Abstract
The presence of the olive tree in Tuscany, Italy, in its forms that have survived to the present day as an essential component of the landscape dates back many centuries. Global change is now threatening it. Therefore, it is important to find markers to enhance the olive tree environment in terms of its resilience. The aim of the research was to investigate the composition of soil bacteriomes in contrasting geochemical environments using a geochemistry approach based on the behavior of the REEs, inherited from parent rock material. Bacteriome assemblages and REE content were analyzed in 48 topsoils developed in six geochemical Tuscan environments. Combined geochemical, geoinformatic, and bioinformatic techniques highlighted the existence of four bacteriome assemblages depending on Light-REEs. Further results showed that the soil bioavailable fraction of REEs was related to parent rock materials, pH, and bacteriome composition. The most abundant bacteria were Microlunatus in graded fluvio-lacustrine soils, Gaiella in graded arenaceous soils, Bradyrizhobium in pyroclastic soils, and Rubrobacter in soils on gentle slopes of calcareous and carbonatic lithologies. This research represents a starting point to define new indicators able to assess the resilience of the olive trees in the Mediterranean landscape and characterize the territory of extra virgin olive oils.
1. Introduction
The soil microbiota plays a key role in fertility, and its composition is influenced by various factors, such as agricultural practices, microclimate, and soil structure [] Also, the mineral components of the soil can affect the composition of the microbiome [,]. Thus, there is a coevolution between soil microbial communities and the mineralogical environment [,].
Both bacteria and fungi coexist in soil with very important roles: they contribute to soil structures and moisture retention through physical and biochemical functions, and they are involved in the nutrient cycles [,,].
Microorganisms, being the main decomposers, are widely distributed in the soil, showing different and complex composition and structure between them (microbial communities). The continuous dynamic changes in the soil microecology, mediated by interactions between plant-microbe-soil communities, involve the regulation of soil ecosystems and therefore the development of the plants themselves. The main functions of the microbiome, including both bacteria and fungi, in the plant-microbe-soil system are: (1) regulating soil physico-chemical properties and fertility; (2) forming mycorrhizal structures with plant roots; (3) decomposing plant and animal residues; (4) participating in soil pollutants fixation and degradation; and (5) inhibiting pathogens and induced systemic resistance to plants [,,].
The present research targeted to study at first the soil bacterial community composition of century-olive orchards in Tuscany (Italy) that are now threatened by global change, especially by prolonged drought during different phenological phases of the plants. In fact, a preliminary and only indicative analysis by quantitative PCR of the 16S and 18S gene sequences was performed, indicating that the number of soil bacterial sequences was 10 times higher than the fungal one (Figure S1). Therefore, we decided to start with monitoring the soil bacteriome.
The general aim of the research was to investigate the composition and diversity of soil bacteria in different contrasting environments, and for this, we used a geochemistry approach based on the behavior of the REEs in the soil bioavailable fraction, which are mostly inherited from the parent rock material. The use of REEs as a natural tracer offers the possibility of further exploring an emerging scientific topic relating to the dependence of some bacteria on specific REEs as enzymatic cofactors [,,]. The REEs were here used as a proxy for different soil mineral assemblages.
REEs, a chemically uniform group due to their similar physicochemical behavior, which promotes their co-existence in nature, consist of 15 elements of the lanthanide series []. All REEs occur in nature but not in pure metal form, although promethium, the rarest, only occurs in trace quantities in natural materials as it has no long-lived or stable isotopes. REEs are widely distributed in the Earth’s crust, with concentrations ranging from 150 to 220 mg/kg []. For example, the mean total crustal abundance of REEs is 169 mg/kg, and Light REE (LREE from La to Eu) are 137.8 mg/kg higher than Heavy REE (HREE from Gd to Lu, 31.34 mg/kg). Some authors [,,], reported that the REE content in soil ranges from 30 to 700 mg/kg and in the topsoil layer reaches 100–200 mg/kg, increasing to 1000 mg/kg because of human activities. In Italy, median REE concentrations in agricultural soils were found to be 0.126 mg/kg and 167.24 mg/kg, bioavailable and total fraction, respectively []. More in depth, for olive groves in Croazia, the REY (REEs plus Yttrium) soil availability, extracted with 1 M NH4NO3 (mobile and short-term-available fraction), was found to be in the range of 0.1 mg/kg to 141 mg/kg []. In Italy, a value of REEs was found to be equal to 0.231 mg/kg for olive grove topsoil samples (extracted with DTPA, a potentially plant available fraction) collected from an Experimental Station of the Agricultural Development Agency (ESA) located in Sicily []. Furthermore, in Central Tuscany (Italy), for centuries-old olive groves, reported values of REEs in the DTPA-bioavailable topsoil fractions ranged from 0.91 mg/kg in claystone to 22.08 mg/kg in sandstone-derived soils [].
In the agricultural sector, the main source of REEs is linked to the use of phosphate fertilizers, and for conventional olive tree cultivation, the recommended dose is approximately 200–250 kg/ha of P2O5 []. Considering that the olive groves in the study area are composed of approximately 300–350 plants per hectare, it can be estimated that in the past, 0.6–0.7 kg of fertilizer per plant per year may have been applied. Our previous investigations carried out in collaboration with the Palermo University laboratory (unpublished data) showed that some commercial fertilizers, such as N:P:K 20:5:10, had a REEs content of about 18 g/kg, whereas the REEs distribution patterns of fertilizer were completely different from those found in the sampled soils.
The use of mixing REEs with fertilizers, currently carried out in paddy soils to improve crop yield, especially in China [] or in Brazilian tropical agroecosystems characterized by low native phosphorus (P) content [], is not a practice used in centenary-olive growing in Tuscany. For the Chianti area (Florence, Tuscany, Central Italy), Pelacani et al. [] found a REEs distribution in olive drupes showing the same patterns as their relative soils, with an exception for Lanthanum, Cerium, and Europium. Indeed, olive drupes from different soils showed variable REE patterns, allowing us to trace their origin [,,]. The Lanthanum anomaly in environmental samples is still poorly understood; however, recent studies have highlighted that biological activity can fractionate light-REE [,,,]. Interesting was that the anomaly for olive trees was probably due to the microbial enzymatic activities related to aerobic methane oxidation []. Moreover, some microorganisms can mediate the mobilization of REEs by exchange and replacement from mineral surfaces, through reductive or oxidative reactions, or by complexing with mineral surfaces. Several studies have shown a close correlation between soil microflora growth, reproduction, colonization, and REEs and in turn, microorganisms can uptake REEs from minerals and immobilize them in the microbial biomass []. Moreover, REEs reflect the mineralogical and geochemical signatures of the basin rocks and indicate the bio-geochemical evolutionary processes of the sedimentary systems [], and the majority of REEs derived from geological parent material vary in concentration as a function of bedrock and source areas [,]. The REE content in soils depends on the stability of primary REE-bearing minerals to weathering, the presence of a clay phase, the soil organic matter content, and therefore, on the soil’s physical-chemical characteristics [,,]. Moreover, in a previously conducted study in the same Chianti area of this work, Pelacani et al. [], by approaching through geostatistics techniques and a machine learning modeling framework, showed that the Lanthanum/Samarium and Lanthanum/Ytterbium spatial distribution models, based on the interaction of the topographic, geochemical, and hydrological variables (geodiversity), might change the response of the biotic components (REE accumulation pattern in olive drupe). Nevertheless, there are still few studies relating changes in REE composition to soil microbial diversity [,].
In Central Italy, extensive EVOO production has been known since the seventh century. At present, about 1.5 million olive trees are cultivated in this area [], and in Tuscany, an indisputable characteristic is the dual role played by the olive tree: a functional plant in the agricultural system for the “high quality” oil it provides and a determining element of the landscape, which guarantees balance between the naturalness of the environment and the degree of anthropization of the territory []. There are still ancient small and medium-sized artisan enterprises that produce EVOO, particularly for its organoleptic and nutraceutical qualities, which, however, are not valued. It is therefore important, to find experimentally valid markers to highlight the link between the olive tree and its environment [,]. In this framework, our research study focused on Tuscan organic olive orchard agro-ecosystems for at least 20 years, that use conservative practices with spontaneous and autochthone herbaceous cover cropping (e.g., Sulla coronaria L., Trifolium, Taraxacum officinale, Crepis Vesicaria, and Plantago lanceolata L.) and recycling pruning residues but grow on different soils developed from geochemically contrasting parent material.
Our hypothesis was that soil bacterial composition was sensitive to changes in REEs along a geochemical gradient and could be used as proxies for soils derived from different parent materials. Through REE content by inductively coupled plasma mass spectrometry and metagenomics by 16S Next Generation Sequencing, we experimented to show the existence of a characteristic soil bacterial community and its contribution to the characterization of the territory of a valuable extra virgin olive oil. To our knowledge, this is the first time that this approach has been applied to old olive grove soils.
2. Material and Methods
2.1. Soil Sampling Strategies and REE Analyses
Soil samples were collected during spring 2021 from 16 olive groves belonging to six geochemical environments of the Tuscany region, Italy (Figure 1): limestone, siltstone, sandstone, shale, volcanic rocks, and conglomeratic deposits; more information on the geology setting of the study area is reported in []. The study areas were (i) Chianti, located in the central part of Tuscany, close to Florence; (ii) Alta Valtiberina, located in the eastern part of Tuscany, close to Anghiari (Arezzo); (iii) Maremma, located in the southeastern part of Tuscany, close to Pitigliano (Grosseto).
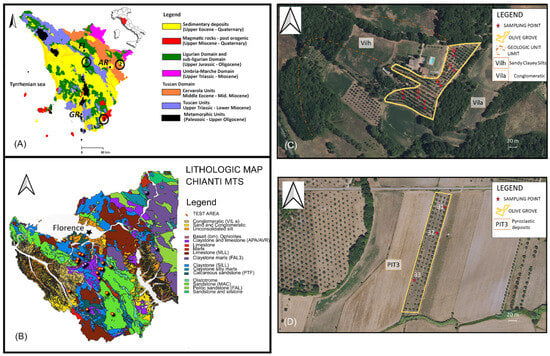
Figure 1.
