Influence of Microbes in Mediating Sorghum Resistance to Sugarcane Aphids
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants and Insects
2.2. Aphid Bioassay, DNA Extraction, and Microbiome Sequencing
2.3. Processing Data and Statistical Analyses
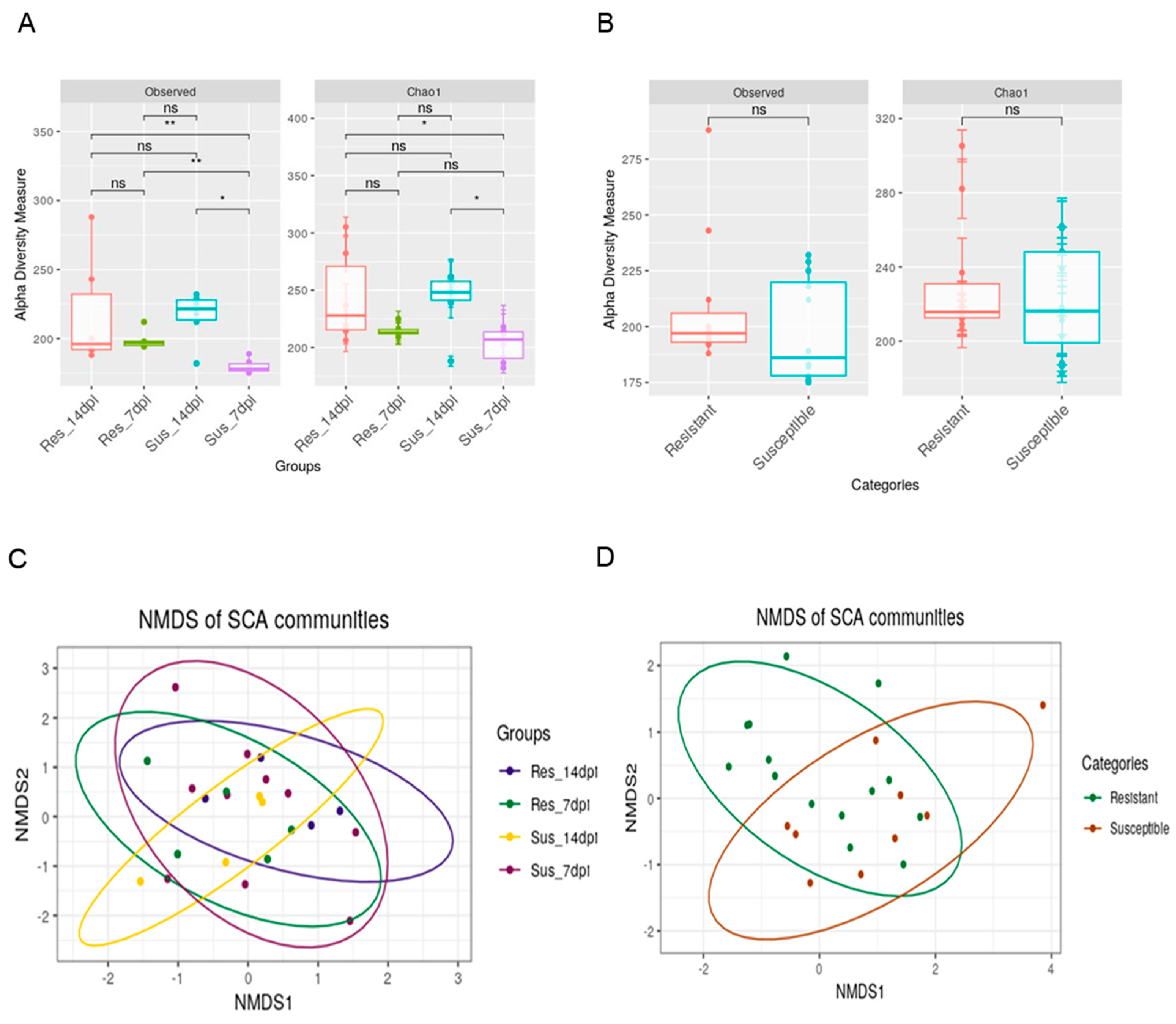
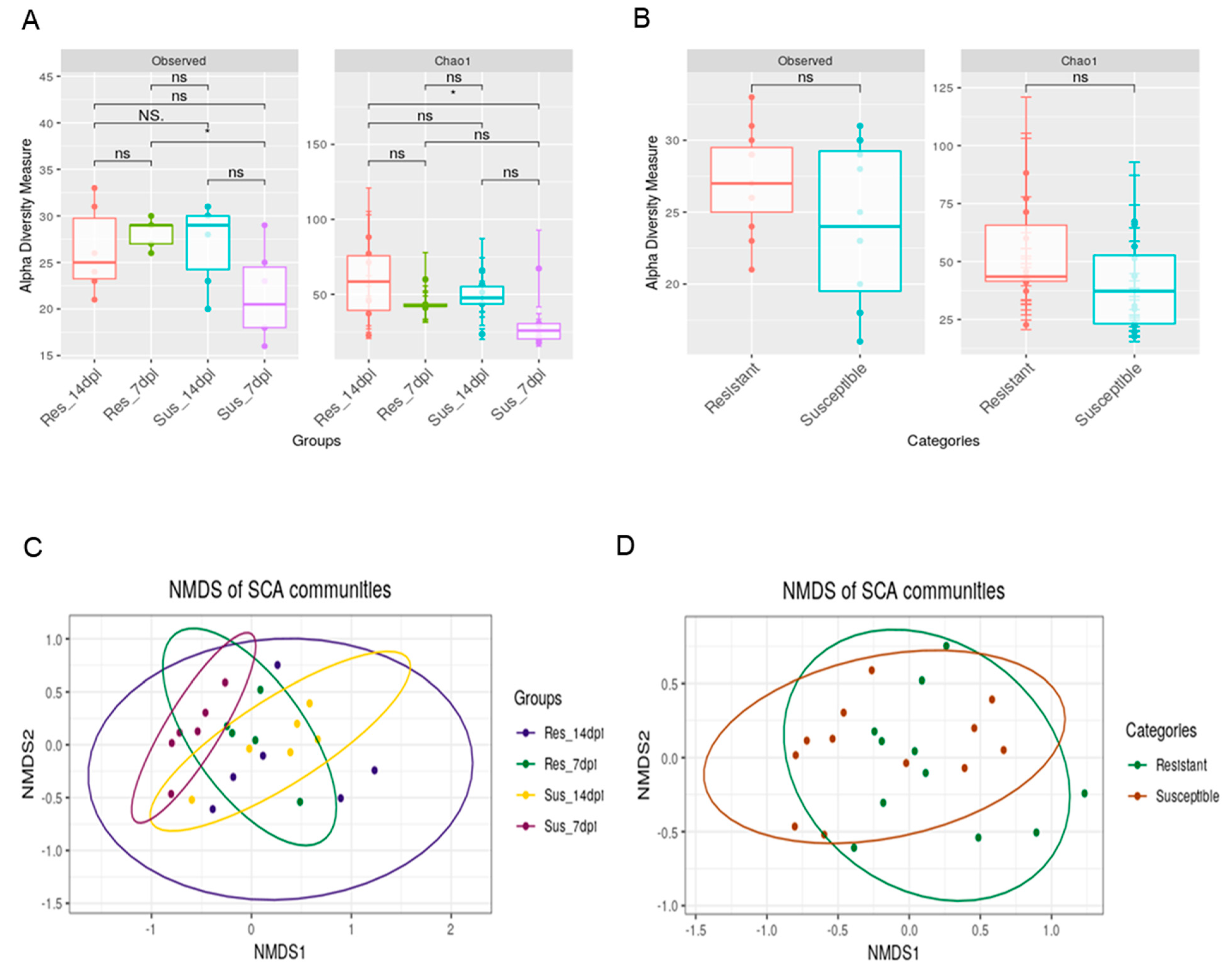
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fukatsu, T. Grand challenges to launching an ideal platform for publishing microbe-insect symbiosis studies. Front. Microbiol. 2019, 10, 2542. [Google Scholar] [CrossRef]
- Armstrong, J.S.; Rooney, W.L.; Peterson, G.C.; Villanueva, R.T.; Brewer, M.J.; Sekula-Ortiz, D. Sugarcane aphid (Hemiptera: Aphididae): Host range and sorghum resistance, including cross-resistance from greenbug sources. J. Econ. Entomol. 2015, 108, 576–582. [Google Scholar] [CrossRef]
- Bowling, R.D.; Brewer, M.J.; Kerns, D.L.; Gordy, J.; Seiter, N.; Elliott, N.E.; Maxson, E. Sugarcane aphid (Hemiptera: Aphididae): A new pest on sorghum in North America. J. Integr. Pest Manag. 2016, 7, 12. [Google Scholar] [CrossRef]
- Medina, R.F.; Armstrong, S.J.; Harrison, K. Genetic population structure of the sugarcane aphid, Melanaphis sacchari. sorghum, sugarcane, and Johnsongrass in continental USA. Entomol. Exp. Et Appl. 2017, 162, 358–365. [Google Scholar] [CrossRef]
- Holt, J.R.; Styer, A.; White, J.A.; Armstrong, J.S.; Nibouche, S.; Costet, L.; Malacrinò, A.; Antwi, J.B.; Wulff, J.; Peterson, G.; et al. Differences in microbiota between two multilocus lineages of the sugarcane aphid (Melanaphis sacchari) in the continental United States. Ann. Entomol. Soc. Am. 2020, 113, 257–265. [Google Scholar] [CrossRef]
- Xoconostle-Cázares, B.; Ramírez-Pool, J.A.; Núñez-Muñoz, L.A.; Calderón-Pérez, B.; Vargas-Hernández, B.Y.; Bujanos-Muñiz, R.; Ruiz-Medrano, R. The Characterization of Melanaphis sacchari microbiota and antibiotic treatment effect on insects. Insects 2023, 14, 807. [Google Scholar] [CrossRef] [PubMed]
- Dadd, R.H. Nutrition: Organisms. Compr. Insect Physiol. Biochem. Pharmacol. 1985, 4, 313–390. [Google Scholar]
