Anatolia: A Hotspot of Avian Genetic Diversity in the Western Palaearctic
Abstract
1. Introduction
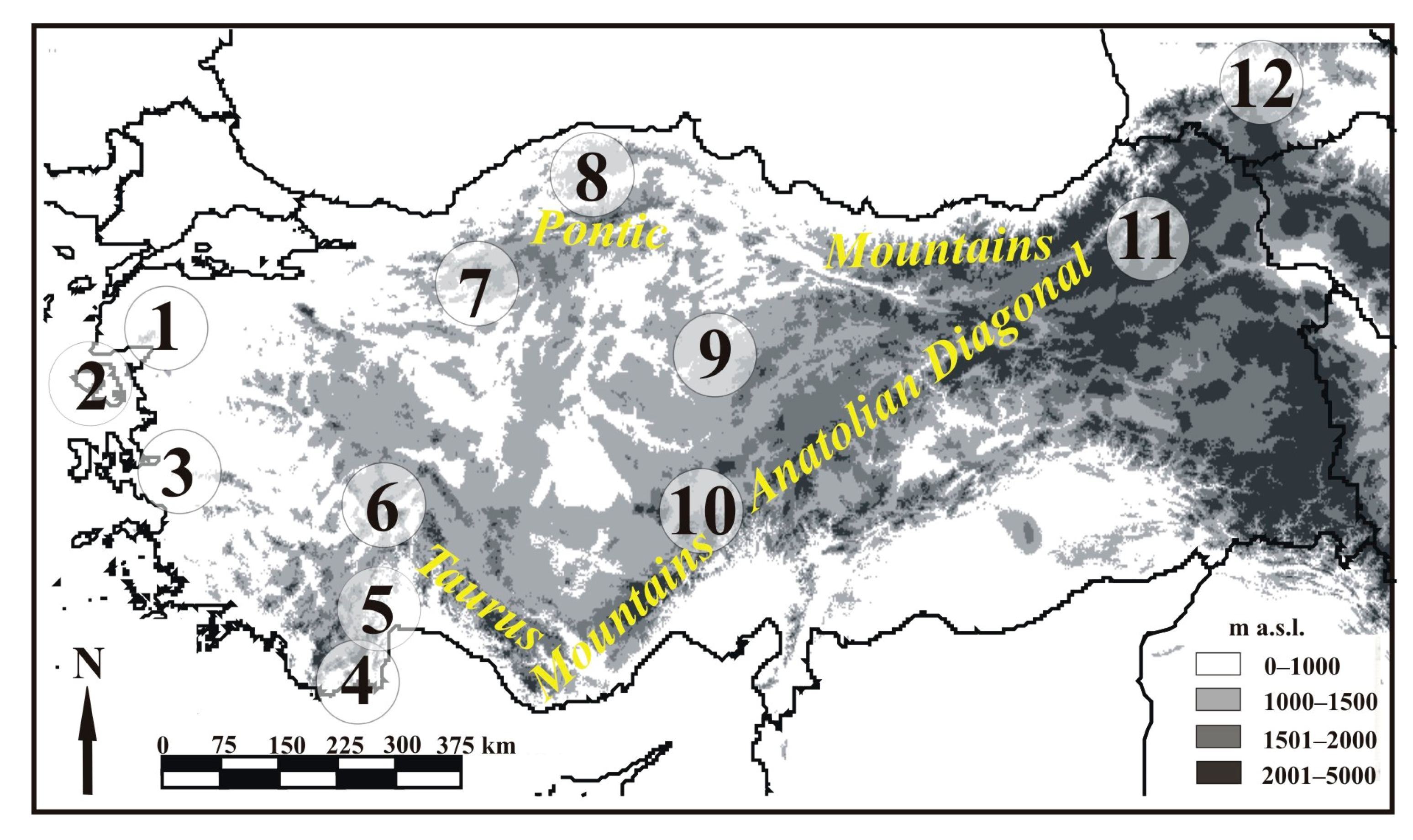
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. DNA Sequencing
2.3. Phylogeographic Analyses
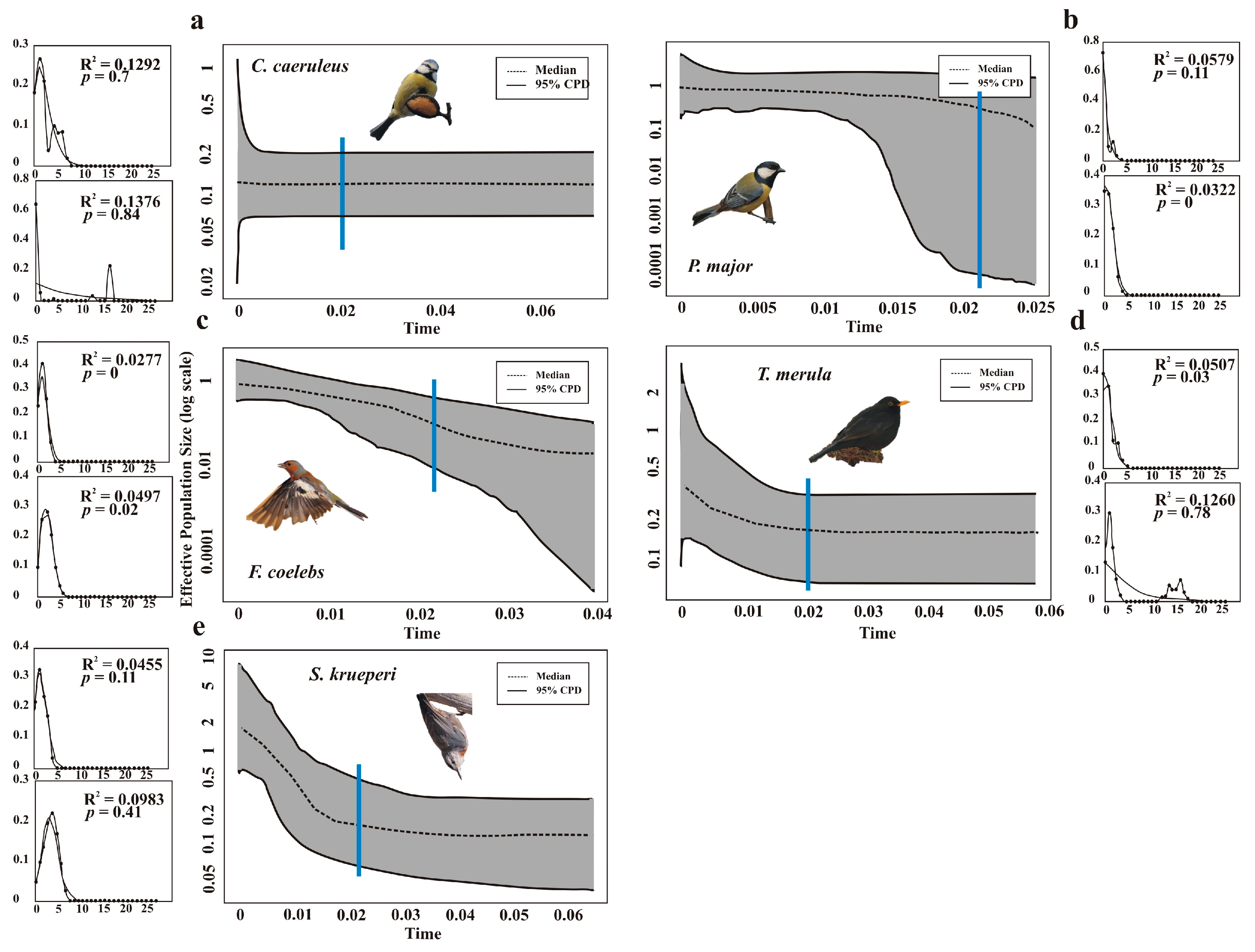
2.4. Demographic History
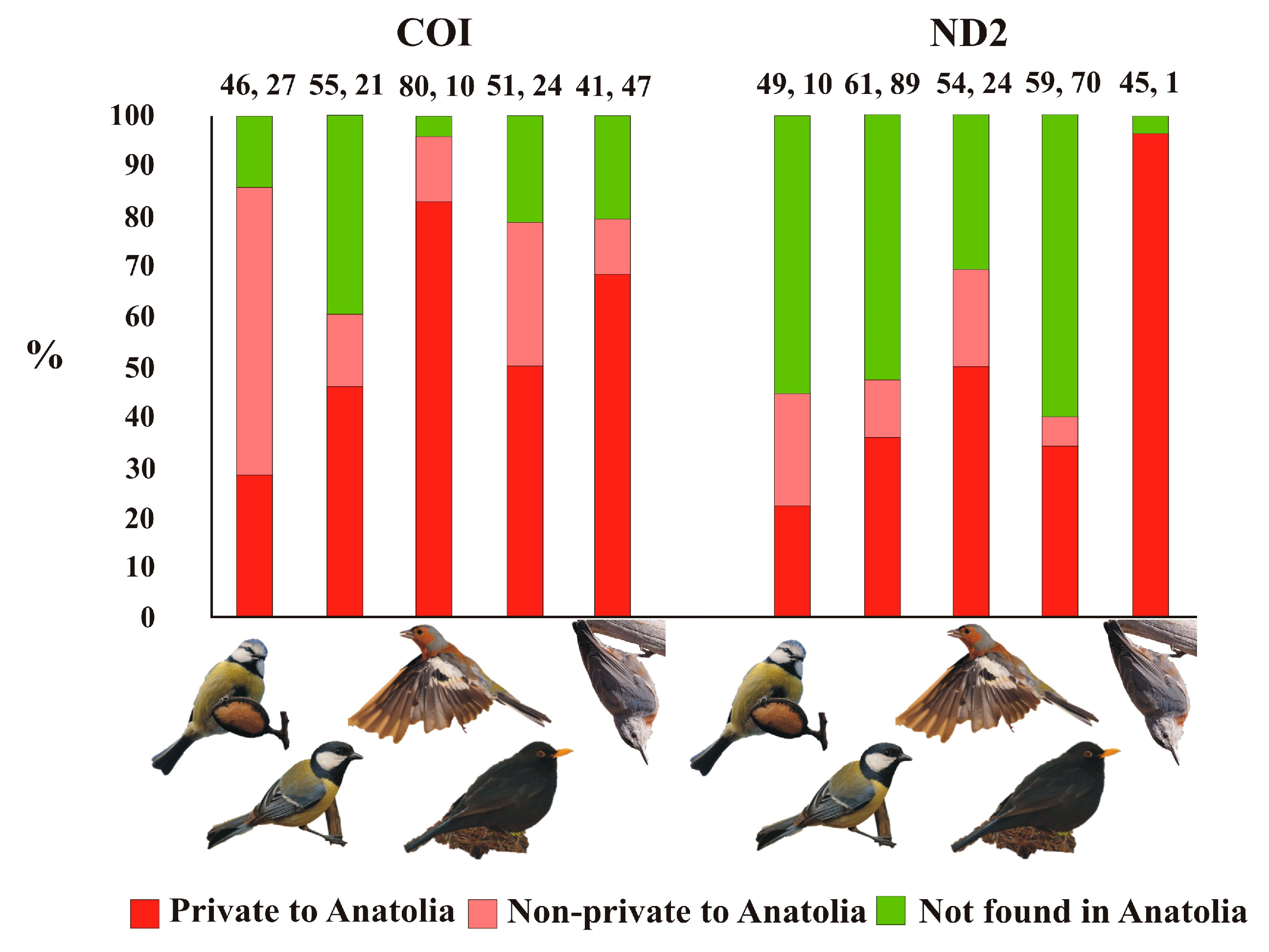
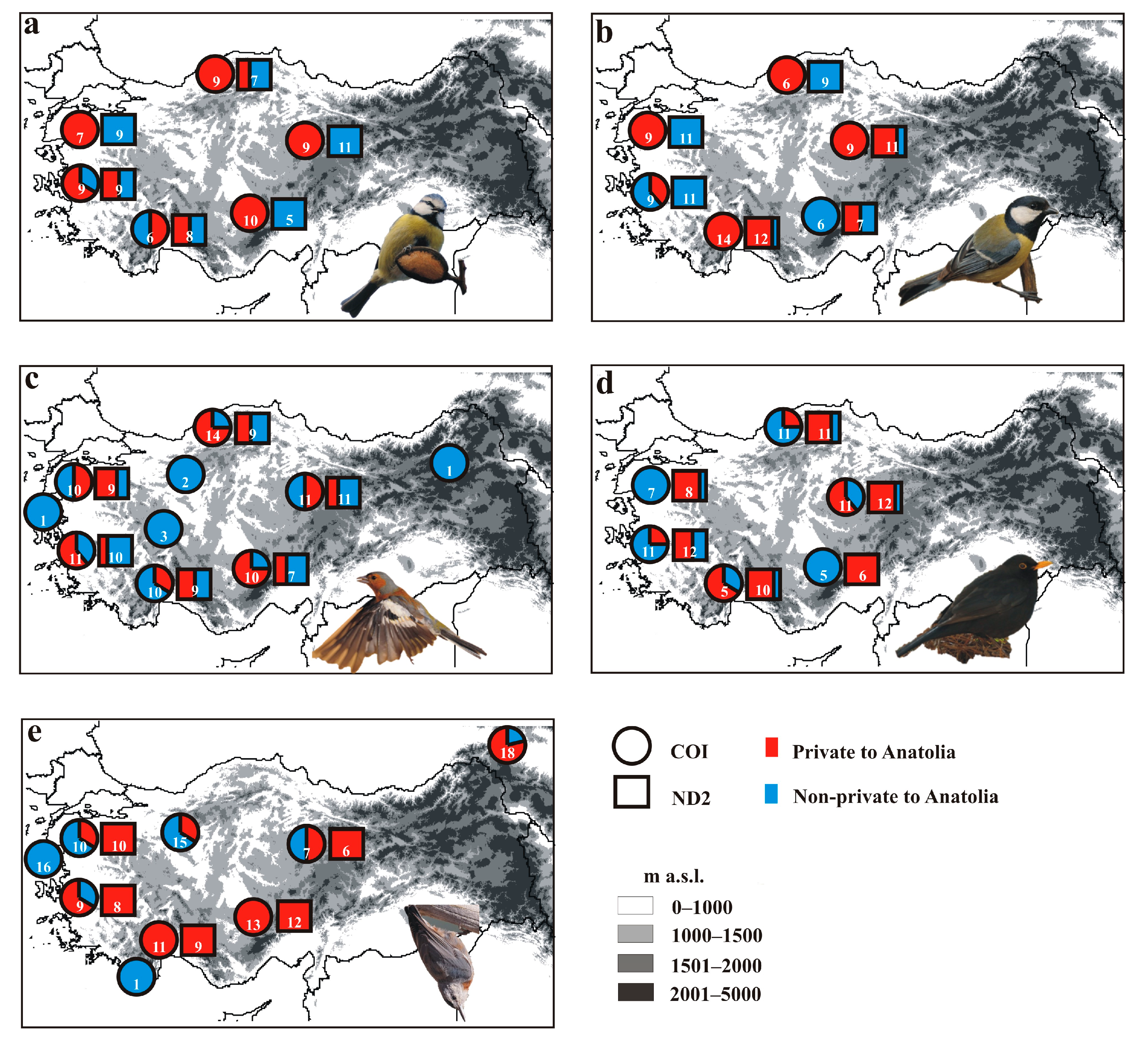
3. Results
4. Discussion
4.1. Genetic Diversity
4.2. Conclusive Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WWF. Living Planet Report 2020—Bending the Curve of Biodiversity Loss; Almond, R.E.A., Grooten, M., Petersen, T., Eds.; WWF: Gland, Switzerland, 2020; ISBN 978-2-940529-99-5. [Google Scholar]
- DeWoody, J.A.; Harder, A.M.; Mathur, S.; Willoughby, J.R. The long-standing significance of genetic diversity in conservation. Mol. Ecol. 2021, 30, 4147–4154. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.J.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Fonseca, G.A.B.d.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Sechrest, W.; Brooks, T.M.; Da Fonseca, G.A.B.; Konstant, W.R.; Mittermeier, R.A.; Purvis, A.; Rylands, A.B.; Gittleman, J.L. Hotspots and the conservation of evolutionary history. Proc. Natl. Acad. Sci. USA 2002, 99, 2067–2071. [Google Scholar] [CrossRef] [PubMed]
- Habel, J.C.; Rasche, L.; Schneider, U.A.; Engler, J.O.; Schmid, E.; Rödder, D.; Meyer, S.T.; Trapp, N.; Sos del Diego, R.; Eggermont, H.; et al. Final countdown for biodiversity hotspots. Conserv. Lett. 2019, 12, 1–9. [Google Scholar] [CrossRef]
- Vié, J.-C.; Hilton-Taylor, C.; Stuart, S.N. (Eds.) Wildlife in a Changing World—An Analysis of the 2008 IUCN Red List of Threatened Species; IUCN: Gland, Switzerland, 2009; ISBN 9782831710631. [Google Scholar]
- Roselaar, C.S.; Sluys, R.; Aliabadian, M.; Mekenkamp, P.G.M. Geographic patterns in the distribution of Palearctic songbirds. J. Ornithol. 2007, 148, 271–280. [Google Scholar] [CrossRef]
- Atalay, I. The effects of mountainous areas on biodiversity: A case study from the northern Anatolian Mountains and the Taurus Mountains. Grazer Schriften Geogr. Raumforsch. 2006, 41, 17–26. [Google Scholar]
- Elibüyük, M.; Yılmaz, E. Türkiye’nin Coğrafi Bölge ve Bölümlerine Göre Yükselti Basamakları ve Eğim Grupları. Coğrafi Bilim. Derg. 2010, 8, 27–55. [Google Scholar] [CrossRef]
- Ansell, S.W.; Stenøien, H.K.; Grundmann, M.; Russell, S.J.; Koch, M.A.; Schneider, H.; Vogel, J.C. The importance of Anatolian mountains as the cradle of global diversity in Arabis alpina, a key arctic-alpine species. Ann. Bot. 2011, 108, 241–252. [Google Scholar] [CrossRef]