(A) Schematic geologic map of the Tuscany Region, Italy. In black numbers are highlighted the different geochemical environments of olive groves where the soil samples were collected, located in: (1) Chianti area (Florence-FI), (2) Alta Valtiberina (Anghiari, Arezzo-AR), and (3) Maremma (Pitigliano, Grosseto-GR); (B) Lithologic Map of the Chianti Mts with the location of the olive groves (TEST AREA); (C) Orthophotos (2023 years, Regione Toscana, Geoscopio WEB GIS) showing the sampling point distribution for the Anghiari olive grove; (D) Orthophotos (2023 years, Regione Toscana, Geoscopio) showing the sampling point distribution for the Pitigliano olive grove.
The olive groves are characterized by different landforms, lithologies and hence soil types (Table 1). The median areal extension of single olive groves is about 0.9 hectares and from each site, three soil samples were collected from the first 30 cm of topsoil, as close as possible to the olive tree roots, for the REEs and metagenomic analysis. These last samples were stored at −20 °C until DNA extraction.

Table 1.
Landform and sampling site descriptions for topsoil collected in 16 Tuscany olive orchards: soil texture, pH, SOM (Soil Organic Matter), soil classification, lithology, aspect, total contents, and fractionation ratios of REE for topsoil developed on 11 lithologies in Tuscany. The soil classification obtained from the Tuscany region soil database was referred to the World Reference Base (WRB-FAO, 2015). The subscripts beside the geological formation indicate the different sampling soils and are not referred to another geological domain (ΣREE [mg/kg]: REE contents in the bioavailable fraction).
The selected olive groves lie on a geomorpho-dynamically stable landform [], where the soil developed from its parent rock material and is not subjected to soil erosion or depositional processes of sediments. Therefore, samples were collected from olive groves lying on the upper part of the relief, where the soil horizons develop from each selected bedrock. If these conditions were not satisfied, the soil hillslope catena concept [] was taken into consideration for sampling design.
REEs include the 15 lanthanides (atomic number 57–71), and, according to their physical and chemical properties, they are usually divided into light (L-REEs = Light Rare Earth Elements from La to Sm), medium (M-REEs from Sm to Gd), and heavy (H-REEs from Gd to Lu) REEs.
Their bioavailable content in soils (Table 1) was analyzed by a quadrupole Inductively Coupled Plasma Mass Spectrometry (ICP-MS; Agilent Technologies 7900 ICPMS-Hachioji, Tokyo, Japan) equipped to remove polyatomic interferences. The reproducibility of measurements was assessed by combining an internal standard normalization by rhodium and an external calibration using AGV-1-certified reference materials. The accuracy (RSD) for all of the estimated elements varied from 2 to 12.9% at the μg g−1 levels of concentration. The variability of the analytical repetitions was <10% at the ng kg−1 levels of concentration. The soils were treated with DTPA at pH 5 [,] to simulate the uptake by the rhizosphere microbiome. Each sample was determined in triplicate. The bioavailable fraction was extracted by reacting 10 g of dried soil with 20 mL of a 5mM DTPA solution at pH 5. The obtained suspension was stirred for 24 h at 25 °C, filtered with Millipore™ membranes (0.45-μm membrane filter), and diluted 50 times.
2.2. Geological Settings
The Chianti study area (Figure 1) is located 30 km in the SW direction of Florence, Tuscany, Italy. The most widely outcropping characterized by olive groves in this area are the sediments belonging to the External Ligurian Domain Mt. Morello units []: Morello Formation, Sillano Fm. [], and Pietraforte Fm., followed by Tuscan Nappe, Macigno Fm., and Palombini Shales (APA-Mt. Gottero Unit), which are part of the Internal Ligurian Domain. A post-orogenic fluvial deposit was also considered in the Chianti area (Plio-Pleistocenic deposits, VILa-Montefiridolfi, Figure 1 and Table 1). The Morello Unit is represented by limestones and shales (Sillano Formation-SILL, late Cretaceous–early Eocene) with interbedded coarse lens-shaped bodies of finely grained quartz-calcareous sandstones (‘Pietraforte’Fm.-PTF); limestones, marly limestones, and sporadically clayey marls and calcarenites (Mt. Morello Fm.-MLL, early-middle Eocene). The inner portion of the Ligurian domain is characterized by shales and silica-rich limestone, quartz-rich siltstones and sandstones, marls, and calcareous turbidites (‘Palombini’ Shales Fm.-APA; early Cretaceous) with interbedded fragments of ophiolitic complexes (basalts-bm, gabbros, serpentinite breccias, and reddish cherts). The Sillano Fm. is a chaotic body [] of mixed rocks, including blocks of different ages and origin, such as varicolored shales (AVR, green, gray, brown, and red), alternating quartz-rich or carbonaceous turbiditic sandstone, siltstones, marls, and calcareous marlstone.
The ‘Macigno’ Sandstones Fm. (‘Macigno’ Fm.-MAC; Chattian-Aquitanian) is the uppermost unit of the Tuscan Nappe with a thickness of up to 1500–2000 m [] of siliciclastic turbidite sandstones and siltstones. Furthermore, the ‘Macigno’ Fm. hosts interbedding up to 50 m thick lenticular successions of hemipelagic marls and very fine/distal turbidites (San Polo Marls–“Marne”).
The lithology belonging to the Middle Pleistocene monogenic conglomerates (VILa), corresponding to the top of fluvio-lacustrine deposits in the Anghiari-Sansepolcro basin [], was selected in Alta Valtiberina (Figure 1), and it is part of the larger Tiberino Basin [].
The Pitigliano Fm. (Maremma, Figure 1) represents the youngest deposit of the Latera volcanic sequence, and it is characterized by a complex sequence comprising airfall, pumice deposit, flow deposit, ignimbrite strongly welded, and pyroclastic flow containing glassy matrix [].
2.3. Extraction of Total DNA and 16S rRNA Gene Sequencing
Metagenomic soil DNA was extracted by the “FastDNA ™ SPIN Kit for Soil” (MP Biomedicals, Santa Ana, CA, USA) from 0.5 g of soil using the FastPrep® instrument [].
The V3-V4 region of the 16S rRNA gene was amplified using specific primers S-D-BACT-0341 and S-D-BACT-0785 [] by using a T-Professional thermal cycler (Biometra, Biomedizinische Analytik GmbH, Gottingen, Germany). The PCR reaction mix (50 µL) contained: 40 ng of template DNA, 1X (plus MgCl2 20 mM) Dream Taq reaction buffer (Thermo Scientific, Carlsbad, CA, USA), 0.05 Units µL−1 of Taq DNA Polymerase (ThermoFisher Scientific), 0.4 µM of each primer, and 0.4 mM of dNTPs. PCR running conditions were: 3 min denaturation at 95 °C, followed by 30 cycles each consisting of 30 s at 95 °C, 30 s at 55 °C, and 30 s at 72 °C, followed by a final extension step at 72 °C for 7 min. PCR products were mixed at equal density ratios. The mixed PCR products were purified with the Qiagen Gel Extraction Kit (Qiagen, Hilden, Germany).
The library preparation and NGS sequencing were performed by Novogene Company (Novogene, Beijing, China) on the Illumina platform on the NovaSeq PE 250 System (Illumina, San Diego, CA, USA).
2.4. Bioinformatic Analysis
2.4.1. Sequencing Data Processing
Paired-end reads were assigned to samples based on their unique barcodes, truncated by cutting off the barcode and primer sequences, and merged using FLASH (V1.2.7) []. Quality filtering on the raw tags was performed to obtain high-quality clean tags [] according to the quality-controlled process Qiime (V1.7.0) [].
The tags were compared with the reference database (Gold database) using the UCHIME algorithm (UCHIME Algorithm) [] to detect chimera sequences; then the chimera sequences were removed [], obtaining, finally, the effective tags.
2.4.2. OTU Cluster and Taxonomic Annotation
Sequence analyses were performed by Uparse software (Uparse, v7.0.1001) [] using all the effective tags. Sequences with ≥97% similarity were assigned to the same OTUs. A representative sequence for each OTU was screened for further annotation.
For each representative sequence, Mothur software (v.1.43.0) was performed against the SSU rRNA database of the SILVA Database [] for species annotation at each taxonomic rank [].
To obtain the phylogenetic relationship of all OTU representative sequences, the MUSCLE alignment software (Version 3.8.31) [] was used to compare multiple sequences rapidly.
OTUs abundance information was normalized using a standard sequence number corresponding to the sample with the fewest sequences. We then selected the top 10 taxa at the Phylum and Genus taxonomic rank for each lithology to obtain the relative abundance of taxa.
2.5. Statistical Analysis
2.5.1. Multivariate Analysis
The relationship between biological assemblages of species and REEs was determined by Canonical Correspondence Analysis (CCA) in R vegan [], to infer information from cross-covariance matrices. An analysis of variance was applied using distance matrices to find the best pattern of REE that describes the community structure. For this purpose, we have employed Adonis as a nonparametric statistical method based on Bray-Curtis distance with 999 permutations, which fits linear models to distance matrices and uses a permutation test with pseudo F-ratios. CCA plot (Figure 4) using only those elements of REEs that were found to be significant (p < 0.01). ANOVA combined with CCA was used to test the effect of REEs. Then the data was visualized on a 3D plot linking weighted average scores with linear combination scores (“orglspider” function in vegan3d-R), where each sample site was connected by segments to the centroid of its group of similarity (Figure 5).
