Biodiversity Assessment of a Mississippi River Backwater Complex Using eDNA Metabarcoding
Abstract
1. Introduction
2. Materials and Methods
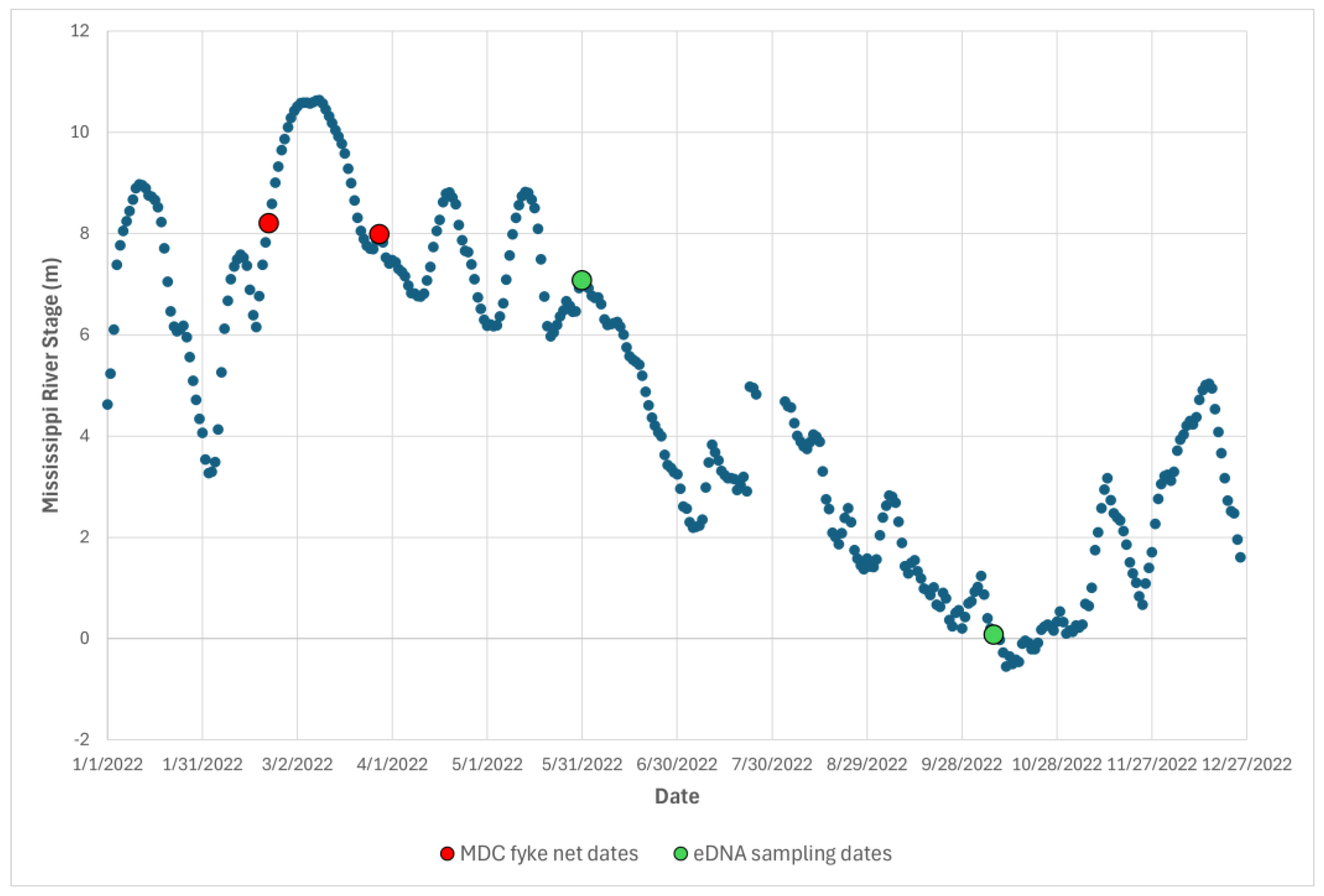
2.1. Study System
2.2. eDNA Field Sampling
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Bioinformatics
2.5. Estimating Species Diversity and Assemblage Structure
3. Results
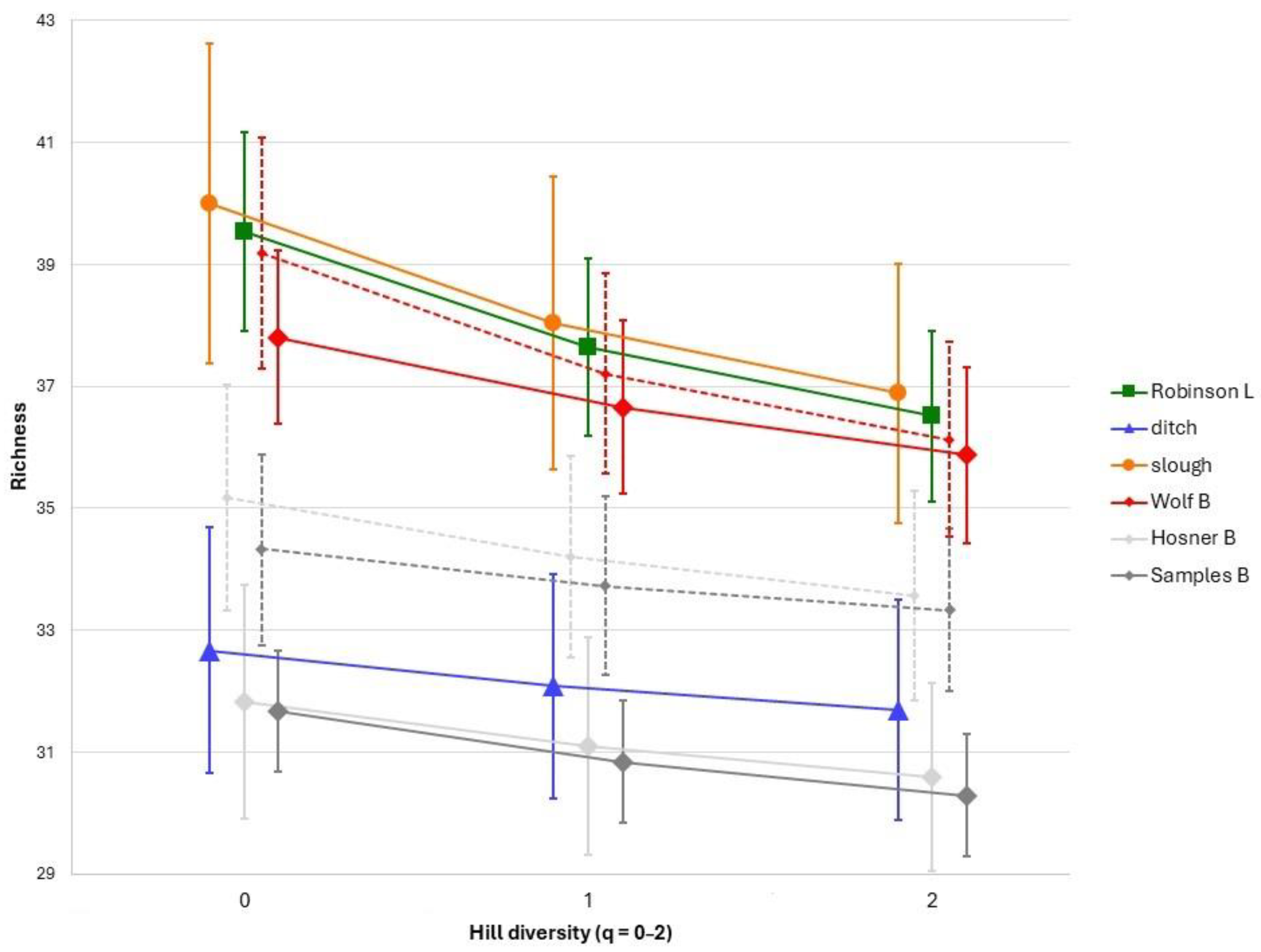
3.1. Species Richness
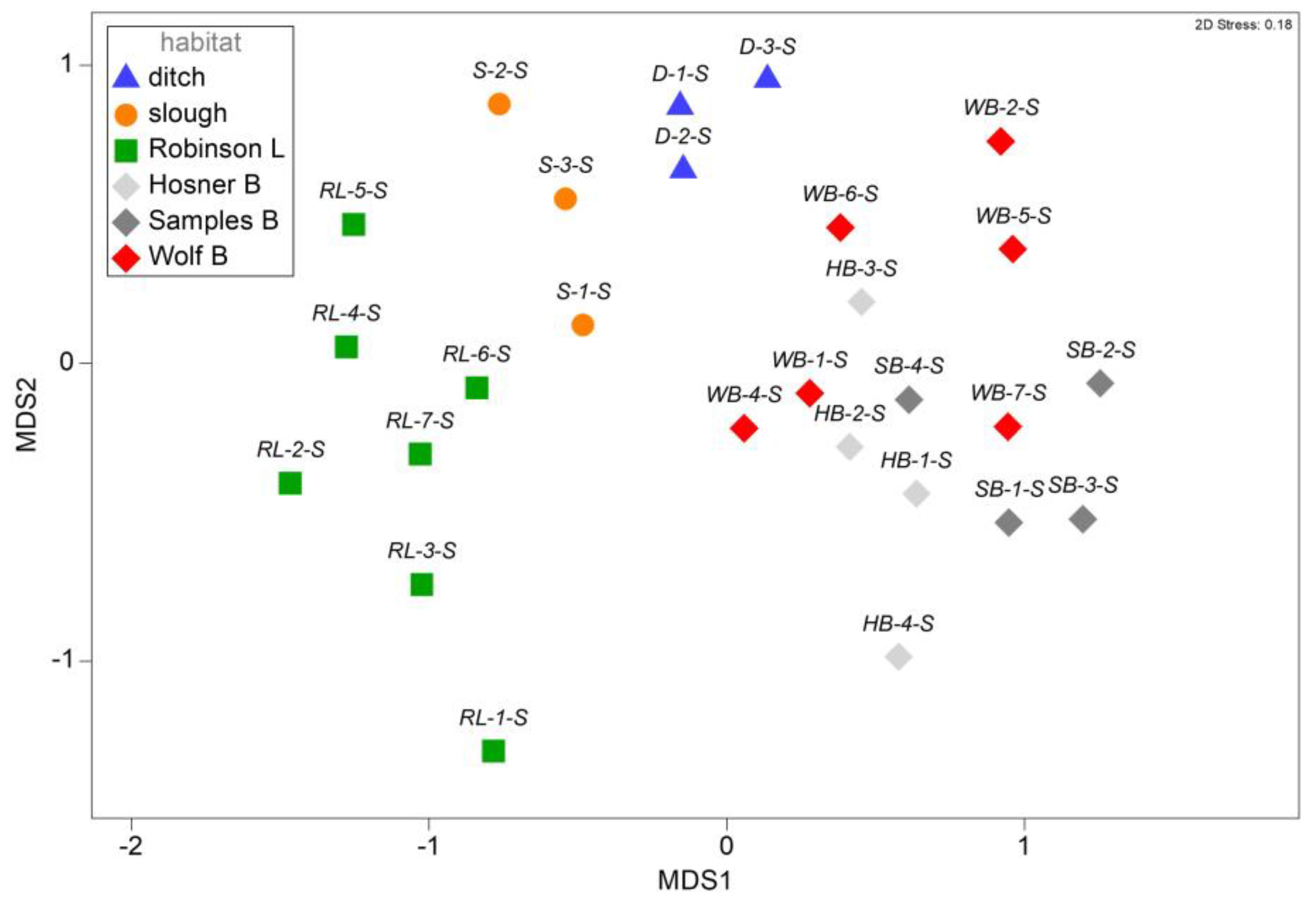
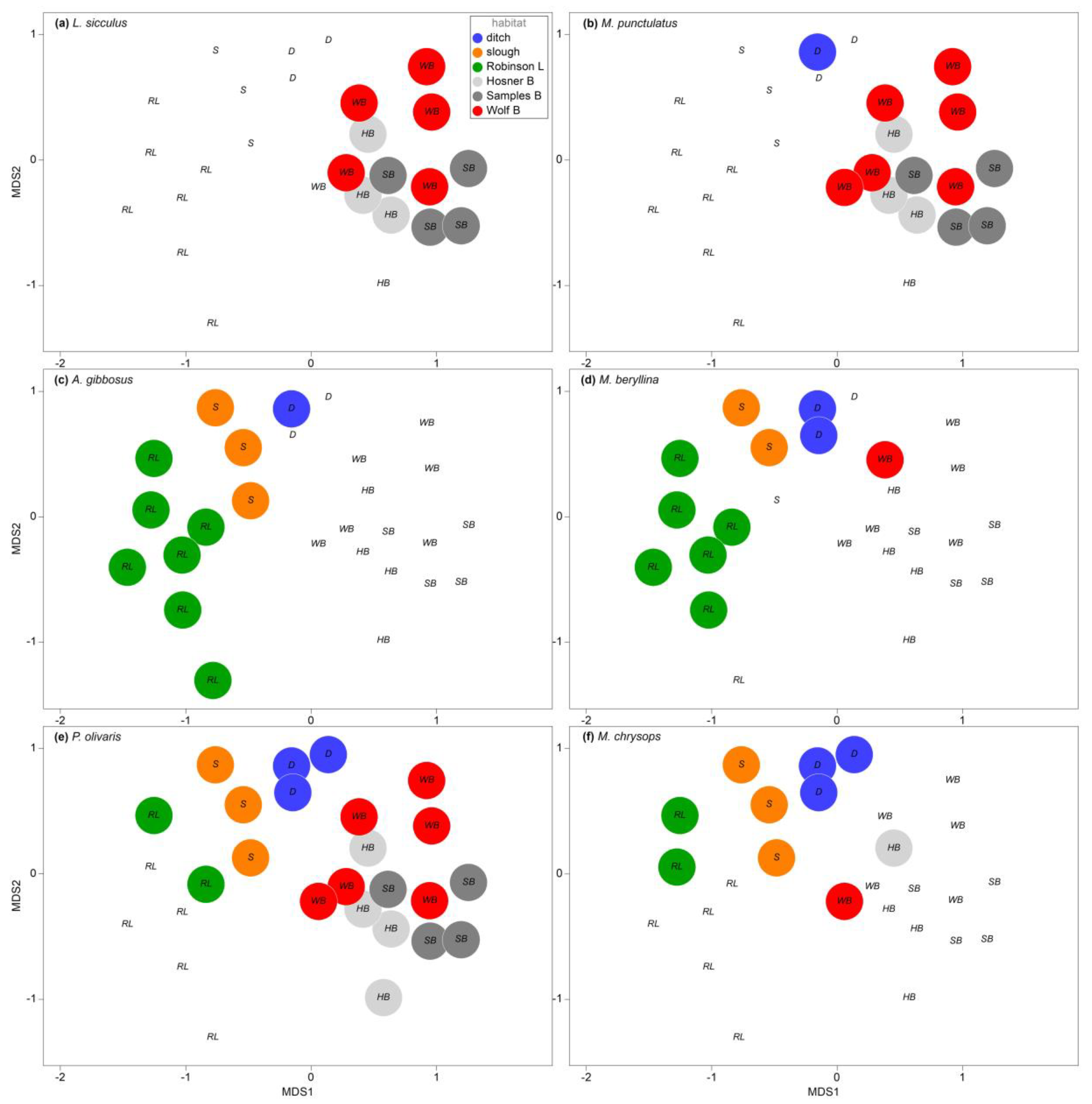
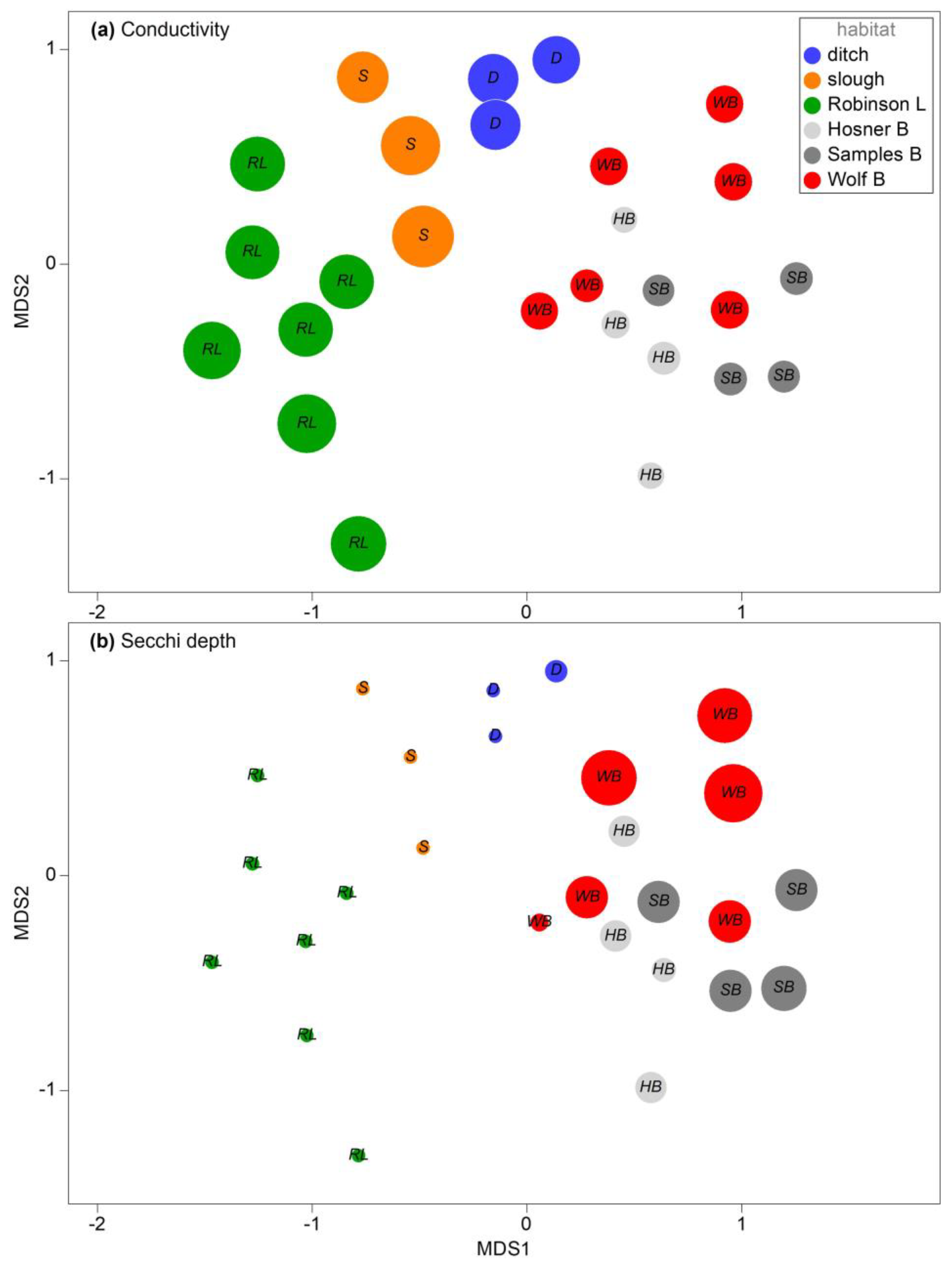
3.2. Assemblage Structure among Spring Samples
3.3. Seasonal and Spatial Variation in Bayou Habitats
3.4. Comparison with Historical and Recent Capture-Based Records of Species Presence
4. Discussion
4.1. Species Diversity
4.2. Assemblage Structure
4.3. Taxonomic Distribution of Diversity
4.4. Invasive and Introduced Species, and Species of Conservation Concern
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jarvis, S.G.; Mackay, E.B.; Risser, H.A.; Feuchtmayr, H.; Fry, M.; Isaac, N.J.B.; Thackeray, S.J.; Henrys, P.A. Integrating freshwater biodiversity data sources: Key challenges and opportunities. Freshw. Biol. 2023, 68, 1479–1488. [Google Scholar] [CrossRef]
- Radinger, J.; Britton, J.R.; Carlson, S.M.; Magurran, A.E.; Alcaraz-Hernández, J.D.; Almodóvar, A.; Benejam, L.; Fernández-Delgado, C.; Nicola, G.G.; Oliva-Paterna, F.J.; et al. Effective monitoring of freshwater fish. Fish Fish. 2019, 20, 729–747. [Google Scholar] [CrossRef]
- Schramm, H.L.; Hatch, J.T.; Hrabik, R.A.; Slack, W.T. Fishes of the Mississippi River. Am. Fish. Soc. Symp. 2016, 84, 53–77. [Google Scholar]
- DuBowy, P.J. Mississippi River Ecohydrology: Past, present and future. Ecohydrol. Hydrobiol. 2013, 13, 73–83. [Google Scholar] [CrossRef]
- Best, J. Anthropogenic stresses on the world’s big rivers. Nat. Geosci. 2019, 12, 7–21. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef] [PubMed]
- Schenekar, T. The current state of eDNA research in freshwater ecosystems: Are we shifting from the developmental phase to standard application in biomonitoring? Hydrobiologia 2023, 850, 1263–1282. [Google Scholar] [CrossRef]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- Schramm, H.L.; Ickes, B.S. The Mississippi River: A Place for Fish the Mississippi River: A Place for Fish. Am. Fish. Soc. Symp. 2016, 84, 3–34. [Google Scholar]
- Pflieger, W.L. The Fishes of Missouri, 2nd ed.; Missouri Department of Conservation: Jefferson City, MI, USA, 1997. [Google Scholar]
- Pierce, S.C.; Kröger, R.; Pezeshki, R. Managing artificially drained low-gradient agricultural headwaters for enhanced ecosystem functions. Biology 2012, 1, 794–856. [Google Scholar] [CrossRef]
- United States Army Corps of Engineers (USACE). Mississippi River and Tributaries Project—Levee System Evaluation Report for the National Flood Insurance Program; United States Army Corps of Engineers: Washington, DC, USA, 2014. [Google Scholar]
- Olson, K.R.; Morton, L.W.; Speidel, D. Missouri Ozark Plateau Headwaters Diversion engineering feat. J. Soil Water Conserv. 2016, 71, 13A–19A. [Google Scholar] [CrossRef]
- Olson, K.R.; Morton, L.W.; Speidel, D. Little river drainage district conversion of big swamp to fertile agricultural land. J. Soil Water Conserv. 2016, 71, 37A–43A. [Google Scholar] [CrossRef]