- IUCN. The IUCN Red List of Threatened Species. Version 2022-2. Available online: https://www.iucnredlist.org (accessed on 27 March 2023).
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004, 359, 183–195; discussion 195. [Google Scholar] [CrossRef]
- Albayrak, T.; Gonzalez, J.; Drovetski, S.V.; Wink, M. Phylogeography and population structure of Krüper’s Nuthatch Sitta krueperi from Turkey based on microsatellites and mitochondrial DNA. J. Ornithol. 2012, 153, 405–411. [Google Scholar] [CrossRef]
- Albayrak, T.; García, J.A.D.; Özmen, Ö.; Karadas, F.; Ateş, D.; Wink, M. Evidence for Genetic Hybridization between Released and Wild Game Birds: Phylogeography and Genetic Structure of Chukar Partridge, Alectoris chukar, in Turkey. Diversity 2022, 14, 571. [Google Scholar] [CrossRef]
- Abellán, P.; Svenning, J. Refugia within refugia—Patterns in endemism and genetic divergence are linked to Late Quaternary climate stability in the Iberian Peninsula. Biol. J. Linn. Soc. 2014, 113, 13–28. [Google Scholar] [CrossRef]
- Kornilios, P.; Ilgaz, Ç.; Kumlutaş, Y.; Giokas, S.; Fraguedakis-Tsolis, S.; Chondropoulos, B. The role of Anatolian refugia in herpetofaunal diversity: An mtDNA analysis of Typhlops vermicularis Merrem, 1820 (Squamata, Typhlopidae). Amphib. Reptil. 2011, 32, 351–363. [Google Scholar] [CrossRef]
- Gür, H. The effects of the Late Quaternary glacial-interglacial cycles on Anatolian ground squirrels: Range expansion during the glacial periods? Biol. J. Linn. Soc. 2013, 109, 19–32. [Google Scholar] [CrossRef]
- Ciplak, B. The analogy between interglacial and global warming for the glacial relicts in a refugium: A biogeographic perspective for conservation of Anatolian Orthoptera. Insect. Ecol. Conserv. 2008, 661, 135–163. [Google Scholar]
- Masseti, M.; Pecchioli, E.; Vernesi, C. Phylogeography of the last surviving populations of Rhodian and Anatolian fallow deer (Dama dama dama L., 1758). Biol. J. Linn. Soc. 2008, 93, 835–844. [Google Scholar] [CrossRef]
- Baker, K.H.; Gray, H.W.I.; Ramovs, V.; Mertzanidou, D.; Akin Pekşen, C.; Bilgin, C.C.; Sykes, N.; Hoelzel, A.R. Strong population structure in a species manipulated by humans since the Neolithic: The European fallow deer (Dama dama dama). Heredity 2017, 119, 16–26. [Google Scholar] [CrossRef]
- Ibiş, O.; Aksöyek, E.; Özcan, S.; Tez, C. A preliminary phylogenetic analysis of golden jackals (Canis aureus) (Canidae: Carnivora: Mammalia) from Turkey based on mitochondrial D-loop sequences. Vertebr. Zool. 2015, 65, 391–397. [Google Scholar] [CrossRef]