2.5.2. Supervised Machine Learning
Prediction of a categorical or numerical response variable using a set of predictor variables is the main objective of supervised machine learning. The categorical variable “Lithologies” was used to train classification and regression trees supervised learning algorithms to recognize REE fractionation patterns associated with different topsoils by an ensemble of bagged decision trees, a flexible nonparametric tool for tree-based algorithms []. Bagging, which stands for bootstrap aggregation, is an ensemble method that reduces the effects of overfitting and improves generalization. The fractionation ratio was used as a predictor variable to characterize soils developed on volcanic rocks, tertiary and quaternary deposits in the Tuscany region (Figure 1). Furthermore, CART was used to identify variables, such as the specific REE and its concentration, associated with a specific soil bacterial composition and, thus, to characterize a particular environment. CART uses training data and creates a model that could predict the value of a class for each test. The CART algorithm uses splitting rules to segment the predictor space into several high-dimensional terminal nodes to minimize the classification error. Recursive binary splitting is used to find the best predictor variable. The class of a new observation is predicted using the mode of the training observations in the terminal node to which the new observation belongs. The number of predictor variables used to fit a predictive model is the major factor contributing to increased model complexity, which may result in decreased accuracy and increased model variance []. In this study, we used 14 geochemical predictors (REEs) and 3017 bacterial predictors (OTUs), considering 11 terminal nodes, equal to the lithologies class (target variable). It was randomly selected from about 30% of the total 9864 OTUs to prevent overfitting in machine learning [].
3. Results
3.1. Geochemistry of REEs
REE contents in the bioavailable fraction ranged from 1.2 to 69.1 ppm in basaltic- and pyroclastic-derived soils, respectively (Table 1). A generally lower content was found in shales- and limestone-derived soils (2.0–4.0 ppm); conversely, a medium to higher content was found in soils derived from sandstone, fluvio, and fluvio-lacustrine deposits (7.0–21.9 ppm).
These results suggested a larger REE delivery to soil from pyroclastic and arenaceous parent materials than calcarenitic and marly parent materials, with the exception of coarse conglomeratic soils of fluvio and fluvio-lacustrine deposits.
REE patterns were calculated by normalizing the measured concentrations to Post-Archean Australian Shale (PAAS) according to Taylor and McLennan []. (La/Sm)n, (La/Yb)n, and (Gd/Yb)n ratios indicated L-REE enrichment over M-REE, L-REE enrichment over H-REE, and M-REE enrichment over H-REE, respectively (Figure S2). A ratio of 1 indicates no fractionation with respect to PAAS, whereas a value greater than 1 denotes enrichment and a value less than 1 denotes depletion. In general, LREE/HREE fractionation is more pronounced in soils developed on pyroclastic sediment (PIT3, volcanic rock) and Macigno Formation rocks (MAC, sandstone). Fractionation for L-REE with respect to M-REE was visible in the pyroclastic sediment, conversely, in sandstone and in other lithologies. MREE/HREE fractionation is evident in all lithologies and mostly in marls, shales (SILL, AVR, and APA), and carbonate sediments (MLL, Monte Morello Formation). Interestingly, a positive fractionation of (Pr/Ce)n is observed in soils developed on shales (APA and SIL; Figure S1 and Table 1).
Although most authors [] pointed to the enrichment of REEs with the increase of finer particles, the correlation between the total content of REEs and the texture of the studied soils (Table 2) is proportional to sand concentration (r > 0.6; r = Pearson coefficient). A negative relationship was found between the total REE content and the silt concentration. There is a significant relationship between the fractionation parameters and the contents of the sandy, silty, and clay fractions. The variation of some fractionation parameters, such as (La/Yb)n, rather reflects the petrographic nature of the source rocks. The lowest values of this parameter are found in soils influenced by basaltic rocks (bm) and developed on conglomerate rocks (Villa). A positive relationship was found between (i) (La/Yb)n, (La/Sm)n, and (La/Gd)n and the sand concentration (r > 0.6); (ii) (Pr/Ce)n and clay concentration (r > 0.5), and a negative linear correlation was found between (La/Yb)n, (La/Sm)n, and (La/Gd)n and the silt concentration (Table 2). A general high correlation (r > 0.7) with a negative relationship was found between the values of pH and (La/Yb)n and (La/Sm)n, whereas no correlation with r > 0.3 was found between pH and (Gd/Yb)n and (Pr/Ce)n (Table 2).

Table 2.
Correlation coefficient for each pair of variables: soil texture, pH, REE, and fractionation ratios of REE for topsoil developed on 11 lithologies of Tuscany.
To address which fractionation ratio of REEs was important for characterizing the topsoil developed in different geological environments, we performed cluster analyses by using a principal component analysis (PCA) []. Figure 2 shows the relationship among soil samples considering the ratios La/Yb and Gd/Yb. Highly clustered samples were found for pyroclastic and arenaceous-derived soil. Furthermore, the component PC1 described the soils as follows: (I) those developed on fluvio-lacustrine deposits and basalts; (II) those related to the soil of the Chianti area developed on fluvio-conglomeratic deposits claystone and carbonatic flysh; and (III) pyroclastic derived soil. The PC2 component highlighted the difference between arenaceous and pyroclastic-derived soils and those derived from calcareous soils, hence limestone, marls, and claystone (IV).
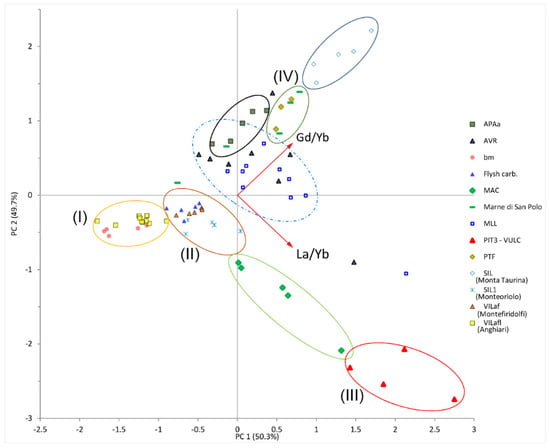
Figure 2.
Correlation among soil samples considering the ratios La/Yb and Gd/Yb. Scores plot for PC1 (50.3% of the total variance) vs. PC2 (49.7%) describe the 73 topsoil samples from each parent rock material, which were color coordinated. The roman numbers in brackets refer to different clusters: (I) conglomeratic (Anghiari, Valtiberina) and basaltic (Chianti); (II) conglomeratic (Montefiridolfi), carbonatic flysh, and claystone in the Chianti area; (III) pyroclastic-derived soils; and (IV) claystone, marls, and siltstone.
3.2. Bacteriome Sequencing
A total of 2,219,018 amplicon sequences from the V3-V4 region of the 16S rRNA gene that clustered into 9864 OTUs (97% nucleotide identity) were obtained from the sequencing analysis. As shown in Figure 3a, the dominant bacterial phyla were Actinobacteria (39.6%), Proteobacteria (21.2%), and Acidobacteria (9.6%); the dominant genera (Figure 3b) were Microlunatus, especially in soils developed on fluvio-lacustrine deposits (VILa1-2-3), Gaiella in arenaceous soil (MAC), Bradyrizhobium in pyroclastic soil (Vulc), and Rubrobacter in carbonate flysch, basalts, and marls soils (Chianti area Marne, bm, Fcar, MLL).
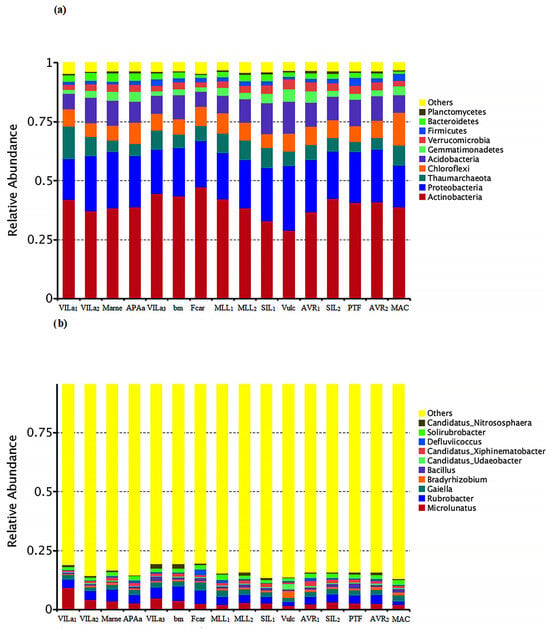
Figure 3.
Principals of 10 groups of bacteria at Phylum (a) and Genus (b) levels for olive grove lithologies (Table 1) in Tuscany. The dominant bacterial phyla were Actinobacteria, Proteobacteria, and Acidobacteria. The dominant genera were Microlunatus in soils developed on fluvio-lacustrine deposits (VILa1-2-3), Gaiella in arenaceous soils (MAC), Bradyrizhobium for pyroclastic (Vulc) soils, and Rubrobacter in carbonate flysch, basalts, and marls soils (Chianti area Marne, bm, Fcar, MLL).
Moreover, the phylogenetic distance between bacterial phyla of the topsoil of different lithologies (Figure S3) highlighted that the bacterial clusters I and II were close in soils developed on a pebbly matrix or of basaltic origin, as also shown by the PCA (Figure 2). Furthermore, cluster III in volcanic soils was distant from cluster IV, which characterizes soils developed from marls and claystone, and relatively close to those in the arenaceous ones (MAC).
3.3. Correlation between Bacteriome and REEs
The fractionation patterns of REEs were used to characterize the topsoil developed on different geologic substrata because they are either inherited from their parent rock materials (source-related) and controlled by natural geochemical processes or influenced by anthropogenic sources. At the same time, we compared the results of PCA soil clustering to the phylogenetic ones (Figure 4).
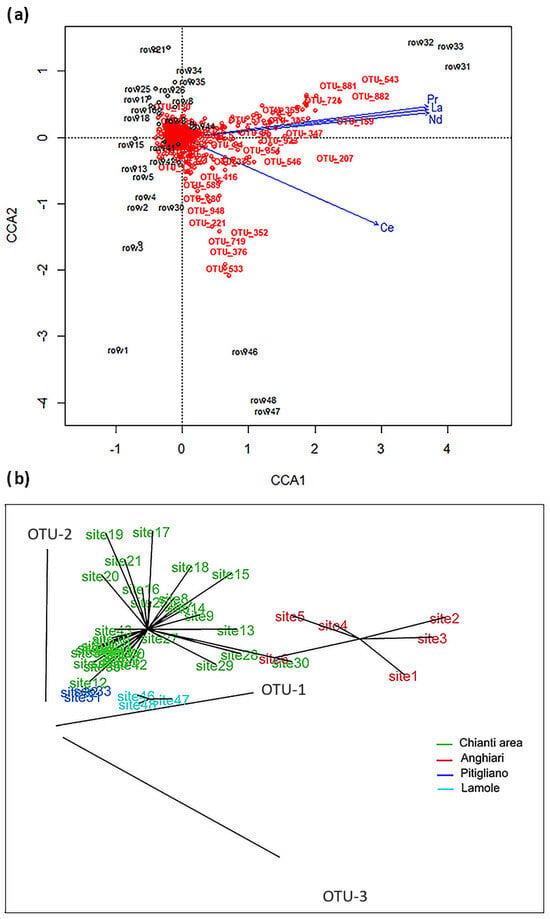
Figure 4.
