Mycorrhizal Fungi of Phalaenopsis japonica (Orchidaceae) and Their Role in Seed Germination and Seedling Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Species and Sampling
2.2. Isolation of Mycorrhizal Fungi
2.3. Molecular Identification of Mycorrhizal Fungi through Sanger Sequencing
2.4. Phylogenetic Analysis
2.5. High-Throughput Sequencing
2.6. Symbiotic Culture
2.6.1. In Vitro Symbiotic Germination
2.6.2. In Vitro Symbiotic Culture of Seedlings
3. Results
3.1. Molecular Identification of Mycorrhizal Fungi through Sanger Sequencing
3.2. Phylogenetic Analysis
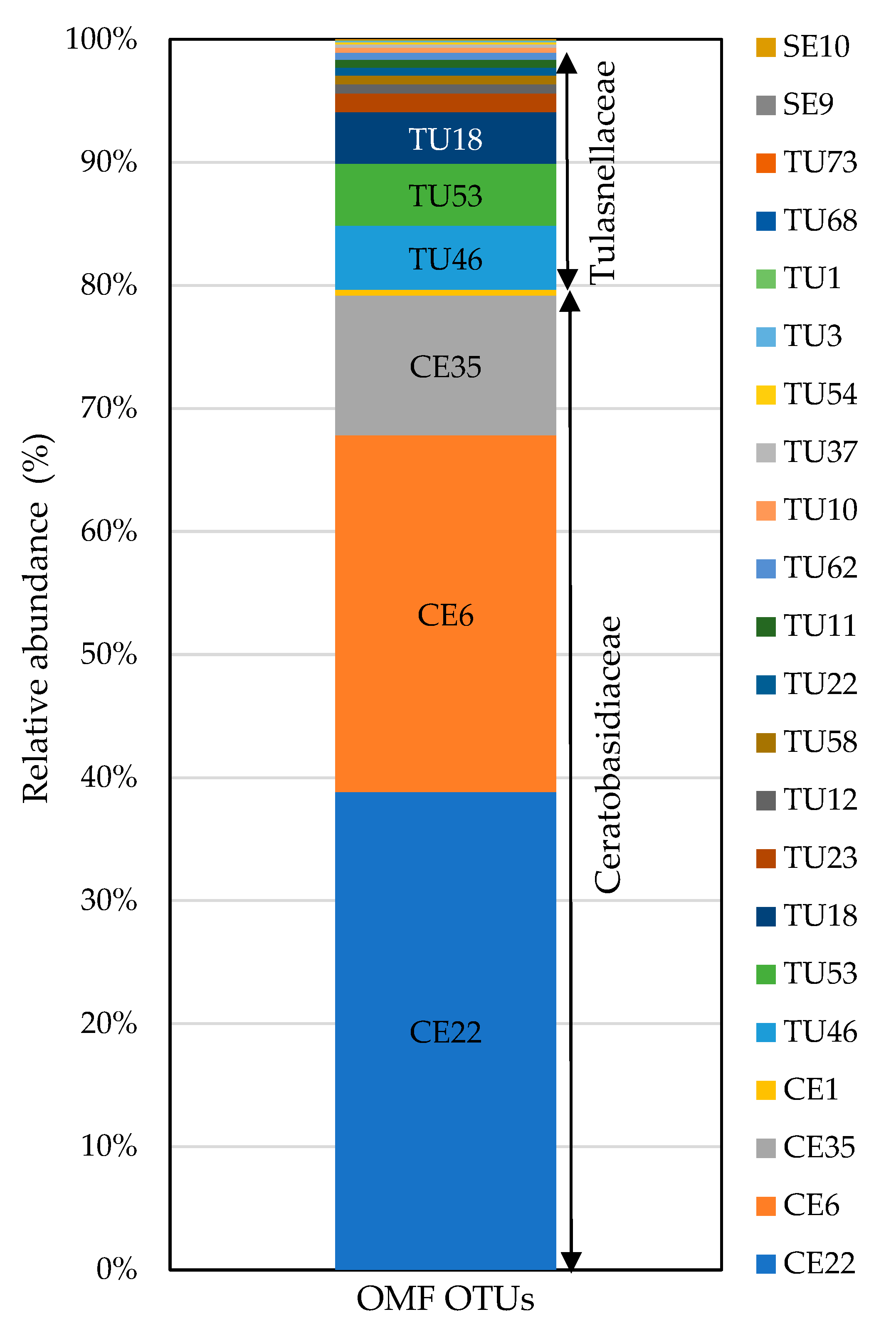
3.3. High-Throughput Sequencing
3.4. Symbiotic Culture
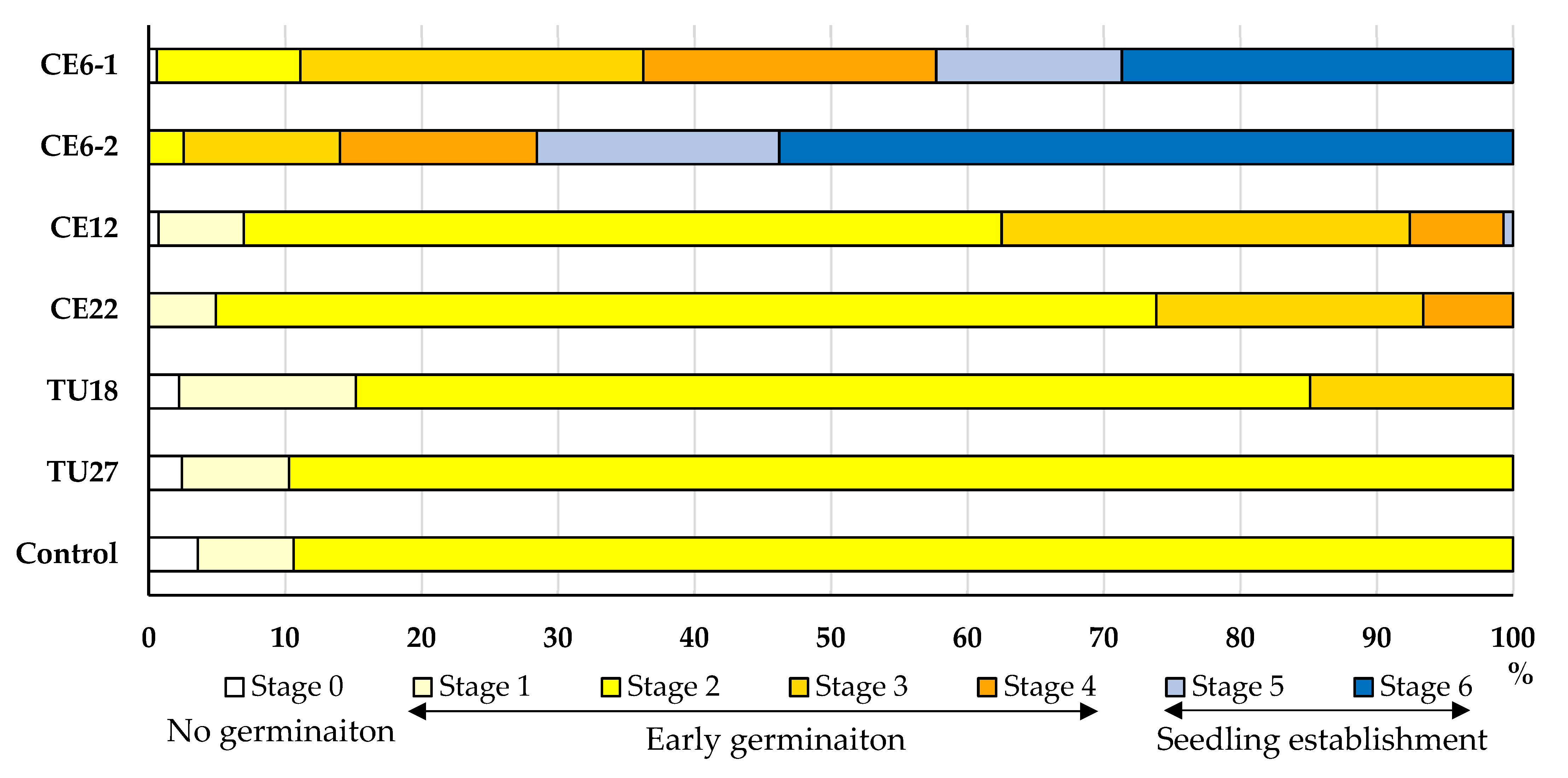
3.4.1. Effects of Fungal Strains on Symbiotic Seed Germination
3.4.2. Effects of Fungal Strains on Seedling Growth
4. Discussion
4.1. Mycorrhizal Fungal Composition of Phalaenopsis japonica
4.2. Fungal Effects on Seed Germination and Seedling Growth
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christenhusz, M.J.M.; Byng, J.W. The number of known plant species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef]
- Zotz, G. The systematic distribution of vascular epiphytes—A critical update. Bot. J. Linn. Soc. 2013, 171, 453–481. [Google Scholar] [CrossRef]
