High Genetic Diversity of Hirudo verbana Carena, 1820 (Annelida: Hirudinea: Hirudinidae) in Romania Confirms That the Balkans Are Refugia Within Refugium
Abstract
1. Introduction
2. Materials and Methods
2.1. Species Sampling and Morphological Examination
2.2. DNA Extraction, Amplification, and Sequencing
2.3. Phylogeographic and Demographic Analyses
2.4. Spatial Analysis and Distribution Modeling
3. Results
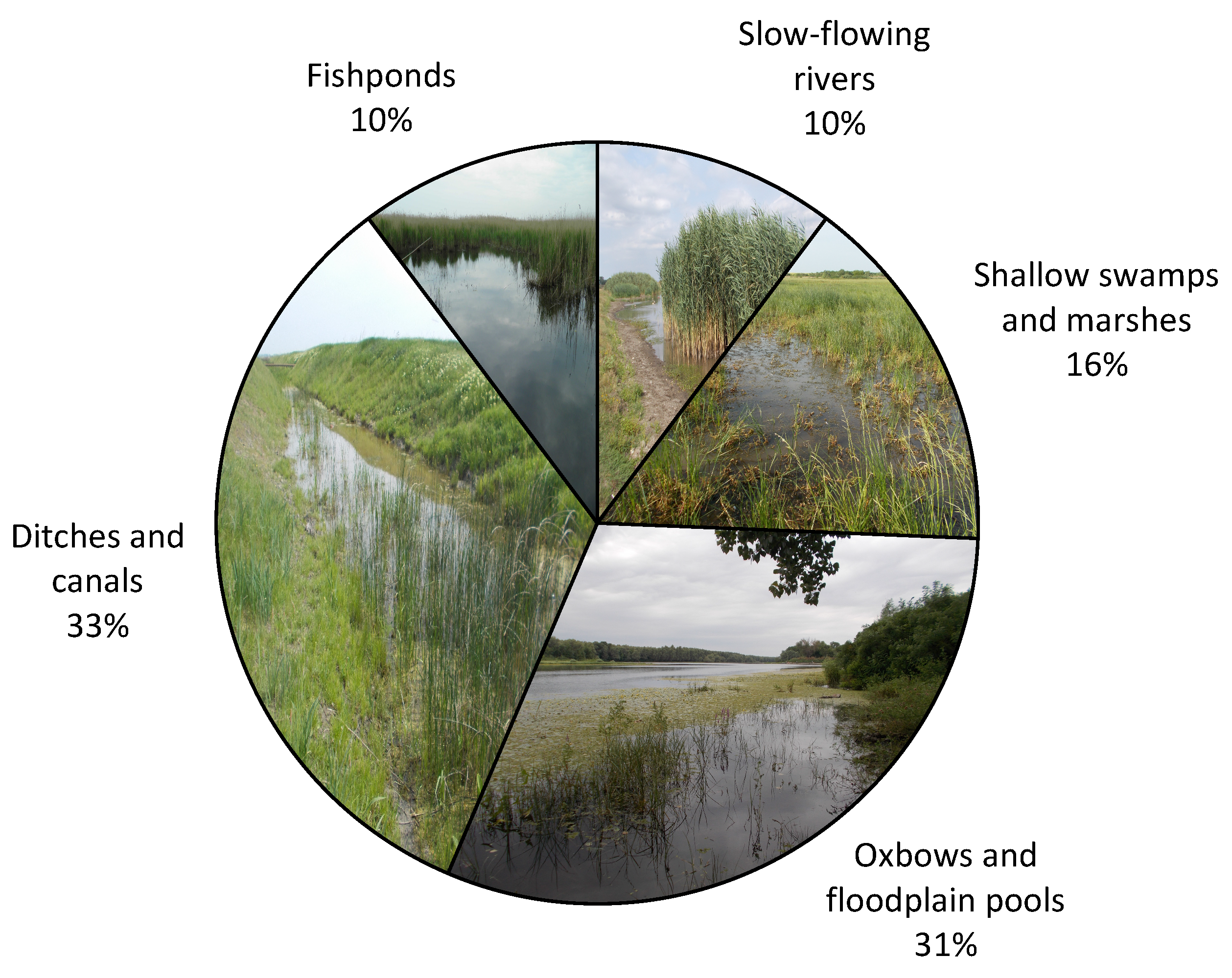
3.1. Distribution of Hirudo verbana in Romania
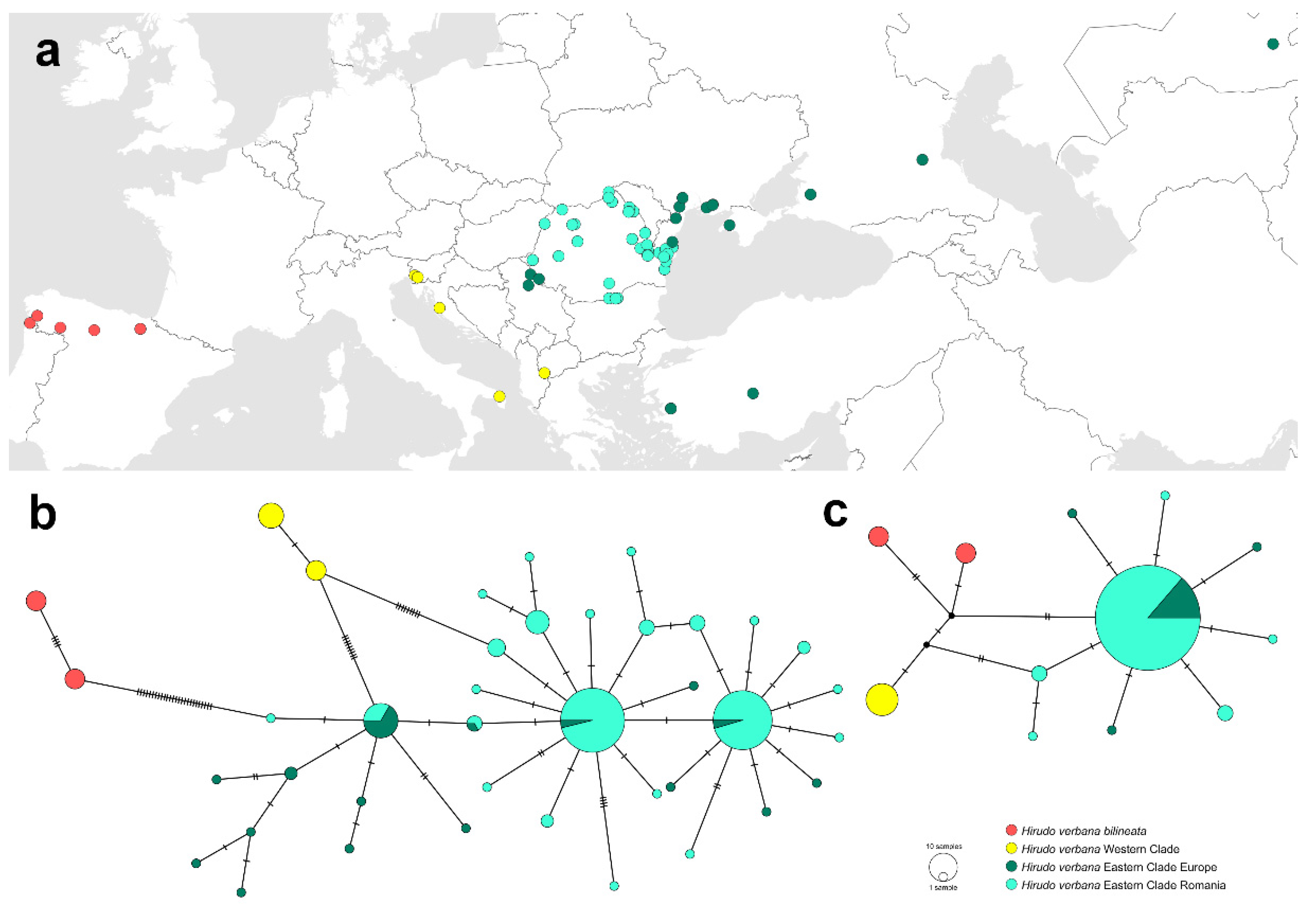
3.2. Phylogenetic and Biogeographic Relationships
3.2.1. Phylogeographic Pattern of Hirudo verbana in Europe
3.2.2. Demographic Parameters of Hirudo verbana in Romania
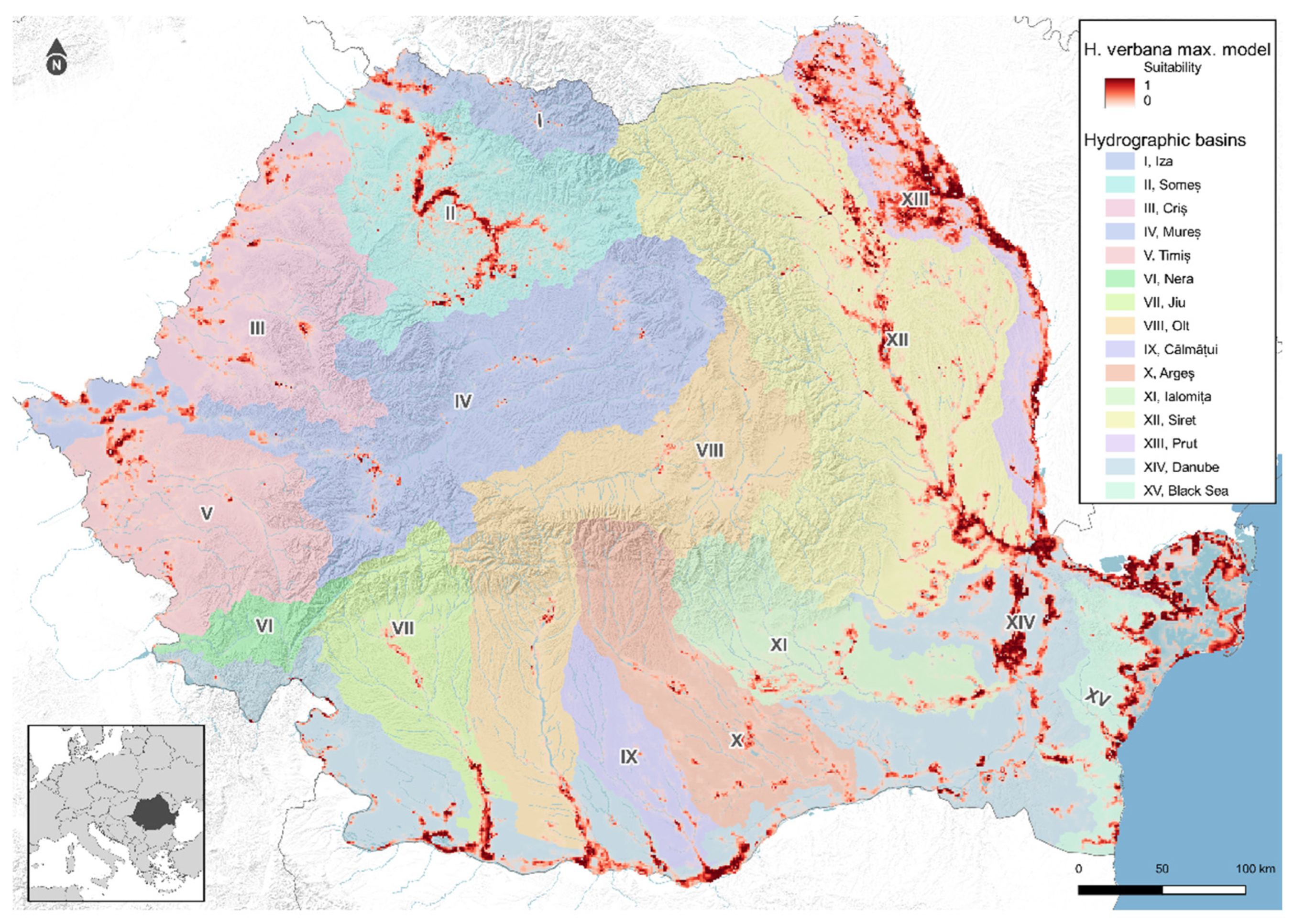
3.3. Spatial Analysis and Distribution Modeling
4. Discussion
4.1. Taxonomic Status
4.2. Distribution of Hirudo verbana in Romania and Neighboring Countries