Relationships between soil bacterial composition and REE content: (a) CCA joint plot between OTUs and significant REEs (Pr: Praseodymium; La: Lanthanum; Nd: Neodymium; Ce: Cerium) in soils (rows) developed on different lithologies of Tuscany. Rows refer to Table 1. (b) Sites of sampled soils grouped by OTUs similarity, clustering in four different bio-geochemical environments of Tuscany. The corresponding lithologies are for Chianti area (green): Marne, APAa, VILa3, bm, Fcar, MLL1-2, SIL1-2, AVR1-2, and PTF; for Anghiari (red) VILa1-2; for Pitigliano (dark blue) Vulc-PIT3, and for Lamole (sky blue) MAC (Table 1).
Two complementary approaches were used to elucidate the relationships between soil bacterial composition and REE content in the analyzed topsoil: a canonical correspondence analysis (CCA) in R vegan [] and CART analyses performed by Salford Predictive Modeler Software (SPM Version 8.3.2, Minitab 19). ANOVA applied to CCA showed a significant (p < 0.01) association between microorganisms and REEs. By Adonis analyses, the CCA1 axis showed that Lanthanum (La), Cerium (Ce), Praseodymium (Pr), and Neodymium (Nd) (p < 0.01) were LREEs that affected the composition of the bacteriome (Figure 4a). Pyroclastic soils (rows 31, 32, 33) were characterized by the highest content of La, Ce, Pr, and Nd, while arenaceous soils (rows 46, 47, 48) were mainly characterized by the Ce content. Considering the OTU relative abundance and REE content of each soil, we observed the following behavior: (i) most of the OTUs were close to the intersection of the axes, representing the core bacteriome []; (ii) some OTUs (i.e., 159, 543, 881, 882), belonging to Thaumarchaeota, Verrucomicrobia, and Chloroflexi, reached high relative abundances in volcanic soils also characterized by the high concentrations of La, Ce, Pr, and Nd; and (iii) other OTUs (376, 533, 719), belonging to Chloroflexi, showed higher values of relative abundances in arenaceous soils (MAC).
The results of the CCA analysis were visualized by the 3D plot (Figure 4b), where each sample site was connected with the centroid of its group of similarity. This plot showed four clusters: conglomeratic derived soil (VILa1-Alta Valtiberina, Anghiari), pyroclastic derived soil (PIT3-Vulc Maremma, Pitigliano), arenaceous soil (MAC-Chianti area, Lamole), and 11 different lithologies in one single cluster (Chianti area).
By using the CART statistical analysis, we further assessed the value of REE contents in soils that could affect the bacteriome composition (OTUs). The cluster results for all selected lithologies of the Tuscany olive grove and, separately, for only the lithologies of the Chianti area were summarized in Figure 5a and 5b, respectively. The Chianti area alone was considered in the model computation because of the presence of a great variety of lithologies as opposed to the monolithologic system of the Pitigliano area. The CART outputs indicated that the L-REE may be used as a proxy for the bacteriome composition. For instance, a Ce content lower than 0.36 ppm was a discriminant factor for soils derived from argillaceous rocks (APA-Palombini shales). This result agreed with the descriptive statistical analyses (Figure S2) where we considered the Pr/Ce ratio. Moreover, in agreement with the CCA results, La content (>12.78 ppm) and Pr content (>1.85 ppm) characterized volcanic soils (VUL-PIT3). During the initial recursive partitioning step of the analysis, the following phyla, Actinobacteria (OTU_95, Rubrobacteria OTU_2) and Thaumarchaeota (Nitrososphaeria OTU_57, Nitrososphaeria OTU_1), depended on the REE contents (Figure 5a) in agreement with CCA results. CART analysis showed that Nitrososphaeriales was a discriminant factor for clustering soils developed on fluvio-lacustrine deposits (VILL), characterized by a high content of gravels, and soils characterized by fine-grained quartz-calcareous sandstones (PTF) and hemipelagic marls (Marne). Furthermore, the Rubrobacteria characterized soils from the carbonate flysch sediment and claystone (APA). The CART analysis of the Chianti area (Figure 5b) showed that Bacteroides (Chitinophagales OTU_14), Protobacteria (Alphaproteobacteria OTU_16, Gammaproteobacteria OTU_17), and Firmicutes (Bacillales OTU_83) were affected by soils developed on different lithologies. Furthermore, considering only the bio-geochemical environment of this area, which is characterized by several different kinds of lithologies (Table 1), the analysis showed that the first discriminant factor was the Ce content. Moreover, Rubrobacteria (OTU_2 Figure 5a) were discriminant for the Chianti area. These findings agreed with the statistical analysis in Figure 3, where the dominant genus in Chianti soil was Rubrobacter.
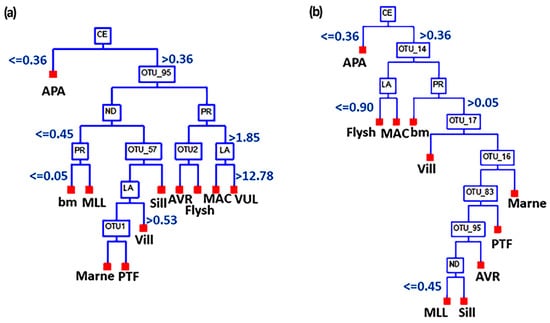
Figure 5.
CART dendrogram grouping biological species assemblages (OTUs) of each site sampled and its REEs content for the selected lithologies of Tuscany (a) and the lithologies of Chianti area (b). The values shown in the figure refer to REE concentration (ppm). The phyla (and classes) of the OTUs listed in the graphs are: (a) Actinobacteria (OTU_95, Rubrobacteria OTU_2) and Thaumarchaeota (Nitrososphaeria OTU_57, Nitrososphaeria OTU_1); (b) Bacteroides (Chitinophagales OTU_14), Protobacteria (Alphaproteobacteria OTU_16, Gammaproteobacteria OUT_17), and Firmicutes (Bacillales OTU_83). For model (a) the accuracy is equal to 97%. The receiver operating characteristics (the area under ROC) curve was 0.87. For model (b), the accuracy is equal to 82% and the ROC value is 0.86.
4. Discussion
The soil bacteriome characterization is one of the major scientific challenges due to its high complexity; therefore, a multidisciplinary approach with different skills and knowledge is required [,]. On the other side, the biological role of REEs has long been underestimated, despite several studies showing how these elements can affect some groups of microorganisms with stimulation or inhibition of their metabolic functions []. For this reason, in the present research, we integrated the biogeochemical approach with bioinformatics and machine learning techniques to investigate the composition and diversity of soil bacteria within contrasting environmental settings but with the same land use and comparable management. Machine learning has proven to be a useful approach for analyzing bacteriome data and predicting outcomes, including human and environmental health (TARA OCEANS project []; Earth Microbiome Project []). For example, machine learning applied to microbial community profiles has been used to predict environmental quality as well as trace evidence in forensics.
Using the REEs as a proxy for different soil mineral assemblages, we showed that the lithologies are closely related to microbial community assemblages (Figure 4). Through the CCA analysis, the existence of four microbial assemblages depending on the LREEs was highlighted, in agreement with [], who found a consistent biotic signature for L-REE with a general depletion with respect to M-REE and H-REE. The found bacteriome assemblages corresponded to geographic ranges with geologically young (pyroclastic–PIT3) and old (Palombini shales–APA) soils and different microclimates. We hypothesized that this result could be due to the long history of weathering at these sites, given that the REE content in the bioavailable fraction of Tuscan olive grove soils is in the range of non-contaminated European soils [,,], varying from 1.2 to 69.1 mg/kg in basaltic- and pyroclastic-derived soils. It is known that soil salinity, temperature, pH, and organic matter individually affect the bacteriome, but they can more significantly modify it if combined with REEs [,]. However, the most sensitive bacterial phyla to REE content belonged to Proteobacteria, Verrucomicrobia, and Firmicutes [,]. Our results from NG sequencing for the considered Tuscany olive groves showed that the dominant phyla were Actinobacteria, Proteobacteria, Acidobacteria, Chloroflexi, Verrucomicrobia, and Firmicutes (Figure 3a). Similar results were found by Fausto et al., [] in soils of mature olive groves in Matera province, Southern Italy, although Firmicutes were not present in consistent numbers in Tuscan olive groves.