- Jenkins, C.N.; Van Houtan, K.S.; Pimm, S.L.; Sexton, J.O. US protected lands mismatch biodiversity priorities. Proc. Natl. Acad. Sci. USA 2015, 112, 5081–5086. [Google Scholar] [CrossRef] [PubMed]
- Noss, R.F.; Platt, W.J.; Sorrie, B.A.; Weakley, A.S.; Means, D.B.; Costanza, J.; Peet, R.K. How global biodiversity hotspots may go unrecognized: Lessons from the North American Coastal Plain. Divers. Distrib. 2015, 21, 236–244. [Google Scholar] [CrossRef]
- Isphording, W.; Fitzpatrick, J., Jr. Geologic and evolutionary history of drainage systems in the Southeastern United States. In Biodiversity of the Southeastern United States Aquatic Communities; Hackney, C., Adams, S., Martin, W., Eds.; John Wiley and Sons: New York, NY, USA, 1992; pp. 19–56. [Google Scholar]
- Cross, F.B.; Mayden, R.L.; Stewart, J.D. Fishes in the western Mississippi drainage. In Zoogeography of North American Freshwater Fishes; Hocutt, C.H., Wiley, E.O., Eds.; Wiley: New York, NY, USA, 1986; pp. 363–412. [Google Scholar]
- Warren, M.L.; Burr, B.M.; Walsh, S.J.; Bart, H.L.; Cashner, R.C.; Etnier, D.A.; Freeman, B.J.; Kuhajda, B.R.; Mayden, R.L.; Robison, H.W.; et al. Diversity, Distribution, and Conservation Status of the Native Freshwater Fishes of the Southern United States. Fisheries 2000, 25, 7–31. [Google Scholar] [CrossRef]
- Sowa, S.P.; Annis, G.; Morey, M.E.; Diamond, D.D. A gap analysis and comprehensive conservation strategy for riverine ecosystems of Missouri. Ecol. Monogr. 2007, 77, 301–334. [Google Scholar] [CrossRef]
- Junk, W.J.; Bayley, P.B.; Sparks, R.E. The flood pulse concept in River-Floodplain Systems. Can. Spec. Publ. Fish. Aquat. Sci. 1989, 106, 110–127. [Google Scholar]
- Phelps, Q.E.; Tripp, S.J.; Herzog, D.P.; Garvey, J.E. Temporary connectivity: The relative benefits of large river floodplain inundation in the lower Mississippi River. Restor. Ecol. 2015, 23, 53–56. [Google Scholar] [CrossRef]
- Allen, Y.; Kimmel, K.; Constant, G. Using Remote Sensing to Assess Alligator Gar Spawning Habitat Suitability in the Lower Mississippi River. North Am. J. Fish. Manag. 2020, 40, 580–594. [Google Scholar] [CrossRef]
- Schramm, H.L.; Eggleton, M.A. Applicability of the flood-pulse concept in a temperate floodplain river ecosystem: Thermal and temporal components. River Res. Appl. 2006, 22, 543–553. [Google Scholar] [CrossRef]
- Humphries, P.; King, A.J.; Koehn, J.D. Fish, flows and flood plains: Links between freshwater fishes and their environment in the Murray-Darling River system, Australia. Environ. Biol. Fishes 1999, 56, 129–151. [Google Scholar] [CrossRef]
- Sparks, R.E. Need for ecosystem management of large rivers and their floodplains. NCASI Tech. Bull. 1999, 2, 507. [Google Scholar] [CrossRef]
- Dettmers, J.M.; Gutreuter, S.; Wahl, D.H.; Soluk, D.A. Patterns in abundance of fishes in main channels of the upper Mississippi River system. Can. J. Fish. Aquat. Sci. 2001, 58, 933–942. [Google Scholar] [CrossRef]
- Mächler, E.; Little, C.J.; Wüthrich, R.; Alther, R.; Fronhofer, E.A.; Gounand, I.; Harvey, E.; Hürlemann, S.; Walser, J.C.; Altermatt, F. Assessing different components of diversity across a river network using eDNA. Environ. DNA 2019, 1, 290–301. [Google Scholar] [CrossRef]