- Chaudhary, R.; Atamian, H.S.; Shen, Z.; Briggs, S.P.; Kaloshian, I. GroEL from the endosymbiont Buchnera aphidicola betrays aphids by triggering plant defenses. Proc. Natl. Acad. Sci. USA 2014, 111, 8919–8924. [Google Scholar] [CrossRef]
- Smith, J.A.; Peccoud, J.; Yourth, C.P. The role of symbiont genetic quality in the evolution of insect host plant use. Front. Microbiol. 2019, 10, 2506. [Google Scholar]
- Chong, R.A.; Moran, N.A. Evolutionary loss and replacement of Buchnera, the obligate endosymbiont of aphids. ISME J. 2018, 12, 898–908. [Google Scholar] [CrossRef]
- Wernegreen, J.J. Endosymbiosis. Curr. Biol. 2012, 22, R555–R561. [Google Scholar] [CrossRef]
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F.; et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229–3236. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.M.; Smith, A.H.; Russell, J.A. Defensive symbiosis in the real world–advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct. Ecol. 2014, 28, 341–355. [Google Scholar] [CrossRef]
- McLean, A.H.; Godfray, H.C.J.; Ellers, J.; Henry, L.M. Host relatedness influences the composition of aphid microbiomes. Environ. Microbiol. Rep. 2019, 11, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schulze-Lefert, P. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Zytynska, S.E.; Tighiouart, K.; Frago, E. Benefits and costs of hosting facultative symbionts in plant-sucking insects: A meta-analysis. Mol. Ecol. 2021, 30, 2483–2494. [Google Scholar] [CrossRef] [PubMed]
- Biere, A.; Bennett, A.E. Three-way interactions among plants, microbes, and insects. Funct. Ecol. 2013, 27, 567–573. [Google Scholar] [CrossRef]
- Khoddami, A.V.; Messina, K.; Vadabalija, V.A.; Venkata, A.; Farahnaky, A.; Blanchard, C.L.; Roberts, T.H. Sorghum in foods: Functionality and potential in innovative products. Crit. Rev. Food Sci. Nutr. 2021, 63, 1170–1186. [Google Scholar] [CrossRef]
- Stamenkovic, O.S.; Siliveru, K.; Veljkovic, V.B.; Bankovic-Ilic, I.B.; Tasic, M.B.; Ciampitti, I.A.; Đalović, I.G.; Mitrović, P.M.; Sikora, V.Š.; Prasad, P.V. Production of biofuels from sorghum. Renew. Sustain. Energy Rev. 2020, 124, 109769. [Google Scholar] [CrossRef]
- Wang, D.; Bean, S.; McLaren, J.; Seib, P.; Madl, R.; Tuinstra, M.; Shi, Y.; Lenz, M.; Wu, X.; Zhao, R. Sorghum grain is a viable feedstock for ethanol production. J. Ind. Microbiol. Biotechnol. 2008, 35, 313–320. [Google Scholar] [CrossRef]