- Özdemir, N.; Dursun, C.; Üzüm, N.; Kutrup, B.; Gül, S. Taxonomic assessment and distribution of common toads (Bufo bufo and B. verrucosissimus) in Turkey based on morphological and molecular data. Amphib. Reptil. 2020, 41, 399–411. [Google Scholar] [CrossRef]
- Lohman, D.J.; Prawiradilaga, D.M.; Meier, R. Improved COI barcoding primers for southeast Asian perching birds (Aves: Passeriformes). Mol. Ecol. Res. 2009, 9, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef] [PubMed]
- Drovetski, S.V.; Zink, R.M.; Rohwer, S.; Fadeev, I.V.; Nesterov, E.V.; Karagodin, I.; Koblik, E.A.; Red’kin, Y.A. Complex biogeographic history of a Holarctic passerine. Proc. Biol. Sci. 2004, 271, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Cramp, S.; Simmons, K.E.K. The Birds of Western Palearctic. Oxford University Press: Oxford, UK, 1977; Volume 1. [Google Scholar]
- Mitchell, D. Birds of Europe, North Africa and the Middle East: An Annotated Checklist; Lynx Edicions: Barcelona, Spain, 2017. [Google Scholar]
- Shirihai, H.; Svensson, L. Handbook of Western Palearctic Birds 1 & 2—Passerines; Helm: London, UK, 2018. [Google Scholar]
- Forcina, G.; Guerrini, M.; Panayides, P.; Hadjigerou, P.; Ahmed Khan, A.; Barbanera, F. Molecular taxonomy and intra-Palaearctic boundary: New insights from the biogeography of the black francolin (Francolinus francolinus) by means of microsatellite DNA. Syst. Biodivers. 2019, 17, 759–772. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.-J.J.; Forster, P.; Röhl, A.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Nabholz, B.; Glémin, S.; Galtier, N. The erratic mitochondrial clock: Variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals. BMC Evol. Biol. 2009, 9, 1–13. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: http://www.R-project.org (accessed on 1 March 2021).
- Ambarlı, D.; Zeydanlı, U.S.; Balkız, Ö.; Aslan, S.; Karaçetin, E.; Sözen, M.; Ilgaz, Ç.; Gürsoy Ergen, A.; Lise, Y.; Demirbaş Çağlayan, S.; et al. An overview of biodiversity and conservation status of steppes of the Anatolian Biogeographical Region. Biodivers. Conserv. 2016, 25, 2491–2519. [Google Scholar] [CrossRef]
- Illera, J.C.; Koivula, K.; Broggi, J.; Päckert, M.; Martens, J.; Kvist, L. A multi-gene approach reveals a complex evolutionary history in the Cyanistes species group. Mol. Ecol. 2011, 20, 4123–4139. [Google Scholar] [CrossRef] [PubMed]