- Gale, S.W.; Fischer, G.A.; Cribb, P.J.; Fay, M.F. Orchid conservation: Bridging the gap between science and practice. Bot. J. Linn. Soc. 2018, 186, 425–434. [Google Scholar] [CrossRef]
- Hinsley, A.; De Boer, H.J.; Fay, M.F.; Gale, S.W.; Gardiner, L.M.; Gunasekara, R.S.; Kumar, P.; Masters, S.; Metusala, D.; Roberts, D.L.; et al. A review of the trade in orchids and its implications for conservation. Bot. J. Linn. Soc. 2018, 186, 435–455. [Google Scholar] [CrossRef]
- Li, T.; Wu, S.; Yang, W.; Selosse, M.A.; Gao, J. How mycorrhizal associations influence the orchid distribution and population dynamics. Front. Plant Sci. 2021, 12, 647114. [Google Scholar] [CrossRef] [PubMed]
- Dearnaley, J.D.; Martos, F.; Selosse, M.A. Orchid mycorrhizas: Molecular ecology, physiology, evolution, and conservation aspects. In Fungal Associations, 2nd ed.; Hock, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 207–230. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: San Diego, CA, USA, 2008. [Google Scholar]
- Yeh, C.M.; Chung, K.; Liang, C.K.; Tsai, W.C. New insights into the symbiotic relationship between orchids and fungi. Appl. Sci. 2019, 9, 585. [Google Scholar] [CrossRef]