- Mou, D.F.; Kundu, P.; Pingault, L.; Puri, H.; Shinde, S.; Louis, J. Monocot crop-aphid interactions: Plant resilience and aphid adaptation. Curr. Opin. Insect Sci. 2023, 57, 101038. [Google Scholar] [CrossRef]
- Grover, S.; Agpawa, E.; Sarath, G.; Sattler, S.E.; Louis, J. Interplay of phytohormones facilitates sorghum tolerance to aphids. Plant Mol. Biol. 2022, 109, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Grover, S.; Puri, H.; Xin, Z.; Sattler, S.E.; Louis, J. Dichotomous role of jasmonic acid in modulating sorghum defense against aphids. Mol. Plant-Microbe Interact. 2022, 35, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Puri, H.; Grover, S.; Pingault, L.; Sattler, S.E.; Louis, J. Temporal transcriptomic profiling elucidates sorghum defense mechanisms against sugarcane aphids. BMC Genom. 2023, 24, 441. [Google Scholar] [CrossRef] [PubMed]
- Bouchet, S.; Olatoye, M.O.; Marla, S.R.; Perumal, R.; Tesso, T.; Yu, J.; Tuinstra, M.; Morris, G.P. Increased power to dissect adaptive traits in global sorghum diversity using a nested association mapping population. Genetics 2017, 206, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Rintala, A.; Pietilä, S.; Munukka, E.; Eerola, E.; Pursiheimo, J.P.; Laiho, A.; Pekkala, S.; and Huovinen, P. Gut microbiota analysis results are highly dependent on the 16S rRNA gene target region, whereas the impact of DNA extraction is minor. J. Biomol. Tech. 2017, 28, 19–30. [Google Scholar] [CrossRef]
- Mukherjee, S.; Huntemann, M.; Ivanova, N.; Kyrpides, N.C.; Pati, A. Large-scale contamination of microbial isolate genomes by Illumina PhiX control. Stand. Genomic Sci. 2015, 10, 1–4. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Salomon, J.D.; Qiu, H.; Feng, D.; Owens, J.; Khailova, L.; Osorio Lujan, S.; Iguidbashian, J.; Chhonker, Y.S.; Murry, D.J.; Riethoven, J.-J.; et al. Piglet cardiopulmonary bypass (CPB) induces intestinal dysbiosis and barrier dysfunction associated with systemic inflammation. Dis. Models Mech. 2023, 16, dmm049742. [Google Scholar] [CrossRef] [PubMed]