- Porlier, M.; Garant, D.; Perret, P.; Charmantier, A. Habitat-linked population genetic differentiation in the blue tit Cyanistes caeruleus. J. Hered. 2012, 103, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Blank, J.; Stauss, M.J.; Tomiuk, J.; Fietz, J.; Segelbacher, G. Habitat type does not affect population genetic structure in sympatric great tits (Parus major) and blue tits (P. caeruleus). J. Negat. Results-Ecol. Evol. Biol. 2007, 4, 1–14. [Google Scholar]
- Suárez, N.M.; Betancor, E.; Klassert, T.E.; Almeida, T.; Hernández, M.; Pestano, J.J. Phylogeography and genetic structure of the Canarian common chaffinch (Fringilla coelebs) inferred with mtDNA and microsatellite loci. Mol. Phylogenet. Evol. 2009, 53, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Recuerda, M.; Illera, J.C.; Blanco, G.; Zardoya, R.; Milá, B. Sequential colonization of oceanic archipelagos led to a species-level radiation in the common chaffinch complex (Aves: Fringilla coelebs). Mol. Phylogenet. Evol. 2021, 164, 107291. [Google Scholar] [CrossRef]
- Voelker, G.; Rohwer, S.; Bowie, R.C.K.; Outlaw, D.C. Molecular systematics of a speciose, cosmopolitan songbird genus: Defi ning the limits of, and relationships among, the Turdus thrushes. Mol. Phylogenet. Evol. 2007, 42, 422–434. [Google Scholar] [CrossRef]
- Zheng, G.M. A Checklist on the Classification and Distribution of the Birds of China, 3rd ed.; Science Press: Beijing, China, 2018. [Google Scholar]
- Stamatis, C.; Suchentrunk, F.; Moutou, K.A.; Giacometti, M.; Haerer, G.; Djan, M.; Vapa, L.; Vukovic, M.; Tvrtković, N.; Sert, H.; et al. Phylogeography of the brown hare (Lepus europaeus) in Europe: A legacy of south-eastern Mediterranean refugia? J. Biogeogr. 2009, 36, 515–528. [Google Scholar] [CrossRef]
- Perktaş, U.; De Silva, T.N.; Quintero, E.; Tavşanoğlu, Ç. Adding ecology into phylogeography: Ecological niche models and phylogeography in tandem reveals the demographic history of the subalpine warbler complex. Bird Study 2019, 66, 234–242. [Google Scholar] [CrossRef]
- Arslan, Y.; Demirtaş, S.; Herman, J.S.; Pustilnik, J.D.; Searle, J.B.; Gündüz, I. The Anatolian glacial refugium and human-mediated colonization: A phylogeographical study of the stone marten (Martes foina) in Turkey. Biol. J. Linn. Soc. 2020, 129, 470–491. [Google Scholar] [CrossRef]
- Ahmadzadeh, F.; Flecks, M.; Carretero, M.A.; Böhme, W.; Ilgaz, C.; Engler, J.O.; James Harris, D.; Üzüm, N.; Rödder, D. Rapid lizard radiation lacking niche conservatism: Ecological diversification within a complex landscape. J. Biogeogr. 2013, 40, 1807–1818. [Google Scholar] [CrossRef]