- Rasmussen, H.N.; Rasmussen, F.N. Orchid mycorrhiza: Implications of a mycophagous lifestyle. Oikos 2009, 118, 334–345. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.Y.; Chen, X.M.; Guo, S.X.; Lee, Y.I. Effect of different mycobionts on symbiotic germination and seedling growth of Dendrobium officinale, an important medicinal orchid. Bot. Stud. 2020, 61, 2. [Google Scholar] [CrossRef]
- Zhang, L.; Rammitsu, K.; Kinoshita, A.; Tokuhara, K.; Yukawa, T.; Ogura-Tsujita, Y. Symbiotic culture of three closely related Dendrobium species reveals a growth bottleneck and differences in mycorrhizal specificity at early developmental stages. Diversity 2022, 14, 1119. [Google Scholar] [CrossRef]
- Jacquemyn, H.; Brys, R.; Merckx, V.S.; Waud, M.; Lievens, B.; Wiegand, T. Coexisting orchid species have distinct mycorrhizal communities and display strong spatial segregation. New Phytol. 2014, 202, 616–627. [Google Scholar] [CrossRef]
- Waud, M.; Busschaert, P.; Lievens, B.; Jacquemyn, H. Specificity and localized distribution of mycorrhizal fungi in the soil may contribute to co-existence of orchid species. Fungal Ecol. 2016, 20, 155–165. [Google Scholar] [CrossRef]
- Govaerts, R.; Bernet, P.; Kratochvil, K.; Gerlach, G.; Carr, G.; Alrich, P.; Pridgeon, A.M.; Pfahl, J.; Campacci, M.A.; Holland, B.D.; et al. World checklist of Orchidaceae. Facilitated by the Royal Botanic Gardens, Kew. 2021. Available online: http://wcsp.science.kew.org/ (accessed on 4 July 2023).