Our results showed that a higher content of bioavailable REEs may have selected bacterial assemblages more suitable to survive in these soils, as indicated by the abundance of bacterial genera (Figure 3b) but also by alfa diversity (Figure S2). In fact, the pyroclastic had the greatest specific number of 275 OUTs, the arenaceous 255 OTUs, the Palombini shales 145, and the carbonate flysh 135 OTUs. In particular, NGS results showed that the dominant phyla were Actinobacteria, Proteobacteria, Acidobacteria, Chloroflexi, Verrucomicrobia, and Firmicutes (Figure 3a). Actinobacteria are described as highly resistant to drought and poor nutrient conditions. Moreover, they are almost ubiquitous in soils and have the function of promoting plant growth thanks to their good adaptability to the plant root surfaces and their ability to counteract many plant pathogens [,,]. Acidobacteria, Verrucomicrobia, Chloroflexi, and Firmicutes include members involved in sulfate- and iron-reduction, those surviving in groundwater, volcanic, or heavy meal-contaminated soils where high REE content has accumulated; thus, these bacteria may be used as indicators of risk assessment [,,]. Among the Proteobacteria, generally divided into five main phylogenetic lineages known as Alpha proteobacteria, Beta proteobacteria, Gamma proteobacteria, Delta proteobacteria, and Epsilon proteobacteria [], Acidobacteria have extensive metabolic versatility and therefore have a dynamic role in soil ecology. For example, they participate in the nutrient biogeochemical cycles and include plant growth promotion rhizobacteria []. Furthermore, oligotrophic chloroflexi are considered omnipresent in soil and involved in various biogeochemical processes. Some members are aerobic thermophiles, demonstrating the ability to live in soil spots with high temperatures and promoting the decomposition of organic matter []. Verrucomicrobia are mostly not cultivable in vitro, but they are involved in the degradation of complex compounds that allow them to survive environmental stresses, characteristics that make them suitable for a possible use in sustainable agricultural systems [,]. Moreover, Firmicutes were found to be discriminant between calcaric-siltstone-derived-soil (PTF) and limestone and claystone-derived-soil of the Chianti area (Figure 5b, OTU 83). This phylum includes bacteria belonging to the genus Bacillus, whether or not endospore-forming, Clostridium, and Lactic Acid Bacteria. Bacillus and Clostridium are able to degrade various carbon substrates, such as polysaccharides and other complex carbon compounds; some are capable of fermentation, and others are involved in the nitrogen cycle. Thanks to their ability to form endospores, they can survive drought for long periods in soil []. The genus Lactobacillus includes aerotolerant anaerobes found in food-rich environments, decaying plants, milk products, and the human gut. Due to these multiple characteristics, they have recently attracted much interest for sustainable agricultural strategies [].
The dominant genera (Figure 3b) were: Microlunatus in soils developed on fluvio-lacustrine deposits (Anghiari VILa1-2, and Montefiridolfi VILa3); Rubrobacter in soil developed from carbonate flysch, basalts, and marls (Chianti area Marne, bm, Fcar, MLL; Table 1); Gaiella and Bradyrhizobium in arenaceous (MAC) and pyroclastic (Pitigliano Vulc) soils, respectively. Overall, these bacterial genera perform important soil metabolic functions, including the major nutrient cycles. In particular, Rubrobacter participates in the dark oxidation of sulfur compounds and thus can affect sulfur availability to plants, but it is also abundant in sunlight-exposed biofilms, in desiccated areas of the Atacama desert [], and in other arid soils [,]. The relative abundance of Rubrobacter was higher in Chianti soils than in Anghiari conglomerate soils. Considering the rising temperature in the Chianti area of the last decade, with temperatures above 35 °C [], we can hypothesize that the higher presence of Rubrobacter could be related to water stress and desiccation conditions in Chianti soils. This hypothesis was also supported by [], who also suggested that Rubrobacter may protect other soil bacteria from heat, salinity, radiation, and desiccation stresses through trehalose synthesis. Moreover, Microlunatus contributes to the decomposition of organic matter; Gaiella might be an indicator for soil PAH contamination; symbiotic Bradyrhizobium is notoriously involved in nitrogen-fixation; and non-symbiotic Bradyrhizobium has been recently described as resistant to heavy metals and REE stress [,,]. It is interesting to note that the volcanic soil (PIT3, Table 1), characterized by minerals such as sanidine and halloysite, showed a bacterial composition predominantly dominated by the genus Bradyrhizobium (Figure 3b). Furthermore, during the pedological investigation, it was possible to detect the presence of Trifolium spp. in this olive grove (Figure 1). It is known in the literature that symbiotic Bradyrhizobium are involved in nitrogen fixation, while non-symbiotic Bradyrhizobium are resistant to heavy metals and REE stress [,,]. A legume symbiotic nitrogen-fixing Bradyrhizobium strain can grow on methanol as a sole carbon source in the presence of La3+, which is essential for the methanol dehydrogenase activity [,]. The presence of Bradyrhizobium is important for biologically significant functions including photosynthesis, nitrogen fixation, heterotrophic denitrification, and the degradation of aromatic compounds [].
The results of CART analysis (Figure 5a,b) led us to think that soils with different REE contents can have a different characteristic bacterial composition. Furthermore, cluster analyses (Figure 2 and Figure S2) showed that the bacterial taxonomic composition was related to soil texture and to the lower fractionation ratio La/Yb in the case of the pebble surface characterizing the conglomeratic-derived-soils (Anghiari, Montefiridolfi). This could be related to the effect of the pebble’s physical properties on bacterial niche differentiation, resulting in different soil community structures [,]. According to the CCA analysis (Figure 4a), a number of specific OTUs were connected to L-REE in soils developed on pyroclastic lithology (i.e., rows 31, 32, 33, VULC-PIT3) and characterized by a high content of La, Nd, Pr, and Ce, while many others were centered on limestone deposits (Marne, APAa, bm, Fcar, MLL, SIL, AVR, PTF, in Chianti) far from all the other L-REE lithologies. The likely selection of bacterial OTUs could be considered as indicators of different olive groves of different Tuscany olive grove soils, i.e., bacterial OTUs belonging to Thaumarchaeota Verrucomicrobia and Chloroflexi might be indicators for pyroclastic soils (Vulc, Pitigliano), since they are considered pioneers in colonizing this type of environment []; Chloroflexi could be indicators of particular arenaceous soils (row 46, 47, 48, MAC, Lamole). More specifically, Thaumarchaeota contributes significantly to nitrification rates in carbon and nitrogen rich soils influenced by perennial grasses. Oligotrophic Chloroflexi are involved in various biogeochemical processes. Some members are aerobic thermophiles, demonstrating the ability to live in soil spots with high temperatures, promoting the decomposition of organic matter []. Verrucomicrobia are mostly not culturable in vitro, but they are involved in the degradation of complex compounds that allow them to survive environmental stresses, a characteristic that makes them suitable for a possible use in sustainable agricultural systems [,]. It is also interesting that Verrucomicrobia and Chloroflexi include members involved in sulfate- and iron-reduction, surviving in groundwater, volcanic, or heavy metal contaminated soils with high REE content; thus, these bacteria may be used as indicators of risk assessment [,,]. The 3D plot of Figure 4b showed four clusters: (i) fluvio-lacustrine deposit (VILa1-2, Anghiari), (ii) arenaceous (MAC, Lamole), (iii) pyroclastic (PIT3-Vulc, Maremma, Pitigliano), and (iv) limestone soils (Chianti area). Based on these results and those obtained from CART analysis, we showed that there is a reciprocal relationship between the bacterial community and the REE content in olive orchard soils of different lithological origins.
Considering the arenaceous soils (MAC, “Tab.1”), they developed on graywacke, a sandstone with ultramafic detritus and a lower clastic carbonate content (less than 2%). These soils, rich in quartz and microcline and poor in albite, have a REE content of 21.9 mg/kg and are therefore higher than limestone and clayey soils (Tab.1). The soil texture is dominated by the sandy fraction (76.8%) with a low clay content (8.1%) that results in a low SOM content, the lowest of the soils investigated, but at the same time they are characterized by a higher bacterial alfa diversity with 255 OTUs more than clayey soils (Tab.1). Our results were in agreement with [], who found that SOC content alone was not an indicator of microbial growth rate or, in our case, bacterial diversity, and that bacteria derived from granitic rocks had higher relative growth, despite having lower SOC, compared to andesite and basalt soils. This could be caused by the higher abundance of short-range order (SRO) mineral phases in those soils [,].
The further agroecological zone highlighted by applying the CCA (Figure 4) was the fluvio-lacustrine deposits (Anghiari, Arezzo). The rounded pebble conglomerate deposits showed an intermediate value content in REE of 13.5 mg/kg, always higher than the limestone and clayey soils of the Chianti area, with the exception of the soils developed on fluvial conglomerates (VILa3, Table 1), resulting from the dismantling of the Chianti Mountains (MAC). These soils showed comparable geochemical behavior of REEs (Figure 2) and the same root of the phylogenetic tree (S10, S210, S270, S280, S70, and S80 Figure S3). This could be related to the effect of the pebble’s physical properties on bacterial niche differentiation, resulting in different soil community structures [,]. Furthermore, cluster analyses (Figure 2) showed that the bacterial taxonomic composition was related to soil texture and to the lower fractionation ratio La/Yb in the case of the pebble surface characterizing the conglomeratic-derived-soils (Anghiari, Montefiridolfi).
The differences between the two conglomerate-derived soils (Anghiari and Montefiridolfi Chianti) were probably related to the climatic characteristics of the two olive groves, highlighted by the presence of Microlunatus genus (Villa1 Figure 3b) in those of Anghiari and Bacillus and Candidatus nitrosospharea in those of Chianti (VILa3 Figure 3). Moreover, Praseodymium was the most important factor to indicate concentrations of Gammaproteobacteria Burkholderiaceae for decision tree approaches (CART analyses) for the conglomerate-derived soil in Chianti area (VILa–Figure 5b). The lower value content of the Gammaproteobacteria was found to be associated with VILa and olive groves cultivated on soil developed on carbonate flysh (Flysh Figure 5). Gammaproteobacteria include fast-growing copiotrophics abundant in soil with sufficient labile substrates. This was also supported by our findings, which highlighted that soil derived from carbonate flysh showed lower alfa diversity (135 OTUs) (Fcar, Figure S4).
This calcaric cambisol (Fcar) was characterized by a SOM value of 2.8%, the highest of the study sites, a clay fraction equal to 20%, and the phyllosilicate clay minerals kaolinite and hydroxy-interlayered vermiculite. Although numerous scientific studies agree on the fact that reducing soil tillage and acting on vegetal cover can increase soil quality [], it is the quality of soil organic compounds linked to vegetal origin that could influence bacterial activity and their temporal priming dynamics []. On the other hand, some authors reported that the addition of complex substrates to the soil can favor the production of more extracellular enzymes in response to the complex substrate accelerating SOC decomposition []. However, there still remains considerable uncertainty about how different types of substrates can influence soil microflora and, therefore, SOC mineralization. The land management of olive groves on calcaric cambisols (Fcar) in the last 20 years has been characterized by no-tillage operations with cover crops and plant residues derived from olive pruning, containing bioactive compounds with antimicrobic and antioxidant activities. This type of management, together with the mineralogical assemblage, could explain the lower alpha diversity (Figure S4) and the high presence of Rubrobacter belonging to the Actinobacteria. These filamentous bacteria are described as highly resistant to drought and poor nutrient conditions but also promoters of plant growth thanks to their good adaptability to the root surfaces [,,]. In particular, Rubrobacter participates in the dark oxidation of sulfur compounds and thus can affect sulfur availability to plants, but it is also abundant in sunlight-exposed biofilms, in desiccated areas of the Atacama desert [], and in other arid soils [,]. The relative abundance of Rubrobacter was higher in Chianti soils than in Anghiari conglomerate soils. It is of some importance to note that traditional Mediterranean olive groves are affected by low SOM content (<2%) due to the recent high temperature that favors SOM mineralization []. Considering the rising temperature in the Chianti area of the last decade, with temperatures above 35 °C [], we can hypothesize that the higher presence of Rubrobacter could be related to water stress and desiccation conditions of these soils. This hypothesis was also supported by [], who also suggested that Rubrobacter may protect other soil bacteria from heat, salinity, radiation, and desiccation stresses through trehalose synthesis.