- Korkmaz, E.M.; Lunt, D.H.; Çiplak, B.; Deǧerli, N.; Başibüyük, H.H. The contribution of Anatolia to European phylogeography: The centre of origin of the meadow grasshopper, Chorthippus parallelus. J. Biogeogr. 2014, 41, 1793–1805. [Google Scholar] [CrossRef]
- Rokas, A.; Atkinson, R.J.; Webster, L.M.I.; Csóka, G.; Stone, G.N. Out of Anatolia: Longitudinal gradients in genetic diversity support an eastern origin for a circum-Mediterranean oak gallwasp Andricus quercustozae. Mol. Ecol. 2003, 12, 2153–2174. [Google Scholar] [CrossRef] [PubMed]
- Özüdoğru, B.; Özgişi, K.; Perktaş, U.; Gür, H. The Quaternary range dynamics of Noccaea iberidea (Brassicaceae), a typical representative of subalpine/alpine steppe communities of Anatolian mountains. Biol. J. Linn. Soc. 2020, 131, 986–1001. [Google Scholar] [CrossRef]
- Hogner, S.; Laskemoen, T.; Lifjeld, J.T.; Porkert, J.; Kleven, O.; Albayrak, T.; Kabasakal, B.; Johnsen, A. Deep sympatric mitochondrial divergence without reproductive isolation in the common redstart Phoenicurus phoenicurus. Ecol. Evol. 2012, 2, 2974–2988. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, L.; Cheng, Y.; Hao, Y.; Xiong, Y.; Song, G.; Qu, Y.; Rheindt, F.E.; Alström, P.; Jia, C.; et al. “Ghost Introgression” as a cause of deep mitochondrial divergence in a bird species complex. Mol. Biol. Evol. 2019, 36, 2375–2386. [Google Scholar] [CrossRef]
- Schrey, A.W.; Grispo, M.; Awad, M.; Cook, M.B.; McCoy, E.D.; Mushinsky, H.R.; Albayrak, T.; Bensch, S.; Burke, T.; Butler, L.K.; et al. Broad-scale latitudinal patterns of genetic diversity among native European and introduced house sparrow (Passer domesticus) populations. Mol. Ecol. 2011, 20, 1133–1143. [Google Scholar] [CrossRef]
- Dubey, S.; Diker, E.; Kurtonur, C.; Vogel, P. Secondary contact zones and hybridizations: The case of the lesser white-toothed shrew (Crocidura suaveolens group, Soricidae). Biol. J. Linn. Soc. 2008, 95, 557–565. [Google Scholar] [CrossRef]
- Fritz, U.; Hundsd, A.K.; Guicking, D.; Wink, M.; Tok, C.V.; Kerim, C.; Buschbom, J. Mitochondrial diversity of European pond turtles (Emys orbicularis) in Anatolia and the Ponto-Caspian Region: Multiple old refuges, hotspot of extant diversification and critically endangered endemics. Org. Divers. Evol. 2009, 9, 100–114. [Google Scholar] [CrossRef]
- Akın, Ç.; Bilgin, C.C.; Beerli, P.; Westaway, R.; Ohst, T.; Litvinchuk, S.N.; Uzzell, T.; Bilgin, M.; Hotz, H.; Guex, G.-D.; et al. Phylogeographic patterns of genetic diversity in eastern Mediterranean water frogs were determined by geological processes and climate change in the Late Cenozoic. J. Biogeogr. 2010, 37, 2111–2124. [Google Scholar] [CrossRef]