- Christenson, E.A. Phalaenopsis—A Monograph; Timber Press: Portland, OR, USA, 2001; pp. 458–465. [Google Scholar]
- Yuan, S.C.; Bolaños-Villegas, P.; Tsao, C.Y.; Chen, F.C. The breeding of Phalaenopsis hybrids. In The Orchid Genome. Compendium of Plant Genomes; Chen, F.C., Chin, S.W., Eds.; Springer: Cham, Switzerland, 2021; pp. 29–40. [Google Scholar] [CrossRef]
- IUCN. IUCN Red List of Threatened Species. Version 2022-2. 2022. Available online: https://www.iucnredlist.org (accessed on 15 July 2023).
- Chang, D.C. The screening of orchid mycorrhizal fungi (OMF) and their applications. In Orchid Biotechnology; Chen, W.H., Chen, H.H., Eds.; World Scientific: Singapore, 2007; pp. 77–98. [Google Scholar]
- Wu, P.H.; Huang, D.D.; Chang, D.C. Mycorrhizal symbiosis enhances Phalaenopsis orchid’s growth and resistance to Erwinia chrysanthemi. Afr. J. Biotechnol. 2011, 10, 10095–10100. [Google Scholar] [CrossRef]
- Izuddin, M.; Srivathsan, A.; Lee, A.L.; Yam, T.W.; Webb, E.L. Availability of orchid mycorrhizal fungi on roadside trees in a tropical urban landscape. Sci. Rep. 2019, 9, 19528. [Google Scholar] [CrossRef]
- Qin, J.; Zhang, W.; Feng, J.Q.; Zhang, S.B. Leafless epiphytic orchids share Ceratobasidiaceae mycorrhizal fungi. Mycorrhiza 2021, 31, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Suárez, J.P.; Weiß, M.; Abele, A.; Garnica, S.; Oberwinkler, F.; Kottke, I. Diverse tulasnelloid fungi form mycorrhizas with epiphytic orchids in an Andean cloud forest. Mycol. Res. 2006, 110, 1257–1270. [Google Scholar] [CrossRef]
- Suárez, J.P.; Weiß, M.; Abele, A.; Oberwinkler, F.; Kottke, I. Members of Sebacinales subgroup B form mycorrhizae with epiphytic orchids in a neotropical mountain rain forest. Mycol. Prog. 2008, 7, 75–85. [Google Scholar] [CrossRef]
- Kottke, I.; Suárez, J.P.; Herrera, P.; Cruz, D.; Bauer, R.; Haug, I.; Garnica, S. Atractiellomycetes belonging to the ‘rust’ lineage (Pucciniomycotina) form mycorrhizae with terrestrial and epiphytic neotropical orchids. Proc. R. Soc. B Biol. 2010, 277, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Valadares, R.B.D.S.; Otero, J.T.; Pereira, M.C.; Cardoso, E.J.B.N. The epiphytic orchids Ionopsis utricularioides and Psygmorchis pusilla associate with different Ceratobasidium lineages at Valle del Cauca, Colombia. Acta Bot. Bras. 2015, 29, 40–44. [Google Scholar] [CrossRef]
- Waud, M.; Busschaert, P.; Ruyters, S.; Jacquemyn, H.; Lievens, B. Impact of primer choice on characterization of orchid mycorrhizal communities using 454 pyrosequencing. Mol. Ecol. Resour. 2014, 14, 679–699. [Google Scholar] [CrossRef]
- Rammitsu, K.; Kajita, T.; Imai, R.; Ogura-Tsujita, Y. Strong primer bias for Tulasnellaceae fungi in metabarcoding: Specific primers improve the characterization of the mycorrhizal communities of epiphytic orchids. Mycoscience 2021, 62, 356–363. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press, Inc.: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Turenne, C.Y.; Sanche, S.E.; Hoban, D.J.; Karlowsky, J.A.; Kabani, A.M. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J. Clin. Microbiol. 1999, 37, 1846–1851. [Google Scholar] [CrossRef] [PubMed]
- Oja, J.; Kohout, P.; Tedersoo, L.; Kull, T.; Kõljalg, U. Temporal patterns of orchid mycorrhizal fungi in meadows and forests as revealed by 454 pyrosequencing. New Phytol. 2015, 205, 1608–1618. [Google Scholar] [CrossRef] [PubMed]