5. Conclusions
This work, although preliminary, highlighted the mutual influence of the soil’s bacterial and lithological components. The machine learning approach is a powerful tool for corroborating results based on bivariate statistics and discovering multivariate relationships between REE and bacteria in soils of different origins whose relationships cannot be detected by bivariate statistics. Coupling metagenomics with geochemical analyses in a machine learning algorithm framework for community profiling can provide a more robust characterization of bacterial communities in relation to environmental features. Here we showed that using a complementary proxy such as REE and bacteriome, through a machine learning approach, it was possible to highlight the bacteriome assembly of a soil related to the parent rock material and land management. In fact, we were able to detect a characteristic bacteriome, which implies the existence of a different metabolic potential, in four Tuscan olive grove soils. Our opinion is that it is necessary to use this combined approach to more comprehensively and concretely characterize a territory whose importance lays on the production of a valuable extra virgin olive oil and on the good agricultural management and environmental conditions hoped for by EU directives and (Common Agricultural Policy-CAP Strategic Plan. This research represents a starting point useful for producers to define new indicators able to highlight the link between the olive groves and the territory in which they are located, but also to improve fertility, resistance to water stress, or assess the resilience of the olive trees in the Mediterranean landscape.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16070427/s1, Figure S1: 16S (341F515R) and 18S (FF390FR1) rRNA Gene Copy Number g-1 soil by qPCR in soils of the studed olive orchards of Tuscany; Figure S2: Figure S2: Comparison of representative fractionation parameters of light, medium, heavy rare earth elements and Ce: (La/Yb)n, (La/Sm)n, (La/Gd)n, (Gd/Yb)n and (Pr/Ce)n for soils developed on different lithologies of Tuscany (Tab.1). Whiskers indicate minimum and maximum values; the lines in boxes-center show median values; Figure S3: Phylogenetic tree and principals 10 groups of bacteria at Phylum level for topsoil of olive groves on different lithologies of Tuscany (Tab.1). The dominant bacterial Phyla were Proteobacteria, Actinobacteria and Acidobacteria; Figure S4: Flower diagram alpha diversity. Each petal in the flower diagram represents for a sample or group, with different colours for different samples or groups. The core number in the center is for the number of OTUs present in all samples, while number in the petalis for the unique OTUs only showing in each sample. For some sites bacterial biodiversity is particularly high, as in the case of R (Lamole) and M (Pitigliano).
Author Contributions
Conceptualization: S.P. and S.T.; Methodology: S.P., S.T., M.T.C. and A.R.; Formal analysis and investigation: A.R., S.P., M.F., M.G., R.R. and L.B.; Data analysis: A.R. and S.P.; Original draft preparation: A.R., S.P. and S.T.; Review and editing: S.P., M.T.C., L.C., G.P. and S.T.; Supervision: S.M. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support provided by Rural Development 2014-2020 for Operational Groups (in the sense of Art 56 of Reg.1305/2013) Tuscany Region, through the Project GeOEVO-App “Enhancement of extra virgin olive oil through geographical traceability and product characterization”.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data are available on request from the authors.
Acknowledgments
The authors thank Leda Acquisti, Clemente Pellegrini Strozzi, Ovidio Mugnaini, Filippo Legnaioli, Paolo Socci, Michele Cencini, Cosimo Gericke, and Gionni Pruneti for their support during the soil survey in the olive groves, and also Filippo Saiano (University of Palermo) for the collaboration on fertilizer REE analysis.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Fausto, C.; Mininni, A.N.; Sofo, A.; Crecchio, C.; Scagliola, M.; Dichio, B.; Xiloyannis, C. Olive orchard microbiome: Characterisation of bacterial communities in soil-plant compartments and their comparison between sustainable and conventional soil management systems. Plant Ecol. Divers. 2018, 11, 597–610. [Google Scholar] [CrossRef]
- Ebrahimi-Zarandi, M.; Etesami, H.; Glick, B.R. Fostering plant resilience to drought with Actinobacteria: Unveiling perennial allies in drought stress tolerance. Plant Stress 2023, 10, 100242. [Google Scholar] [CrossRef]
- Pošćić, F.; Žanetić, M.; Fiket, Ž.; Furdek Turk, M.; Mikac, N.; Bačić, N.; Lučić, M.; Romić, M.; Bakić, H.; Jukić Špika, M. Accumulation and partitioning of rare earth elements in olive trees and extra virgin olive oil from Adriatic coastal region. Plant Soil 2020, 448, 133–151. [Google Scholar] [CrossRef]
- Festa, A.; Pini, G.A.; Dilek, Y.; Codegone, G. Mélanges and mélange-forming processes: A historical overview and new concepts. Int. Geol. Rev. 2010, 52, 1040–1105. [Google Scholar] [CrossRef]
- Lotti, B. Descrizione Geologica dell’Umbria; Istituto Poligrafico e Zecca dello Stato: Roma, Italy, 1926. [Google Scholar]
- Hibi, Y.; Asai, K.; Arafuka, H.; Hamajima, M.; Iwama, T.; Kawai, K. Molecular structure of La3+-induced methanol dehydrogenase-like protein in Methylobacterium radiotolerans. J. Biosci. Bioeng. 2011, 111, 547–549. [Google Scholar] [CrossRef]
- Laveuf, C.; Cornu, S. A review on the potentiality of rare earth elements to trace pedogenetic processes. Geoderma 2009, 154, 1–12. [Google Scholar] [CrossRef]
- Pang, X.; Li, D.; Peng, A. Application of rare-earth elements in the agriculture of China and its environmental behavior in soil. Environ. Sci. Pollut. Res. 2002, 9, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Kalaimurugan, D.; Sivasankar, P.; Lavanya, K.; Shivakumar, M.S.; Venkatesan, S. Antibacterial and Larvicidal Activity of Fusarium proliferatum (YNS2) Whole Cell Biomass Mediated Copper Nanoparticles. J. Clust. Sci. 2019, 30, 1071–1080. [Google Scholar] [CrossRef]
- Li, X.; Qu, Z.; Zhang, Y.; Ge, Y.; Sun, H. Soil fungal community and potential function in different forest ecosystems. Diversity 2022, 14, 520. [Google Scholar] [CrossRef]
- Srikamwang, C.; Onsa, N.E.; Sunanta, P.; Sangta, J.; Chanway, C.P.; Thanakkasaranee, S.; Sommano, S.R. Role of microbial volatile organic compounds in promoting plant growth and disease resistance in horticultural production. Plant Signal. Behav. 2023, 18, 2227440. [Google Scholar] [CrossRef]
- Pastawan, V.; Suganuma, S.; Mizuno, K.; Wang, L.; Tani, A.; Mitsui, R.; Nakamura, K.; Shimada, M.; Hayakawa, T.; Fitriyanto, N.A. Regulation of lanthanide-dependent methanol oxidation pathway in the legume symbiotic nitrogen-fixing bacterium Bradyrhizobium sp. strain Ce-3. J. Biosci. Bioeng. 2020, 130, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Picone, N.; den Camp, H.J.M.O. Role of rare earth elements in methanol oxidation. Curr. Opin. Chem. Biol. 2019, 49, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Henderson, P. General geochemical properties and abundances of the rare earth elements. In Developments in Geochemistry; Elsevier: Amsterdam, The Netherlands, 1984; Volume 2, pp. 1–32. [Google Scholar]
- Kamenopoulos, S.N.; Agioutantis, Z.; Komnitsas, K. Framework for sustainable mining of rare earth elements. In Rare Earths Industry; Elsevier: Amsterdam, The Netherlands, 2016; pp. 111–120. [Google Scholar]
- Bohn, H.L.; Myer, R.A.; O’Connor, G.A. Soil Chemistry; John Wiley & Sons: Hoboken, NJ, USA, 2002. [Google Scholar]
- Li, X.; Chen, Z.; Chen, Z.; Zhang, Y. A human health risk assessment of rare earth elements in soil and vegetables from a mining area in Fujian Province, Southeast China. Chemosphere 2013, 93, 1240–1246. [Google Scholar] [CrossRef]