- Rees, H.C.; Maddison, B.C.; Middleditch, D.J.; Patmore, J.R.M.; Gough, K.C. The detection of aquatic animal species using environmental DNA—A review of eDNA as a survey tool in ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Harrison, J.B.; Sunday, J.M.; Rogers, S.M. Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc. R. Soc. B Biol. Sci. 2019, 286, 20191409. [Google Scholar] [CrossRef] [PubMed]
- Nevers, M.B.; Przybyla-Kelly, K.; Shively, D.; Morris, C.C.; Dickey, J.; Byappanahalli, M.N. Influence of sediment and stream transport on detecting a source of environmental DNA. PLoS ONE 2020, 15, e0244086. [Google Scholar] [CrossRef]
- Turner, C.R.; Uy, K.L.; Everhart, R.C. Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biol. Conserv. 2015, 183, 93–102. [Google Scholar] [CrossRef]
- Cilleros, K.; Valentini, A.; Allard, L.; Dejean, T.; Etienne, R.; Grenouillet, G.; Iribar, A.; Taberlet, P.; Vigouroux, R.; Brosse, S. Unlocking biodiversity and conservation studies in high-diversity environments using environmental DNA (eDNA): A test with Guianese freshwater fishes. Mol. Ecol. Resour. 2019, 19, 27–46. [Google Scholar] [CrossRef]
- García-Machado, E.; Laporte, M.; Normandeau, E.; Hernández, C.; Côté, G.; Paradis, Y.; Mingelbier, M.; Bernatchez, L. Fish community shifts along a strong fluvial environmental gradient revealed by eDNA metabarcoding. Environ. DNA 2022, 4, 117–134. [Google Scholar] [CrossRef]
- Lecaudey, L.A.; Schletterer, M.; Kuzovlev, V.V.; Hahn, C.; Weiss, S.J. Fish diversity assessment in the headwaters of the Volga River using environmental DNA metabarcoding. Aquat. Conserv. Mar. Freshw. Ecosyst. 2019, 29, 1785–1800. [Google Scholar] [CrossRef]
- Kačergytė, I.; Petersson, E.; Arlt, D.; Hellström, M.; Knape, J.; Spens, J.; Żmihorski, M.; Pärt, T. Environmental DNA metabarcoding elucidates patterns of fish colonisation and co-occurrences with amphibians in temperate wetlands created for biodiversity. Freshw. Biol. 2021, 66, 1915–1929. [Google Scholar] [CrossRef]
- United States Geological Survey. Floodplain Elevation Data. Available online: https://apps.nationalmap.gov/viewer/ (accessed on 12 August 2024).
- Williams, K.E.; Huyvaert, K.P.; Piaggio, A.J. No filters, no fridges: A method for preservation of water samples for eDNA analysis. BMC Res. Notes 2016, 9, 298. [Google Scholar] [CrossRef] [PubMed]
- Spens, J.; Evans, A.R.; Halfmaerten, D.; Knudsen, S.W.; Sengupta, M.E.; Mak, S.S.T.; Sigsgaard, E.E.; Hellström, M. Comparison of capture and storage methods for aqueous macrobial eDNA using an optimized extraction protocol: Advantage of enclosed filter. Methods Ecol. Evol. 2017, 8, 635–645. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Turner, C.R.; Deiner, K.; Klymus, K.E.; Thomsen, P.F.; Murphy, M.A.; Spear, S.F.; McKee, A.; Oyler-McCance, S.J.; Cornman, R.S.; et al. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 2016, 7, 1299–1307. [Google Scholar] [CrossRef]
- Kumar, G.; Farrell, E.; Reaume, A.M.; Eble, J.A.; Gaither, M.R. One size does not fit all: Tuning eDNA protocols for high- and low-turbidity water sampling. Environ. DNA 2022, 4, 167–180. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef] [PubMed]
- Deagle, B.E.; Kirkwood, R.; Jarman, S.N. Analysis of Australian fur seal diet by pyrosequencing prey DNA in faeces. Mol. Ecol. 2009, 18, 2022–2038. [Google Scholar] [CrossRef] [PubMed]