- İpekdal, K.; Burban, C.; Sauné, L.; Battisti, A.; Kerdelhué, C. From refugia to contact: Pine processionary moth hybrid zone in a complex biogeographic setting. Ecol. Evol. 2020, 10, 1623–1638. [Google Scholar] [CrossRef]
- Dubey, S.; Cosson, J.-F.; Vohralík, V.; Krystufek, B.; Diker, E.; Vogel, P.; Krys, B.; Dubey, S.; Cosson, J.-F.; Vohrali, V.; et al. Molecular evidence of Pleistocene bidirectional faunal exchange between Europe and the Near East: The case of the bicoloured shrew (Crocidura leucodon, Soricidae). J. Evol. Biol. 2007, 20, 1799–1808. [Google Scholar] [CrossRef]
- Stratakis, M.; Koutmanis, I.; Ilgaz, Ç.; Jablonski, D.; Kukushkin, O.V.; Crnobrnja-Isialovic, J.; Carretero, M.A.; Liuzzi, C.; Kumlutaş, Y.; Lymberakis, P.; et al. Evolutionary divergence of the smooth snake (Serpentes, Colubridae): The role of the Balkans and Anatolia. Zool. Scr. 2022, 51, 310–329. [Google Scholar] [CrossRef]
- Şekercioĝlu, Ç.H.; Anderson, S.; Akçay, E.; Bilgin, R.; Can, Ö.E.; Semiz, G.; Tavşanoĝlu, Ç.; Yokeş, M.B.; Soyumert, A.; Ipekdal, K.; et al. Turkey’s globally important biodiversity in crisis. Biol. Conserv. 2011, 144, 2752–2769. [Google Scholar] [CrossRef]
- World Bank. The World Bank Annual Report 2022: Helping Countries Adapt to a Changing World; World Bank: Washington, DC, USA, 2022. [Google Scholar]
- Aliabadian, M.; Beentjes, K.K.; Roselaar, C.S.; van Brandwijk, H.; Beentjes, K. DNA barcoding of Dutch birds. Zookeys 2013, 365, 25–48. [Google Scholar] [CrossRef]
- Bilgin, R.; Ebeoǧlu, N.; Inak, S.; Kırpık, M.A.; Horns, J.J.; Şekercioğlu, Ç.H. DNA barcoding of birds at a migratory hotspot in eastern Turkey highlights continental phylogeographic relationships. PLoS ONE 2016, 11, 1–17. [Google Scholar] [CrossRef]
- Dimitriou, A.C.; Forcina, G.; Papazoglou, C.; Panayides, P.; Guerrini, M.; Crabtree, A.; Barbanera, F.; Sfenthourakis, S. DNA barcoding of Bird species in Cyprus: A tool for conservation purposes. Bird Conserv. Int. 2017, 27, 483–494. [Google Scholar] [CrossRef]
- Gohli, J.; Leder, E.H.; Garcia-Del-Rey, E.; Johannessen, L.E.; Johnsen, A.; Laskemoen, T.; Popp, M.; Lifjeld, J.T. The evolutionary history of Afrocanarian blue tits inferred from genomewide SNPs. Mol. Ecol. 2015, 24, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.M.; Drovetski, S.V.; Zink, R.M. Matching loci surveyed to questions asked in phylogeography. Proc. R. Soc. B Biol. Sci. 2016, 283, 12–14. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, A.; Rindal, E.; Ericson, P.G.P.; Zuccon, D.; Kerr, K.C.R.; Stoeckle, M.Y.; Lifjeld, J.T. DNA barcoding of Scandinavian birds reveals divergent lineages in trans-Atlantic species. J. Ornithol. 2010, 151, 565–578. [Google Scholar] [CrossRef]