- Liang, T.; Zhang, S.; Wang, L.; Kung, H.-T.; Wang, Y.; Hu, A.; Ding, S. Environmental biogeochemical behaviors of rare earth elements in soil–plant systems. Environ. Geochem. Health 2005, 27, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, M.; Albanese, S.; Morris, G.; Ladenberger, A.; Andersson, M.; Cannatelli, C.; Lima, A.; De Vivo, B. REE concentrations in agricultural soil in Sweden and Italy: Comparison of weak MMI® extraction with near total extraction data. Appl. Geochem. 2015, 63, 22–36. [Google Scholar] [CrossRef]
- Barbera, M.; Saiano, F.; Tutone, L.; Massenti, R.; Pisciotta, A. The pattern of rare earth elements like a possible helpful tool in traceability and geographical characterization of the soil-olive system (Olea europaea L.). Plants 2022, 11, 2579. [Google Scholar] [CrossRef]
- Pelacani, S.; Maerker, M.; Tommasini, S.; Moretti, S. Combining biodiversity and geodiversity on landscape scale: A novel approach using rare earth elements and spatial distribution models in an agricultural Mediterranean landscape. Ecol. Indic. 2024, 158, 111583. [Google Scholar] [CrossRef]
- Ramos, S.J.; Dinali, G.S.; de Carvalho, T.S.; Chaves, L.C.; Siqueira, J.O.; Guilherme, L.R.G. Rare earth elements in raw materials and products of the phosphate fertilizer industry in South America: Content, signature, and crystalline phases. J. Geochem. Explor. 2016, 168, 177–186. [Google Scholar] [CrossRef]
- Bispo, F.H.A.; de Menezes, M.D.; Fontana, A.; de Souza Sarkis, J.E.; Gonçalves, C.M.; de Carvalho, T.S.; Curi, N.; Guilherme, L.R.G. Rare earth elements (REEs): Geochemical patterns and contamination aspects in Brazilian benchmark soils. Environ. Pollut. 2021, 289, 117972. [Google Scholar] [CrossRef]
- Pelacani, S.; Tommasini, S.; Ungaro, F.; Falcone, E.; Saiano, F.R.E. Of Eafevooatcs, Tuscany Olive Groves IX, Italiana CNdSC (2017) Rare Earth Elements Analysis For Extra Virgin Olive Oil Assessment: The Case Study Of Tuscany Olive Groves, Italy. In Proceedings of the XXVI Congresso Nazionale della Società Chimica Italiana, Paestum, Italy, 10–14 September 2017. [Google Scholar]
- Barrat, J.-A.; Bayon, G.; Lalonde, S. Calculation of cerium and lanthanum anomalies in geological and environmental samples. Chem. Geol. 2023, 615, 121202. [Google Scholar] [CrossRef]
- Pol, A.; Barends, T.R.M.; Dietl, A.; Khadem, A.F.; Eygensteyn, J.; Jetten, M.S.M.; Op den Camp, H.J.M. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ. Microbiol. 2014, 16, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Semrau, J.D.; DiSpirito, A.A.; Gu, W.; Yoon, S. Metals and methanotrophy. Appl. Environ. Microbiol. 2018, 84, e02289-17. [Google Scholar] [CrossRef] [PubMed]
- Anguita-Maeso, M.; Olivares-García, C.; Haro, C.; Imperial, J.; Navas-Cortés, J.A.; Landa, B.B. Culture-dependent and culture-independent characterization of the olive xylem microbiota: Effect of sap extraction methods. Front. Plant Sci. 2020, 10, 1708. [Google Scholar] [CrossRef] [PubMed]
- Zaharescu, D.G.; Burghelea, C.I.; Dontsova, K.; Presler, J.K.; Maier, R.M.; Huxman, T.; Domanik, K.J.; Hunt, E.A.; Amistadi, M.K.; Gaddis, E.E. Ecosystem composition controls the fate of rare earth elements during incipient soil genesis. Sci. Rep. 2017, 7, 43208. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, W.; Zhang, Y.; Chen, C.; Wu, W.; Zhang, T.C. Microbial communities in rare earth mining soil after in-situ leaching mining. Sci. Total Environ. 2021, 755, 142521. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Cohen, D.R.; Rutherford, N.F.; Zissimos, A.M.; Morisseau, E.G. Reflections of the geological characteristics of Cyprus in soil rare earth element patterns. Appl. Geochem. 2015, 56, 80–93. [Google Scholar] [CrossRef]
- Silva, C.M.C.A.C.; Barbosa, R.S.; Nascimento, C.W.A.D.; Silva, Y.J.A.B.D.; Silva, Y.J.A.B.D. Geochemistry and spatial variability of rare earth elements in soils under different geological and climate patterns of the Brazilian Northeast. Rev. Bras. Ciência Solo 2018, 42, e0170342. [Google Scholar] [CrossRef]
- Foley, N.; Ayuso, R. REE enrichment in granite-derived regolith deposits of the Southeastern United States: Prospective source rocks and accumulation processes. In British Columbia Geological Survey; Symposium on Strategic and Critical Materials Proceedings, 13–14 November 2015, Victoria, British Columbia, Ministry of Energy and Mines, 2015; Simandl, G.J., Neetz, M., Eds.; Volume 3, pp. 131–138.
- Johannesson, K.H.; Lyons, W.B.; Stetzenbach, K.J.; Byrne, R.H. The solubility control of rare earth elements in natural terrestrial waters and the significance of PO43− and CO32− in limiting dissolved rare earth concentrations: A review of recent information. Aquat. Geochem. 1995, 1, 157–173. [Google Scholar] [CrossRef]
- Zhou, W.; Han, G.; Liu, M.; Song, C.; Li, X. Geochemical distribution characteristics of rare earth elements in different soil profiles in Mun River Basin, Northeast Thailand. Sustainability 2020, 12, 457. [Google Scholar] [CrossRef]
- Brandl, H.; Barmettler, F.; Castelberg, C.; Fabbri, C. Microbial mobilization of rare earth elements (REE) from mineral solids: A mini review. AIMS Microbiol. 2016, 3, 190–204. [Google Scholar]
- Su, H.; Zhang, D.; Antwi, P.; Xiao, L.; Liu, Z.; Deng, X.; Asumadu-Sakyi, A.B.; Li, J. Effects of heavy rare earth element (yttrium) on partial-nitritation process, bacterial activity and structure of responsible microbial communities. Sci. Total Environ. 2020, 705, 135797. [Google Scholar] [CrossRef]
- Cantini, C.; Cimato, A.; Sani, G. Morphological evaluation of olive germplasm present in Tuscany region. Euphytica 1999, 109, 173–181. [Google Scholar] [CrossRef]
- Zorini, L.O.; Polidori, R. Economic and environmental aspects of the Tuscan olive production. Georgofili 2010, 7 (Suppl. S2), 17–47. [Google Scholar]
- Agnoletti, M.; Conti, L.; Frezza, L.; Santoro, A. Territorial analysis of the agricultural terraced landscapes of Tuscany (Italy): Preliminary results. Sustainability 2015, 7, 4564–4581. [Google Scholar] [CrossRef]
- Nanni, P. Vite e olivo nelle proprietà dei Medici nel Quattrocento. Medicea 2008, 1, 12–19. [Google Scholar]
- Rohdenburg, H. Methods for the analysis of agro-ecosystems in central Europe, with emphasis on geoecological aspects. Catena 1989, 16, 1–57. [Google Scholar] [CrossRef]
- Ruhe, R.V.; Walker, P.H. Hillslope models and soil formation. I. Open systems. Int. Soc. Soil. Sci. Trans. 1968, 4, 551–560. [Google Scholar]
- Ehlers, L.J.; Luthy, R.G. Peer reviewed: Contaminant bioavailability in soil and sediment. Environ. Sci. Technol. 2003, 37, 295A–302A. [Google Scholar] [CrossRef]
- Feng, M.H.; Shan, X.Q. A comparison of the rhizosphere-based method with DTPA, EDTA, CaCl2 and NaNO3 estraction methods for predition of bioavailability of metals in soil to barley. Environ. Pollut. 2005, 137, 231–240. [Google Scholar] [CrossRef]
- Bortolotti, V. Nota illustrativa della carta della distribuzione geografica della Formazione di Monte Morello (Alberese). Arti Graf. Pacini Mariotti 1964, 83, 155–190. [Google Scholar]
- Principi, G.; de Luca-Cardillo, M. Nuovi dati preliminari sulla coltre alloctona a Nord di Prato (Firenze). Boll. Della Soc. Geol. Ital. 1975, 94, 1199–1206. [Google Scholar]
- Merla, G. Macigno del Chianti. Studi Illustrativi della Carta Geologica d’Italia. Formazioni Geol. 1969, 2, 65–77. [Google Scholar]
- Cattuto, C.; Cencetti, C.; Fisauli, M.; Gregori, L. I bacini pleistocenici di Anghiari e Sansepolcro nell’alta valle del Tevere. Il Quat. 1995, 8, 119–128. [Google Scholar]
- Varekamp, J.C. The geology of the Vulsinian area, Lazio, Italy. Bull. Volcanol. 1980, 43, 489–503. [Google Scholar] [CrossRef]
- Ceccherini, M.T.; Ascher, J.; Pietramellara, G.; Mocali, S.; Viti, C.; Nannipieri, P. The effect of pharmaceutical waste-fungal biomass, treated to degrade DNA, on the composition of eubacterial and ammonia oxidizing populations of soil. Biol. Fertil. Soils 2007, 44, 299–306. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’hara, B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. R package, version 1.15-1. Vegan: Community Ecology Package. 2008. Available online: http://vegan.r-forge.r-project.org/ (accessed on 1 July 2024).