- Klymus, K.E.; Richter, C.A.; Thompson, N.; Hinck, J.E. Metabarcoding of environmental DNA samples to explore the use of uranium mine containment ponds as a water source for wildlife. Diversity 2017, 9, 54. [Google Scholar] [CrossRef]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F.; et al. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef]
- Nakagawa, H.; Yamamoto, S.; Sato, Y.; Sado, T.; Minamoto, T.; Miya, M. Comparing local- and regional-scale estimations of the diversity of stream fish using eDNA metabarcoding and conventional observation methods. Freshw. Biol. 2018, 63, 569–580. [Google Scholar] [CrossRef]
- Mathon, L.; Valentini, A.; Guérin, P.E.; Normandeau, E.; Noel, C.; Lionnet, C.; Boulanger, E.; Thuiller, W.; Bernatchez, L.; Mouillot, D.; et al. Benchmarking bioinformatic tools for fast and accurate eDNA metabarcoding species identification. Mol. Ecol. Resour. 2021, 21, 2565–2579. [Google Scholar] [CrossRef] [PubMed]
- Alberdi, A.; Aizpurua, O.; Gilbert, M.T.P.; Bohmann, K. Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods Ecol. Evol. 2018, 9, 134–147. [Google Scholar] [CrossRef]
- McColl-Gausden, E.F.; Weeks, A.R.; Coleman, R.A.; Robinson, K.L.; Song, S.; Raadik, T.A.; Tingley, R. Multispecies models reveal that eDNA metabarcoding is more sensitive than backpack electrofishing for conducting fish surveys in freshwater streams. Mol. Ecol. 2021, 30, 3111–3126. [Google Scholar] [CrossRef]
- Chao, A.; Jost, L. Coverage-based rarefaction and extrapolation: Standardizing samples by completeness rather than size. Ecology 2012, 93, 2533–2547. [Google Scholar] [CrossRef]
- Chao, A.; Gotelli, N.J.; Hsieh, T.C.; Sander, E.L.; Ma, K.H.; Colwell, R.K.; Ellison, A.M. Rarefaction and extrapolation with Hill numbers: A framework for sampling and estimation in species diversity studies. Ecol. Monogr. 2014, 84, 45–67. [Google Scholar] [CrossRef]
- Roswell, M.; Dushoff, J.; Winfree, R. A conceptual guide to measuring species diversity. Oikos 2021, 130, 321–338. [Google Scholar] [CrossRef]
- Hsieh, T.; Ma, K.; Chao, A. iNEXT: Interpolation and Extrapolation for Species Diversity, R package version 3.0.1; R Core Team: Vienna, Austria, 2024. [Google Scholar]
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods; PRIMER-E: Plymouth, UK, 2008. [Google Scholar]
- Mathieu, C.; Hermans, S.M.; Lear, G.; Buckley, T.R.; Lee, K.C.; Buckley, H.L. A systematic review of sources of variability and uncertainty in eDNA data for environmental monitoring. Front. Ecol. Evol. 2020, 8, 135. [Google Scholar] [CrossRef]
- Beng, K.C.; Corlett, R.T. Applications of Environmental DNA (eDNA) in Ecology and Conservation: Opportunities, Challenges and Prospects; Springer: Dordrecht, The Netherlands, 2020; Volume 29, ISBN 0123456789. [Google Scholar]
- Missouri Department of Conservation (MDC). Missouri Species and Communities of Conservation Concern Checklist; Missouri Department of Conservation: Jefferson City, MO, USA, 2022. [Google Scholar]
- Geheber, A.D.; Piller, K.R. Spatio-temporal patterns of fish assemblage structure in a coastal plain stream: Appropriate scales reveal historic tales. Ecol. Freshw. Fish 2012, 21, 627–639. [Google Scholar] [CrossRef]
- Zbinden, Z.D.; Geheber, A.D.; Lehrter, R.J.; Matthews, W.J. Multifaceted assessment of stream fish alpha and beta diversity using spatial models. Hydrobiologia 2022, 849, 1795–1820. [Google Scholar] [CrossRef]