- Kerr, K.C.R.; Birks, S.M.; Kalyakin, M.V.; Red’kin, Y.A.; Koblik, E.A.; Hebert, P.D. Filling the gap-COI barcode resolution in eastern Palearctic birds. Front. Zool. 2009, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mondino, A.; Crovadore, J.; Ursenbacher, S.; Lefort, F. COI sequences of a mock community for study of the diet of the green whip snake. unpublished. Available online: https://www.uniprot.org/uniprotkb/A0A899J8S3/publications (accessed on 5 May 2024).
- Päckert, M.; Bader-Blukott, M.; Künzelmann, B.; Sun, Y.; Hsu, Y.; Kehlmaier, C.; Albrecht, F.; Cobo, J.C.; Martens, J. A revised phylogeny of nuthatches (Aves, Passeriformes, Sitta) reveals insight in intra- and interspecific diversification patterns in the Palearctic. Vertebr. Zool. 2020, 70, 241–262. [Google Scholar] [CrossRef]
- Päckert, M.; Martens, J.; Sun, Y.H.; Strutzenberger, P. The phylogenetic relationships of Przevalski’s Finch Urocynchramus pylzowi, the most ancient Tibetan endemic passerine known to date. Ibis 2016, 158, 530–540. [Google Scholar] [CrossRef]
- Perktaş, U.; Gür, H.; Sağlam, I.K.; Quintero, E. Climate-driven range shifts and demographic events over the history of Krüper’s Nuthatch Sitta krueperi. Bird Study 2015, 62, 14–28. [Google Scholar] [CrossRef]
- Rodrigues, P.; Lopes, R.J.; Reis, S.; Resendes, R.; Ramos, J.A.; Tristão da Cunha, R. Genetic diversity and morphological variation of the common chaffinch Fringilla coelebs in the Azores. J. Avian Biol. 2014, 45, 167–178. [Google Scholar] [CrossRef]
- Rodrigues, P.; Lopes, R.J.; Resendes, R.; Ramos, J.A.; Tristão da Cunha, R. Genetic Diversity of the Azores Blackbirds Turdus merula Reveals Multiple Founder Events. Acta Ornithol. 2016, 51, 221–234. [Google Scholar] [CrossRef]
- Schäffer, S.; Zachos, F.E.; Koblmüller, S. Opening the treasure chest: A DNA-barcoding primer set for most higher taxa of Central European birds and mammals from museum collections. PLoS ONE 2017, 12, 1–14. [Google Scholar] [CrossRef]
- Schindel, D.E.; Stoeckle, M.Y.; Milensky, C.M.; Trizna, M.; Schmidt, B.K.; Gebhard, C.A.; Graves, G.R. Project description: DNA Barcodes of Bird Species in the National Museum of Natural History, Smithsonian Institution, USA. Zookeys 2011, 152, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Stoeckle, M.Y. All Birds Barcoding Initiative. unpublished. Available online: https://www.ncbi.nlm.nih.gov/nuccore/JN801388 (accessed on 5 May 2024).