- Wasimuddin, W.; Schlaeppi, K.; Ronchi, F.; Leib, S.L.; Erb, M.; Ramette, A. Evaluation of primer pairs for microbiome profiling from soils to humans within the One Health framework. Mol. Ecol. Resour. 2020, 20, 1558–1571. [Google Scholar] [CrossRef]
- Xu, S.; Jiang, L.; Qiao, G.; Chen, J. Diversity of bacterial symbionts associated with Myzus persicae (Sulzer) (Hemiptera: Aphididae: Aphidinae) was revealed by 16S rRNA Illumina sequencing. Microb. Ecol. 2021, 81, 784–794. [Google Scholar] [CrossRef] [PubMed]
- McKnight, D.T.; Huerlimann, R.; Bower, D.S.; Schwarzkopf, L.; Alford, R.A.; Zenger, K.R. Methods for normalizing microbiome data: An ecological perspective. Methods Ecol. Evol. 2019, 10, 389–400. [Google Scholar] [CrossRef]
- Chiu, C.H.; Wang, Y.T.; Walther, B.A.; Chao, A. An improved nonparametric lower bound of species richness via a modified good–turing frequency formula. Biometrics 2014, 70, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Dastogeer, K.M.; Tumpa, F.H.; Sultana, A.; Akter, M.A.; Chakraborty, A. Plant microbiome—An account of the factors that shape community composition and diversity. Curr. Plant Biol. 2020, 23, 100161. [Google Scholar] [CrossRef]
- Douglas, A.E. Symbiotic Interactions; Oxford University Press: Oxford, UK, 1994. [Google Scholar]
- Douglas, A.E. Reproductive failure and the free amino acid pools in pea aphids (Acyrthosiphon pisum) lacking symbiotic bacteria. J. Insect Physiol. 1996, 42, 247–255. [Google Scholar] [CrossRef]
- Wang, Q.; Yuan, E.; Ling, X.; Zhu-Salzman, K.; Guo, H.; Ge, F.; Sun, Y. An aphid facultative symbiont suppresses plant defence by manipulating aphid gene expression in salivary glands. Plant Cell Environ. 2020, 43, 2311–2322. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, Y.; Xu, S.; Wang, J.; Zhang, Z.; Wang, M.Q.; Turlings, T.C.J.; Zhang, P.; Zhou, A. Mealybug salivary microbes inhibit induced plant defenses. Pest Manag. Sci. 2023, 79, 4034–4047. [Google Scholar] [CrossRef]
- Chung, S.H.; Rosa, C.; Scully, E.D.; Peiffer, M.; Tooker, J.F.; Hoover, K.; Luthe, D.S.; Felton, G.W. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Natl. Acad. Sci. USA 2013, 110, 15728–15733. [Google Scholar] [CrossRef]