- Lee, V.M.; Berkman, L.K.; Geheber, A.D.; Landwer, B.; Ludwig, E.J.; Duvernell, D.D. Putting eDNA to the test: A field comparison of eDNA metabarcoding to established protocols for assessing biodiversity in Missouri’s Ozark Highland streams. Environ. DNA 2024, 6, e510. [Google Scholar] [CrossRef]
- Hallam, J.; Clare, E.L.; Jones, J.I.; Day, J.J. Biodiversity assessment across a dynamic riverine system: A comparison of eDNA metabarcoding versus traditional fish surveying methods. Environ. DNA 2021, 3, 1247–1266. [Google Scholar] [CrossRef]
- Euclide, P.T.; Lor, Y.; Spear, M.J.; Tajjioui, T.; Vander Zanden, J.; Larson, W.A.; Amberg, J.J. Environmental DNA metabarcoding as a tool for biodiversity assessment and monitoring: Reconstructing established fish communities of north-temperate lakes and rivers. Divers. Distrib. 2021, 27, 1966–1980. [Google Scholar] [CrossRef]
- Miya, M. Environmental DNA Metabarcoding: A Novel Method for Biodiversity Monitoring of Marine Fish Communities. Ann. Rev. Mar. Sci. 2022, 14, 161–185. [Google Scholar] [CrossRef]
- Koel, T.M. Spatial Variation in Fish Species Richness of the Upper Mississippi River System. Trans. Am. Fish. Soc. 2004, 133, 984–1003. [Google Scholar] [CrossRef]
- Dembkowski, D.J.; Miranda, L.E. Hierarchy in factors affecting fish biodiversity in floodplain lakes of the Mississippi Alluvial Valley. Environ. Biol. Fishes 2012, 93, 357–368. [Google Scholar] [CrossRef]
- Harris, P.M.; Roe, K.J.; Mayden, R.L. A Mitochondrial DNA Perspective on the Molecular Systematics of the Sunfish Genus Lepomis (Actinopterygii: Centrarchidae). Copeia 2005, 2005, 340–346. [Google Scholar] [CrossRef]
- Near, T.J.; Koppelman, J.B. Species diversity, phylogeny and phylogeography of Centrarchidae. In Centrarchid Fishes: Diversity, Biology, and Conservation; Cooke, S.J., Philipp, D.P., Eds.; Wiley-Blackwell: Chichester, UK, 2009; pp. 1–38. ISBN 9781405171229. [Google Scholar]
- Schönhuth, S.; Vukić, J.; Šanda, R.; Yang, L.; Mayden, R.L. Phylogenetic relationships and classification of the Holarctic family Leuciscidae (Cypriniformes: Cyprinoidei). Mol. Phylogenet. Evol. 2018, 127, 781–799. [Google Scholar] [CrossRef] [PubMed]
- Pukk, L.; Kanefsky, J.; Heathman, A.L.; Weise, E.M.; Nathan, L.R.; Herbst, S.J.; Sard, N.M.; Scribner, K.T.; Robinson, J.D. eDNA metabarcoding in lakes to quantify influences of landscape features and human activity on aquatic invasive species prevalence and fish community diversity. Divers. Distrib. 2021, 27, 2016–2031. [Google Scholar] [CrossRef]
- Rojahn, J.; Gleeson, D.M.; Furlan, E.; Haeusler, T.; Bylemans, J. Improving the detection of rare native fish species in environmental DNA metabarcoding surveys. Aquat. Conserv. Mar. Freshw. Ecosyst. 2021, 31, 990–997. [Google Scholar] [CrossRef]
- Solomon, L.E.; Phelps, Q.E.; Herzog, D.P.; Kennedy, C.J.; Taylor, M.S. Juvenile alligator gar movement patterns in a disconnected floodplain habitat in southeast Missouri. Am. Midl. Nat. 2013, 169, 336–344. [Google Scholar] [CrossRef]
- Smith, N.G.; Daugherty, D.J.; Brinkman, E.L.; Wegener, M.G.; Kreiser, B.R.; Ferrara, A.M.; Kimmel, K.D.; David, S.R. Advances in Conservation and Management of the Alligator Gar: A Synthesis of Current Knowledge and Introduction to a Special Section. North Am. J. Fish. Manag. 2020, 40, 527–543. [Google Scholar] [CrossRef]