- Tietze, D.T.; Päckert, M.; Martens, J.; Lehmann, H.; Sun, Y.-H. Complete phylogeny and historical biogeography of true rosefinches (Aves: Carpodacus). Zool. J. Linn. Soc. 2013, 169, 215–234. [Google Scholar] [CrossRef]
- Zink, R.M. Natural selection on mitochondrial DNA in Parusand its relevance for phylogeographic studies. Proc. R. Soc. B Biol. Sci. 2005, 272, 71–78. [Google Scholar] [CrossRef]

| COI | S | Haplotypes | Hd | π (× 10−3) | ND2 | S | Haplotypes | Hd | π (× 10−3) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | h (%) | ph | N | h (%) | ph | |||||||
| Cyanistes caeruleus | ||||||||||||
| Anatolia | 46 | 9 | 12 (86) | 4 | 0.82 ± 0.04 | 3.35 ± 0.47 | 49 | 17 | 4 (44) | 2 | 0.35 ± 0.08 | 4.57 ± 1.16 |
| WP* | 27 | 12 | 10 (71) | 9 | 0.86 ± 0.04 | 5.46 ± 0.4 | 10 | 23 | 7 (78) | 5 | 0.91 ± 0.08 | 10.27 ± 1.64 |
| Total WP | 73 | 14 | 14 | 0.85 ± 0.02 | 5.90 ± 0.40 | 59 | 23 | 9 | 0.48 ± 0.08 | 5.72 ± 1.06 | ||
| Parus major | ||||||||||||
| Anatolia | 55 | 9 | 7 (54) | 6 | 0.27 ± 0.08 | 1.00 ± 0.33 | 61 | 16 | 16 (44) | 13 | 0.65 ± 0.07 | 1.00 ± 1.00 |
| WP* | 21 | 24 | 9 (69) | 6 | 0.76 ± 0.08 | 1.27 ± 2.37 | 89 | 31 | 23 (64) | 19 | 0.61 ± 0.06 | 1.11 ± 0.18 |
| Total WP | 76 | 26 | 13 | 0.42 ± 0.07 | 5.34 ± 1.52 | 150 | 39 | 36 | 0.63 ± 0.01 | 1.08 ± 0.13 | ||
| Fringilla coelebs | ||||||||||||
| Anatolia | 80 | 21 | 22 (95) | 19 | 0.77 ± 0.05 | 1.85 ± 0.20 | 54 | 20 | 24 (67) | 18 | 0.91 ± 0.03 | 2.03 ± 0.21 |
| WP* | 10 | 3 | 4 (17) | 1 | 0.64 ± 0.15 | 1.46 ± 0.47 | 22 | 28 | 18 (50) | 12 | 0.97 ± 0.03 | 3.87 ± 0.98 |
| Total WP | 90 | 21 | 23 | 0.75 ± 0.05 | 1.90 ± 0.20 | 76 | 41 | 36 | 0.92 ± 0.02 | 2.62 ± 0.37 | ||
| Turdus merula | ||||||||||||
| Anatolia | 51 | 11 | 11 (79) | 7 | 0.61 ± 0.07 | 1.69 ± 0.33 | 59 | 25 | 14 (40) | 12 | 0.87 ± 0.03 | 6.05 ± 1.10 |
| WP* | 24 | 11 | 7 (50) | 3 | 0.76 ± 0.06 | 2.28 ± 0.48 | 69 | 39 | 23 (66) | 21 | 0.86 ± 0.03 | 4.72 ± 0.67 |
| Total | 75 | 17 | 14 | 0.67 ± 0.05 | 1.91 ± 0.28 | 128 | 46 | 35 | 0.92 ± 0.01 | 5.90 ± 0.68 | ||
| Sitta krueperi | ||||||||||||
| Anatolia | 115 | 16 | 15 (79) | 13 | 0.78 ± 0.02 | 2.25 ± 0.17 | 48 | 17 | 27 (96) | 27 | 0.96 ± 0.02 | 3.37 ± 0.20 |
| Lesvos Island | 20 | 0 | 5 | 0 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0 | |||||
| Caucasus | 28 | 4 | 5 (26) | 4 | 0.38 ± 0.11 | 0.74 ± 0.24 | 1 | 1 | 1 (4) | 1 | ||
| Total | 163 | 18 | 19 | 0.77 ± 0.01 | 2.94 ± 0.13 | 49 | 20 | 28 | 0.96 ± 0.02 | 3.49 ± 0.22 | ||
| COI | ND2 | |||
|---|---|---|---|---|
| Cyanistes caeruleus | 0.57 * | 0.49 * | ||
| Parus major | 0.27 * | 0.01 | ||
| Fringilla coelebs | 0.04 | 0.57 * | ||
| Turdus merula | 0.31 * | 0.34 * | ||
| Lesvos | Caucasus | Lesvos | Caucasus | |
| Sitta krueperi | 0.29 * | 0.53 * | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albayrak, T.; Tunçel, T.; Öğe, P.; Tietze, D.T.; Forcina, G. Anatolia: A Hotspot of Avian Genetic Diversity in the Western Palaearctic. Diversity 2024, 16, 339. https://doi.org/10.3390/d16060339
Albayrak T, Tunçel T, Öğe P, Tietze DT, Forcina G. Anatolia: A Hotspot of Avian Genetic Diversity in the Western Palaearctic. Diversity. 2024; 16(6):339. https://doi.org/10.3390/d16060339
Chicago/Turabian StyleAlbayrak, Tamer, Tuğba Tunçel, Pınar Öğe, Dieter Thomas Tietze, and Giovanni Forcina. 2024. "Anatolia: A Hotspot of Avian Genetic Diversity in the Western Palaearctic" Diversity 16, no. 6: 339. https://doi.org/10.3390/d16060339
APA StyleAlbayrak, T., Tunçel, T., Öğe, P., Tietze, D. T., & Forcina, G. (2024). Anatolia: A Hotspot of Avian Genetic Diversity in the Western Palaearctic. Diversity, 16(6), 339. https://doi.org/10.3390/d16060339