- Rammitsu, K.; Abe, S.; Abe, T.; Kotaka, N.; Kudaka, M.; Kudaka, N.; Kinoshita, A.; Ogura-Tsujita, Y. The endangered epiphytic orchid Dendrobium okinawense has a highly specific mycorrhizal association with a single Tulasnellaceae fungus. J. For. Res. 2021, 26, 215–221. [Google Scholar] [CrossRef]
- McCormick, M.; Burnett, R.; Whigham, D. Protocorm-supporting fungi are retained in roots of mature Tipularia discolor orchids as mycorrhizal fungal diversity increases. Plants 2021, 10, 1251. [Google Scholar] [CrossRef]
- Rammitsu, K.; Yamamoto, N.; Chamara, R.M.S.R.; Minobe, M.; Kinoshita, A.; Kotaka, N.; Ogura-Tsujita, Y. The epiphytic orchid Vanda falcata is predominantly associated with a single Tulasnellaceae fungus in adulthood, and Ceratobasidiaceae fungi strongly induce its seed germination in vitro. Plant Species Biol. 2023, 38, 306–318. [Google Scholar] [CrossRef]
- Alrich, P.; Higgins, W. Phalaenopsis japonica. Phytotaxa 2014, 161, 67. [Google Scholar]
- Shim, Y.J.; Park, Y.S.; Jang, R.H.; Yoon, Y.J.; Kim, S.R.; Han, S.H. The development of habitat suitability index model of class I endangered wildlife, Sedirea japonica. J. Korean Isl. 2020, 32, 153–172. [Google Scholar] [CrossRef]
- Kim, Y.K.; Jo, S.J.; Kim, K.J. Phylogenetic position of Neofinetia and Sedirea (Orchidaceae) and their species identification using the chloroplast matK and the nuclear ITS sequences. Korean J. Pl. Taxon. 2014, 44, 39–50. [Google Scholar] [CrossRef][Green Version]
- Izumitsu, K.; Hatoh, K.; Sumita, T.; Kitade, Y.; Morita, A.; Tanaka, C.; Gafur, A.; Ohta, A.; Kawai, M.; Yamanaka, T.; et al. Rapid and simple preparation of mushroom DNA directly from colonies and fruiting bodies for PCR. Mycoscience 2012, 53, 396–401. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice, and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evol. 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 5 May 2023).
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open-source tool for metagenomics. PeerJ 2016, 4, 2584. [Google Scholar] [CrossRef]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE QIIME release for fungi. Version 16.10.2022. UNITE Community 2022. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Tokuhara, K.; Mii, M. Micropropagation of Phalaenopsis and Doritaenopsis by culturing shoot tips of flower stalk buds. Plant Cell Rep. 1993, 13, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.M.; Wang, C.L.; Chen, X.M.; Zhou, Y.Q.; Wang, Y.Q.; Luo, A.X.; Liu, Z.H.; Guo, S.X. In vitro seed germination and seedling growth of an endangered epiphytic orchid, Dendrobium officinale, endemic to China using mycorrhizal fungi (Tulasnella sp.). Sci. Hortic. 2014, 165, 62–68. [Google Scholar] [CrossRef]
- Saha, D.; Rao, A.N. Studies on endophytic mycorrhiza of some selected orchids of Arunachal Pradesh–1. Isolation and identification. Bull. Arunachal Forest Res. 2006, 22, 9–16. [Google Scholar]
- Idris, N.A.; Zuhir, Z.M.; Radzuan, N.A.M.; Muda, N.S.; Rosli, R.I. In vitro response of fungi isolated from orchids in BRIS, Setiu wetland and mangrove in Morib, to different concentrations of lead. Malays. Appl. Biol. 2019, 48, 229–233. [Google Scholar]
- Situmorang, N.; Kasiamdari, R.S. Isolation and identification of Rhizoctonia associated with Phalaenopsis amabilis (L.) Blume roots. In Proceedings of the ICBB, The International Conference on Bioscience and Biotechnology, Yogyakarta, Indonesia, 11–12 October 2011; Volume 1, pp. 39–44. [Google Scholar]