- Adams, S.R.; Inebnit, T.E.; Lewis, L.C.; Naus, C.J.; Kluender, E. Spawning Ecology and Spawning Site Fidelity of Alligator Gar, Atractosteus spatula, in the Fourche LaFave River: Implications for River-Floodplain Management and Alligator Gar Conservation Spawning Ecology and Spawning Site Fidelity of Alligator Gar. Southeast. Fishes Counc. Proc. 2024, 1, 64. [Google Scholar]
- Morrison, R.R.; Simonson, K.; McManamay, R.A.; Carver, D. Degradation of floodplain integrity within the contiguous United States. Commun. Earth Environ. 2023, 4, 215. [Google Scholar] [CrossRef]

| Length | Width | Spring Depth (m) | Fall Depth (m) | |||||
|---|---|---|---|---|---|---|---|---|
| Habitat Complex | Sites | (km) | (m) | Mean | Max | Mean | Max | Habitats |
| Wolf Bayou complex-oxbow lake bayou complex | Wolf Bayou | 1.9 | 100 | 2.5 | 7.9 | 2 | 6.7 | Steep banks, little vegetation, submerged structure extensive in places, canopy cover limited along shorelines. |
| Hosner Bayou | 0.9 | 50 | 1.5 | 3.3 | 1.2 | 2.4 | ||
| Samples Bayou | 0.7 | 50 | 1.7 | 4.3 | 1 | 2.1 | ||
| Robinson Bayou-expanse of low relief floodplain | Robinson Lake | 2 | 200 | 0.6 | 1.2 | 0.3 | 0.5 | Shallow banks, extensive flooding into vegetated habitat (in spring), very little submerged structure, canopy cover extensive within flooded vegetation (in spring). |
| Black Island margin-slough and ditch complex | Ditch | >5 | 10 | 0.9 | 1.7 | dry | dry | Steep banks, vegetation along shoreline, no submerged structure, canopy cover extensive along slough but absent along ditch. |
| Slough | 5 | 30 | 0.8 | 1.4 | 0.5 | 0.6 | ||
| Abundance | Dependence | Percent of Total Read Count | Spring | Fall | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family | Species | Slough | Ditch | Robin-son Lake | Wolf Bayou | Hosner Bayou | Samples Bayou | Wolf Bayou | Hosner Bayou | Samples Bayou | MDC Surveys (Years Recorded) | |||
| Amiidae | Amia ocellicauda | U | B | 3.43% | 100% | 100% | 100% | 100% | 75% | 100% | 83% | 100% | 100% | 40; 79; 22 |
| Anguillidae | Anguilla rostrata | U | 0.004% | 33% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | ||
| Aphredoderidae | Aphredoderus gibbosus | U | B | 0.13% | 100% | 33% | 100% | 0% | 0% | 0% | 0% | 0% | 50% | 40; 79; 22 |
| Atherinopsidae | Labidesthes sicculus | U | F | 0.33% | 0% | 0% | 0% | 83% | 75% | 100% | 100% | 100% | 100% | 66; 79; 94 |
| Menidia beryllina | C | F | 0.14% | 67% | 67% | 86% | 17% | 0% | 0% | 17% | 50% | 0% | 78; 22 | |
| Catostomidae | Carpiodes spp. (carpio, cyprinus) | A/U | F | 0.05% | 100% | 100% | 43% | 50% | 25% | 50% | 50% | 75% | 50% | 79 |
| Ictiobus spp. (bubalus, cyprinellus, niger) | A/C/U | B | 10.36% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 40; 79 | |
| Moxostoma macrolepidotum | U | F | 0.00% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 79 b | |
| Centrarchidae | Centrarchus macropterus | P | B | 0.00% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 40 |
| Lepomis cyanellus | U | B | 2.35% | 67% | 67% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 66; 22 | |
| Lepomis gulosus | U | B | 0.89% | 100% | 100% | 100% | 100% | 100% | 100% | 83% | 75% | 100% | 40; 66; 22 | |
| Lepomis humilis | U | B | 6.89% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 79; 94; 22 | |
| Lepomis macrochirus | C | B | 12.3% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 40; 66; 79; 94; 22 | |
