A.; Giovos, I.; Doumpas, N.; Batjakas, I.E. Penaeus aztecus Ives, 1891 (Decapoda, Dendrobranchiata, Penaeidae): On the Range Expansion in Sicilian Waters and on the First Record from Albanian Coast. Int. J. Fish. Aquat. Stud. 2018, 6, 468–471. [Google Scholar]
- Mulas, A.; Bellodi, A.; Cau, A.; Cannas, R.; Marongiu, M.F.; Pesci, P.; Porcu, C.; Follesa, M.C. First Records of Penaeus aztecus Ives, 1891 (Decapoda Penaeidae) in Sardinian Waters (Central-Western Mediterranean). Biol. Mar. Mediterr. 2019, 26, 360–361. [Google Scholar]
- Hamida, O.B.A.B.H.; Hamida, N.B.H.; Chouikh, A.S.; Abidi, D.; Missaoui, H. Sur l’expansion de La Crevette Royale Grise Penaeus aztecus (Ives, 1904) En Mediterranee et Sa Premiere Observation au Nord de La Tunisie. INSTM Bull. Mar. Freshw. Sci. 2020, 47, 29–36. [Google Scholar]
- El-Deeb, R.S.; Sarhan, M.; Khafage, A.R.; Abdel Razek, F.A.; Abdel-Wahab, M.; Omar, H.A. Occurrence of Penaeus aztecus, Ives, 1891 (Crustacea: Decapoda: Penaeidae) in the Coastal Water of Alexandria, Egypt. Egypt. J. Aquat. Res. 2020, 46, 303–309. [Google Scholar] [CrossRef]
- Abdulrraziq, A.; Abdulghani, A.; Ibrahim, S.; Zava, B.; Deidun, A. First Record of the Northern Brown Shrimp Penaeus aztecus Ives, 1891 (Crustacea, Decapoda, Penaeidae) from Libyan Waters. BioInvasions Rec. 2021, 10, 287–294. [Google Scholar] [CrossRef]
- Froglia, C.; Scanu, M. Notes on the Spreading of Penaeus aztecus Ives 1891 (Decapoda, Penaeidae) in the Mediterranean Sea and on Its Repeated Misidentifications in the Region. Biology 2023, 12, 793. [Google Scholar] [CrossRef]
- Cheminée, A.; Le Direach, L.; Rouanet, E.; Astruch, P.; Goujard, A.; Blanfuné, A.; Bonhomme, D.; Chassaing, L.; Jouvenel, J.-Y.; Ruitton, S.; et al. All shallow coastal habitats matter as nurseries for Mediterranean juvenile fish. Sci. Rep. 2021, 11, 14631. [Google Scholar] [CrossRef] [PubMed]
- Pinna, M.; Zangaro, F.; Specchia, V. Assessing benthic macroinvertebrate communities’ spatial heterogeneity in Mediterranean transitional waters through eDNA metabarcoding. Sci. Rep. 2024, 14, 17890. [Google Scholar] [CrossRef]
- Verdiell-Cubedo, D.; Oliva-Paterna, F.J.; Ruiz-Navarro, A.; Torralva, M. Assessing the nursery role for marine fish species in a hypersaline coastal lagoon (Mar Menor, Mediterranean Sea). Mar. Biol. Res. 2013, 9, 739–748. [Google Scholar] [CrossRef]
- Newton, A.; Brito, A.C.; Icely, J.D.; Derolez, V.; Clara, I.; Angus, S.; Schernewski, G.; Inácio, M.; Lillebø, A.I.; Sousa, A.I.; et al. Assessing, quantifying and valuing the ecosystem services of coastal lagoons. J. Nat. Conserv. 2018, 44, 50–65. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).