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Ghannam, R.B.; Techtmann, S.M. Machine learning applications in microbial ecology, human microbiome studies, and environmental monitoring. Comput. Struct. Biotechnol. J. 2021, 19, 1092–1107. [Google Scholar] [CrossRef]
- Taylor, S.R.; McLennan, S.M. The Continental Crust: Its Composition and Evolution. Blackwell Scientific Publications: Hoboken, NJ, USA, 1985; 312p. [Google Scholar]
- Hotelling, H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 1933, 24, 417. [Google Scholar] [CrossRef]
- Bacci, G.; Ceccherini, M.T.; Bani, A.; Bazzicalupo, M.; Castaldini, M.; Galardini, M.; Giovannetti, L.; Mocali, S.; Pastorelli, R.; Pantani, O.L.; et al. Exploring the dynamics of bacterial community composition in soil: The panbacteriome approach. Antonie Van Leeuwenhoek 2015, 107, 785–797. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher-Jenull, J.; Ceccherini, M.T.; Pietramellara, G.; Renella, G.; Schloter, M. Beyond microbial diversity for predicting soil functions: A mini review. Pedosphere 2020, 30, 5–17. [Google Scholar] [CrossRef]
- Sharifi, R.; Ryu, C.M. Chapter Six—Chatting With a Tiny Belowground Member of the Holobiome: Communication Between Plants and Growth-Promoting Rhizobacteria. In Advances in Botanical Research; Becard, G., Ed.; Academic Press: Cambridge, MA, USA, 2017; Volume 82, pp. 135–160. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, Z.; Pan, H.; Bai, Y.; Hu, Y.; Jin, S. Effects of rare earth elements on bacteria in rhizosphere, root, phyllosphere and leaf of soil–rice ecosystem. Sci. Rep. 2022, 12, 2089. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A. Structure and function of the global ocean microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Brioschi, L.; Steinmann, M.; Lucot, E.; Pierret, M.-C.; Stille, P.; Prunier, J.; Badot, P.-M. Transfer of rare earth elements (REE) from natural soil to plant systems: Implications for the environmental availability of anthropogenic REE. Plant Soil 2013, 366, 143–163. [Google Scholar] [CrossRef]
- Fedele, L.; Plant, J.A.; De Vivo, B.; Lima, A. The rare earth element distribution over Europe: Geogenic and anthropogenic sources. Geochemistry: Exploration, Environment. Analysis 2008, 8, 3–18. [Google Scholar]
- Loell, M.; Reiher, W.; Felix-Henningsen, P. Contents and bioavailability of rare earth elements in agricultural soils in Hesse (Germany). J. Plant Nutr. Soil Sci. 2011, 174, 644–654. [Google Scholar] [CrossRef]
- Luo, Y.; Yuan, H.; Zhao, J.; Qi, Y.; Cao, W.-W.; Liu, J.-M.; Guo, W.; Bao, Z.-H. Multiple factors influence bacterial community diversity and composition in soils with rare earth element and heavy metal co-contamination. Ecotoxicol. Environ. Saf. 2021, 225, 112749. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.; Liu, W.; Chen, Y.; Chen, W.; Zhao, L.; Ding, Q.; Wang, S.; Tang, Y.-T.; Zhang, T.; Qiu, R.-L. Structure, Variation, and Co-occurrence of Soil Microbial Communities in Abandoned Sites of a Rare Earth Elements Mine. Environ. Sci. Technol. 2016, 50, 11481–11490. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, W.; Yang, Y.; Ma, J.; Li, S.; Wen, Z. Soil characteristics and microbial community response in rare earth mining areas in southern Jiangxi Province, China. Environ. Sci. Pollut. Res. 2021, 28, 56418–56431. [Google Scholar] [CrossRef]
- Battistuzzi, F.U.; Hedges, S.B. A Major Clade of Prokaryotes with Ancient Adaptations to Life on Land. Mol. Biol. Evol. 2009, 26, 335–343. [Google Scholar] [CrossRef]
- Prathyusha, A.M.V.N.; Bramhachari, P.V. Chapter 16—Novel Perspectives of Biotic and Abiotic Stress Tolerance Mechanism in Actinobacteria. In New and Future Developments in Microbial Biotechnology and Bioengineering; Singh, B.P., Gupta, V.K., Passari, A.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 235–244. [Google Scholar] [CrossRef]
- Gupta, A.; Dutta, A.; Sarkar, J.; Panigrahi, M.K.; Sar, P. Low-abundance members of the Firmicutes facilitate bioremediation of soil impacted by highly acidic mine drainage from the Malanjkhand copper project, India. Front. Microbiol. 2018, 9, 2882. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Zheng, C.; Chen, M.; Du, W.; Xu, X. Geobiochemistry characteristics of rare earth elements in soil and ground water: A case study in Baotou, China. Sci. Rep. 2020, 10, 11740. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ullah, F.; Ahmad, R.; Shah, S.U.A.; Khan, A.; Adnan, M. Response of soil proteobacteria to biochar amendment in sustainable agriculture-a mini review. J. Soil Plant Environ. 2022, 1, 16–30. [Google Scholar] [CrossRef]
- Kalam, S.; Basu, A.; Ahmad, I.; Sayyed, R.Z.; El-Enshasy, H.A.; Dailin, D.J.; Suriani, N.L. Recent understanding of soil acidobacteria and their ecological significance: A critical review. Front. Microbiol. 2020, 11, 580024. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Bu, L.; Zhang, M.; Yuan, J.; Zhang, Y.; Wei, G.; Wang, H. Soil bacteria with distinct diversity and functions mediates the soil nutrients after introducing leguminous shrub in desert ecosystems. Glob. Ecol. Conserv. 2021, 31, e01841. [Google Scholar] [CrossRef]
- Baliyarsingh, B.; Dash, B.; Nayak, S.; Nayak, S.K. Soil verrucomicrobia and their role in sustainable agriculture. In Advances in Agricultural and Industrial Microbiology; Microbial Diversity and Application in Agroindustry; Springer: Berlin/Heidelberg, Germany, 2022; Volume 1, pp. 105–124. [Google Scholar]
- Dash, B.; Nayak, S.; Pahari, A.; Nayak, S.K. Verrucomicrobia in soil: An agricultural perspective. In Frontiers in Soil and Environmental Microbiology; CRC Press: Boca Raton, FL, USA, 2020; pp. 37–46. [Google Scholar]
- Aislabie, J.; Deslippe, J.R.; Dymond, J. Soil microbes and their contribution to soil services. In Ecosystem Services in New Zealand; Conditions and Trends Manaaki Whenua Press: Lincoln, New Zealand, 2013; Volume 1, pp. 143–161. [Google Scholar]
- Murindangabo, Y.T.; Kopecký, M.; Perná, K.; Nguyen, T.G.; Konvalina, P.; Kavková, M. Prominent use of lactic acid bacteria in soil-plant systems. Appl. Soil Ecol. 2023, 189, 104955. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Robinson, C.K.; Barnum, T.; Fricke, W.F.; Davila, A.F.; Jedynak, B.; McKay, C.P.; DiRuggiero, J. Colonization patterns of soil microbial communities in the Atacama Desert. Microbiome 2013, 1, 28. [Google Scholar] [CrossRef] [PubMed]
- Bachar, A.; Soares, M.I.M.; Gillor, O. The effect of resource islands on abundance and diversity of bacteria in arid soils. Microb. Ecol. 2012, 63, 694–700. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, Y.; Valverde, A.; Pierneef, R.E.; Cowan, D.A. Differences in Precipitation Regime Shape Microbial Community Composition and Functional Potential in Namib Desert Soils. Microb. Ecol. 2022, 83, 689–701. [Google Scholar] [CrossRef]
- Salinger, M.J.; Baldi, M.; Grifoni, D.; Jones, G.; Bartolini, G.; Cecchi, S.; Messeri, G.; Dalla Marta, A.; Orlandini, S.; Dalu, G.A.; et al. Seasonal differences in climate in the Chianti region of Tuscany and the relationship to vintage wine quality. Int. J. Biometeorol. 2015, 59, 1799–1811. [Google Scholar] [CrossRef]
- Mocali, S.; Kuramae, E.E.; Kowalchuk, G.A.; Fornasier, F.; Priori, S. Microbial functional diversity in vineyard soils: Sulfur metabolism and links with grapevine plants and wine quality. Front. Environ. Sci. 2020, 8, 75. [Google Scholar] [CrossRef]
- Noisangiam, R.; Nuntagij, A.; Pongsilp, N.; Boonkerd, N.; Denduangboripant, J.; Ronson, C.; Teaumroong, N. Heavy metal tolerant Metalliresistens boonkerdii gen. nov., sp. nov., a new genus in the family Bradyrhizobiaceae isolated from soil in Thailand. Syst. Appl. Microbiol. 2010, 33, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Gao, M.; Sun, X.; Wang, Y.; Yuan, C.; Sun, H. Nationwide distribution of polycyclic aromatic hydrocarbons in soil of China and the association with bacterial community. J. Environ. Sci. 2023, 128, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Avontuur, J.R.; Wilken, P.M.; Palmer, M.; Coetzee, M.P.A.; Stępkowski, T.; Venter, S.N.; Steenkamp, E.T. Complex evolutionary history of photosynthesis in Bradyrhizobium. Microb. Genom. 2023, 9, 001105. [Google Scholar] [CrossRef] [PubMed]
- Certini, G.; Campbell, C.D.; Edwards, A.C. Rock fragments in soil support a different microbial community from the fine earth. Soil Biology and Biochemistry 2004, 36, 1119–1128. [Google Scholar] [CrossRef]
- Huang, L.; Bao, W.; Hu, H.; Eissenstat, D.M.; Li, F. Influences of rock fragment content and vegetation on soil microbial communities. Catena 2023, 225, 107018. [Google Scholar] [CrossRef]
- Willms, I.M.; Bolz, S.H.; Yuan, J.; Krafft, L.; Schneider, D.; Schöning, I.; Schrumpf, M.; Nacke, H. The ubiquitous soil verrucomicrobial clade ‘Candidatus Udaeobacter’ shows preferences for acidic pH. Environ. Microbiol. Rep. 2021, 13, 878–883. [Google Scholar] [CrossRef] [PubMed]
- Finley, B.K.; Mau, R.L.; Hayer, M.; Stone, B.W.; Morrissey, E.M.; Koch, B.J.; Rasmussen, C.; Dijkstra, P.; Schwartz, E.; Hungate, B.A. Soil minerals affect taxon-specific bacterial growth. ISME J. 2022, 16, 1318–1326. [Google Scholar] [CrossRef]
- Mikutta, R.; Kleber, M.; Jahn, R. Poorly crystalline minerals protect organic carbon in clay subfractions from acid subsoil horizons. Geoderma 2005, 128, 106–115. [Google Scholar] [CrossRef]
- Lal, R. Digging deeper: A holistic perspective of factors affecting soil organic carbon sequestration in agroecosystems. Glob. Chang. Biol. 2018, 24, 3285–3301. [Google Scholar] [CrossRef]
- Hamer, U.; Marschner, B. Priming effects in different soil types induced by fructose, alanine, oxalic acid and catechol additions. Soil Biol. Biochem. 2005, 37, 445–454. [Google Scholar] [CrossRef]
- Fontaine, S.; Mariotti, A.; Abbadie, L. The priming effect of organic matter: A question of microbial competition? Soil Biol. Biochem. 2003, 35, 837–843. [Google Scholar] [CrossRef]
- Aguilera-Huertas, J.; Lozano-García, B.; González-Rosado, M.; Parras-Alcántara, L. Effects of management and hillside position on soil organic carbon stratification in Mediterranean centenary olive grove. Agronomy 2021, 11, 650. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).