| Lepomis marginatus | P | B | 6.13% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | ||
| Lepomis megalotis | U | B | 0.43% | 100% | 100% | 71% | 100% | 100% | 100% | 83% | 100% | 100% | 79; 94; 22 | |
| Lepomis microlophus | U | B | 0.003% | 0% | 0% | 0% | 0% | 0% | 25% | 0% | 0% | 0% | ||
| Lepomis miniatus | U | B | 0.01% | 0% | 0% | 14% | 0% | 0% | 0% | 0% | 0% | 0% | ||
| Lepomis symmetricus | U | B | 0.02% | 0% | 0% | 86% | 67% | 25% | 0% | 17% | 0% | 0% | ||
| Micropterus punctulatus | P | B | 0.24% | 33% | 33% | 0% | 100% | 75% | 100% | 100% | 50% | 100% | ||
| Micropterus nigricans | U | B | 0.71% | 100% | 100% | 86% | 100% | 100% | 100% | 100% | 75% | 100% | 66; 79; 94 | |
| Pomoxis annularis | C | B | 0.33% | 100% | 100% | 100% | 83% | 100% | 75% | 100% | 75% | 100% | 40; 79; 22 | |
| Pomoxis nigromaculatus | U | B | 0.52% | 100% | 100% | 100% | 83% | 100% | 75% | 83% | 75% | 75% | 40; 22 | |
| Clupeidae | Alosa chrysochloris | C | F | 0.01% | 67% | 0% | 0% | 33% | 0% | 0% | 0% | 25% | 0% | |
| Dorosoma cepedianum | A | B | 4.37% | 100% | 100% | 100% | 100% | 100% | 75% | 100% | 100% | 100% | 40; 66; 79 | |
| Dorosoma petenense | U | B | 0.25% | 67% | 100% | 57% | 67% | 0% | 0% | 100% | 100% | 50% | ||
| Esocidae | Esox americanus | P | B | 0.01% | 0% | 0% | 29% | 0% | 0% | 0% | 0% | 0% | 0% | 79 b |
| Fundulidae | Fundulus spp. (notatus, olivaceus) | P | B | 0.87% | 100% | 100% | 100% | 100% | 100% | 100% | 83% | 100% | 100% | 66; 79; 94 |
| Ictaluridae | Ameiurus spp. (melas, natalis) | U | B | 0.01% | 0% | 0% | 57% | 0% | 0% | 0% | 0% | 0% | 0% | 40; 79; 22 |
| Ictalurus punctatus | C | 0.21% | 100% | 100% | 100% | 100% | 75% | 75% | 100% | 100% | 100% | 79 b | ||
| Noturus gyrinus | U | B | 0.02% | 0% | 0% | 14% | 50% | 0% | 50% | 50% | 25% | 50% | 66; 79; 94 | |
| Pylodictis olivaris | A | 0.35% | 100% | 100% | 29% | 100% | 100% | 100% | 83% | 100% | 100% | |||
| Lepisosteidae | Atractosteus spatula | U | B | 0.004% | 0% | 0% | 0% | 50% | 0% | 0% | 17% | 0% | 0% | |
| Lepisosteus oculatus | U | B | 1.49% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 22 | |
| Lepisosteus osseus | C | B | 0.15% | 67% | 100% | 57% | 100% | 100% | 75% | 100% | 100% | 100% | ||
| Lepisosteus platostomus | C | B | 3.22% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 40; 79; 22 | |
| Leuciscidae | Hybognathus hayi a | U | B | 0.00% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 79 b |
| Hybognathus nuchalis | C | F | 0.0002% | 33% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | ||
| Notemigonus crysoleucas | P | B | 0.01% | 0% | 0% | 57% | 0% | 25% | 0% | 33% | 25% | 0% | 40; 79; 22 | |
| Notropis atherinoides | A | F | 0.002% | 33% | 0% | 43% | 0% | 0% | 0% | 0% | 0% | 0% | ||
| Opsopoeodus emiliae | P | B | 0.28% | 100% | 0% | 86% | 67% | 100% | 75% | 83% | 100% | 100% | 79 | |
| Paranotropis shumardi a | C | F | 0.00% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 79 | |
| Moronidae | Morone chrysops | C | F | 0.07% | 100% | 100% | 29% | 17% | 25% | 0% | 33% | 50% | 0% | 79 b |
| Percidae | Etheostoma asprigene | U | B | 0.68% | 100% | 100% | 86% | 83% | 100% | 100% | 67% | 75% | 100% | 66;79 |
| Etheostoma chlorosoma | P | B | 0.40% | 100% | 0% | 86% | 83% | 75% | 75% | 67% | 100% | 100% | 79 | |