- Smith, D.L.; Goodman, R.M. Rhizobium symbiosis: Nod factors in perspective. Plant Cell 1999, 11, 2235–2248. [Google Scholar]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Garcia, A.V.; Charrier, A.; Schikora, A.; Bigeard, J.; Pateyron, S.; de Tauzia-Moreau, M.L.; Evrard, A.; Mithöfer, A.; Martin-Magniette, M.L.; Virlogeux-Payant, I.; et al. Salmonella enterica flagellin is recognized via FLS2 and activates PAMP-triggered immunity in Arabidopsis thaliana. Mol. Plant 2014, 7, 657–674. [Google Scholar] [CrossRef] [PubMed]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.T.; Weigel, D.; Kemen, E.M. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef]
- Pronk, L.J.; Bakker, P.A.; Keel, C.; Maurhofer, M.; Flury, P. The secret life of plant-beneficial rhizosphere bacteria: Insects as alternative hosts. Environ. Microbiol. 2022, 24, 3273–3289. [Google Scholar] [CrossRef] [PubMed]
- White, J.F., Jr.; Torres, M.S.; Sullivan, R.F.; Jabbour, R.E.; Chen, Q.; Tadych, M.; Irizarry, I.; Bergen, M.S.; Havkin-Frenkel, D.; Belanger, F.C. Occurrence of Bacillus amyloliquefaciens as a systemic endophyte in vanilla orchids. Microsc. Res. Tech. 2014, 77, 874–885. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Oliver, K.M.; Moran, N.A.; Hunter, M.S. The variation in resistance to parasitism in aphids is due to symbionts rather than the host genotype. Proc. Natl. Acad. Sci. USA 2005, 102, 12795–12800. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikuze, E.; Cromwell, S.; Ayayee, P.; Louis, J. Influence of Microbes in Mediating Sorghum Resistance to Sugarcane Aphids. Diversity 2024, 16, 85. https://doi.org/10.3390/d16020085
Ikuze E, Cromwell S, Ayayee P, Louis J. Influence of Microbes in Mediating Sorghum Resistance to Sugarcane Aphids. Diversity. 2024; 16(2):85. https://doi.org/10.3390/d16020085
Chicago/Turabian StyleIkuze, Edith, Stephanie Cromwell, Paul Ayayee, and Joe Louis. 2024. "Influence of Microbes in Mediating Sorghum Resistance to Sugarcane Aphids" Diversity 16, no. 2: 85. https://doi.org/10.3390/d16020085
APA StyleIkuze, E., Cromwell, S., Ayayee, P., & Louis, J. (2024). Influence of Microbes in Mediating Sorghum Resistance to Sugarcane Aphids. Diversity, 16(2), 85. https://doi.org/10.3390/d16020085