- Warcup, J.H. The mycorrhizal relationships of Australian orchids. New Phytol. 1981, 87, 371–381. [Google Scholar] [CrossRef]
- Ogura-Tsujita, Y.; Yokoyama, J.; Miyoshi, K.; Yukawa, T. Shifts in mycorrhizal fungi during the evolution of autotrophy to mycoheterotrophy in Cymbidium (Orchidaceae). Am. J. Bot. 2012, 99, 1158–1176. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Song, X.; Meng, Q.; Zhu, J.; Zhao, Y.; Yu, W. Influence of host tree species on isolation and communities of mycorrhizal and endophytic fungi from roots of a tropical epiphytic orchid, Dendrobium sinense (Orchidaceae). Mycorrhiza 2017, 27, 709–718. [Google Scholar] [CrossRef]
- Ventre Lespiaucq, A.; Jacquemyn, H.; Rasmussen, H.N.; Mendez, M. Temporal turnover in mycorrhizal interactions: A proof of concept with orchids. New Phytol. 2021, 230, 1690–1699. [Google Scholar] [CrossRef]




| Site | Locality | No. of Populations | Host Tree Species | No. of Individuals | No. of DNA Samples | |
|---|---|---|---|---|---|---|
| Roots | Isolates | |||||
| S1 | Fukuoka Pref. * | 1 | Quercus glauca | 4 | 5 | 4 |
| S2 | Miyazaki Pref. * | 2 | Unknown | 5 | 13 | 9 |
| S3 | Yakushima, Kagoshima Pref. | 1 | Ilex integra | 2 | 3 | 1 |
| S4 | Tokunoshima, Kagoshima Pref. | 4 | Pinus luchuensis | 9 | 17 | − |
| Unknown | 12 | 20 | 2 | |||
| Family | Fungal OTU | Host Orchid | DDBJ Accession No. | NBRC Accession No. |
|---|---|---|---|---|
| Ceratobasidiaceae | CE6-1 | P. japonica | LC746361 | NBRC 116432 |
| CE6-2 | P. japonica | LC746362 | NBRC 116431 | |
| CE12 | P. japonica | LC746363 | NBRC 116433 | |
| CE22 | P. japonica | LC597348 | NBRC 114917 | |
| Tulasnellaceae | TU18 | Staurochilus lutchuensis | LC597353 | NBRC 114910 |
| TU27 | Dendrobium officinale | LC683202 | NBRC 115262 |
| Fungal Family | OTUs from Sanger Sequencing | OTUs from HTS |
|---|---|---|
| Ceratobasidiaceae | CE6, CE12, CE22, CE35 | CE1, CE6, CE22, CE35 |
| Tulasnellaceae | TU11, TU12, TU18, TU53, TU54 | TU1, TU3, TU10, TU11, TU12, TU18, TU22, TU23, TU37, TU46, TU53, TU54, TU58, TU62, TU68, TU73 |
| Serendipitaceae | - | SE9, SE10 |
| Total OTUs | 9 | 22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chamara, R.M.S.R.; Rammitsu, K.; Minobe, M.; Kinoshita, A.; Kotaka, N.; Yukawa, T.; Ogura-Tsujita, Y. Mycorrhizal Fungi of Phalaenopsis japonica (Orchidaceae) and Their Role in Seed Germination and Seedling Development. Diversity 2024, 16, 218. https://doi.org/10.3390/d16040218
Chamara RMSR, Rammitsu K, Minobe M, Kinoshita A, Kotaka N, Yukawa T, Ogura-Tsujita Y. Mycorrhizal Fungi of Phalaenopsis japonica (Orchidaceae) and Their Role in Seed Germination and Seedling Development. Diversity. 2024; 16(4):218. https://doi.org/10.3390/d16040218
Chicago/Turabian StyleChamara, R. M. S. Ruwan, Kento Rammitsu, Mutsumi Minobe, Akihiko Kinoshita, Nobuhiko Kotaka, Tomohisa Yukawa, and Yuki Ogura-Tsujita. 2024. "Mycorrhizal Fungi of Phalaenopsis japonica (Orchidaceae) and Their Role in Seed Germination and Seedling Development" Diversity 16, no. 4: 218. https://doi.org/10.3390/d16040218
APA StyleChamara, R. M. S. R., Rammitsu, K., Minobe, M., Kinoshita, A., Kotaka, N., Yukawa, T., & Ogura-Tsujita, Y. (2024). Mycorrhizal Fungi of Phalaenopsis japonica (Orchidaceae) and Their Role in Seed Germination and Seedling Development. Diversity, 16(4), 218. https://doi.org/10.3390/d16040218