| Etheostoma gracile | U | F | 0.01% | 33% | 0% | 86% | 33% | 0% | 0% | 17% | 0% | 0% | 66; 22 | |
| Percina caprodes | U | 0.004% | 0% | 33% | 0% | 50% | 0% | 0% | 0% | 0% | 25% | |||
| Percina shumardi | U | F | 0.001% | 33% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 79 b | |
| Poeciliidae | Gambusia affinis | U | B | 1.59% | 100% | 100% | 100% | 83% | 50% | 25% | 33% | 75% | 100% | 40; 66; 79; 94; 22 |
| Polyodontidae | Polyodon spathula | C | F | 0.01% | 0% | 33% | 43% | 67% | 0% | 0% | 83% | 0% | 25% | 79 |
| Sciaenidae | Aplodinotus grunniens | A | F | 0.85% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| Cyprinidae | Cyprinus carpio | I | B | 6.89% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 40 |
| Xenocyprididae | Ctenopharyngodon idella | I | F | 1.45% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| Hypophthalmichthys molitrix | I | F | 8.54% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 22 | |
| Hypophthalmichthys nobilis | I | F | 20.6% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | ||
| Mylopharyngodon piceus | I | 2.36% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |||
| Source | df | SS | MS | Pseudo-F | P (perm) | Unique Perms |
|---|---|---|---|---|---|---|
| Habitat | 5 | 2284.2 | 456.83 | 6.8944 | 0.0001 | 9897 |
| Res | 21 | 1391.5 | 66.261 | |||
| Total | 26 | 3675.7 |
| Secchi (m) | Cond (µS/cm) | DO (mg/L) | Temp (°C) | |||||
|---|---|---|---|---|---|---|---|---|
| Sites | Sp | Fa | Sp | Fa | Sp | Fa | Sp | Fa |
| Wolf Bayou | 1.1 | 1 | 309 | 309 | 6.6 | 4.8 | 24 | 23 |
| Hosner Bayou | 0.7 | 0.7 | 261 | 310 | 8.4 | 6.9 | 27 | 23 |
| Samples Bayou | 1 | 0.8 | 281 | 300 | 7.5 | 5.5 | 26 | 20 |
| Robinson Lake | 0.5 | 0.1 | 465 | 339 | 5.5 | 5.9 | 28 | 15 |
| Ditch | 0.5 | NA | 406 | NA | 5.8 | NA | 24 | NA |
| Slough | 0.5 | 0.1 | 485 | 361 | 7.7 | 6.6 | 26 | 16 |
| Source | df | SS | MS | Pseudo-F | P (perm) | Unique Perms |
|---|---|---|---|---|---|---|
| Bayou | 2 | 364.9 | 182.44 | 2.81 | 0.0025 | 9941 |
| Season | 1 | 146.1 | 146.07 | 2.25 | 0.055 | 9957 |
| Bayou × Season | 2 | 180.1 | 90.05 | 1.39 | 0.19 | 9940 |
| Res | 22 | 1429.5 | 64.98 | |||
| Total | 27 | 2119.6 |
| Groups | t | P (perm) | Unique Perms | P (MC) |
|---|---|---|---|---|
| HB, SB | 1.28 | 0.14 | 9954 | 0.17 |
| HB, WB | 1.73 | 0.013 | 9947 | 0.028 |
| SB, WB | 1.76 | 0.022 | 9959 | 0.028 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ludwig, E.J.; Lee, V.M.; Berkman, L.K.; Geheber, A.D.; Duvernell, D.D. Biodiversity Assessment of a Mississippi River Backwater Complex Using eDNA Metabarcoding. Diversity 2024, 16, 495. https://doi.org/10.3390/d16080495
Ludwig EJ, Lee VM, Berkman LK, Geheber AD, Duvernell DD. Biodiversity Assessment of a Mississippi River Backwater Complex Using eDNA Metabarcoding. Diversity. 2024; 16(8):495. https://doi.org/10.3390/d16080495
Chicago/Turabian StyleLudwig, Eric J., Veronica M. Lee, Leah K. Berkman, Aaron D. Geheber, and David D. Duvernell. 2024. "Biodiversity Assessment of a Mississippi River Backwater Complex Using eDNA Metabarcoding" Diversity 16, no. 8: 495. https://doi.org/10.3390/d16080495
APA StyleLudwig, E. J., Lee, V. M., Berkman, L. K., Geheber, A. D., & Duvernell, D. D. (2024). Biodiversity Assessment of a Mississippi River Backwater Complex Using eDNA Metabarcoding. Diversity, 16(8), 495. https://doi.org/10.3390/d16080495