4.3. Phylogenetic and Biogeographic Relationships
4.4. Conservation Status of the Mediterranean Medicinal Leech
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sawyer, R.T. Why we need to save the medicinal leech. Oryx 1981, 16, 165–168. [Google Scholar] [CrossRef]
- Whitaker, I.S.; Rao, J.; Izadi, D.; Butler, P.E. Historical Article: Hirudo medicinalis: Ancient origins of the use of medicinal leeches throughout history. Br. J. Oral Maxillofac. Surg. 2004, 42, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J.M.; Kutschera, U. Medicinal leeches: Historical use, ecology, genetics and conservation. Freshw. Rev. 2011, 4, 21–41. [Google Scholar] [CrossRef]
- Trontelj, P.; Utevsky, S.Y. Celebrity with a neglected taxonomy: Molecular systematics of the medicinal leech (genus Hirudo). Mol. Phylogenet. Evol. 2005, 34, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Utevsky, S.Y.; Trontelj, P. A new species of the medicinal leech (Oligochaeta, Hirudinida, Hirudo) from Transcaucasia and an identification key for the genus Hirudo. Parasitol. Res. 2005, 98, 61–66. [Google Scholar] [CrossRef]
- Moquin-Tandon, A. Monographie de la famille des Hirudinées, 2nd ed.; Ballière: Paris, France, 1846. [Google Scholar] [CrossRef]
- Sawyer, R.T. Leech Biology and Behaviour. Vol. II. Feeding Biology, Ecology, and Systematics; Clarendon Press: Oxford, UK, 1986. [Google Scholar]
- Hechtel, F.O.P.; Sawyer, R.T. Toward a taxonomic revision of the medicinal leech Hirudo medicinalis Linnaeus, 1758 (Hirudinea: Hirudinidae): Re-description of Hirudo troctina Johnston, 1816 from North Africa. J. Nat. Hist. 2002, 36, 1269–1289. [Google Scholar] [CrossRef]
- Nesemann, H.; Neubert, E. Annelida, Clitellata: Branchiobdellida, Acanthobdellea, Hirudinea. In Süßwasserfauna von Mitteleuropa; Schwoerbel, J., Zwick, P., Eds.; Spektrum Akademischer Verlag: Berlin/Heidelberg, Germany, 1999; Band 6/2; pp. 1–178. [Google Scholar]
- Siddall, M.E.; Trontelj, P.; Utevsky, S.Y.; Nkamany, M.; Macdonald, K.S. Diverse molecular data demonstrate that commercially available medicinal leeches are not Hirudo medicinalis. Proc. R. Soc. B 2007, 274, 1481–1487. [Google Scholar] [CrossRef]
- Trontelj, P.; Sotler, M.; Verovnik, R. Genetic differentiation between two species of the medicinal leech, Hirudo medicinalis and the neglected H. verbana, based on random-amplified polymorphic DNA. Parasitol. Res. 2004, 94, 118–124. [Google Scholar] [CrossRef]
- Călinescu, R.; Antonescu, C.; Bănărescu, P.; Botoșeneanu, L.; Coteț, P.; Decu, V.; Doniță, N.; Negrea, Ș.; Pleșa, C.; Tălpeanu, M. Biogeografia României; Editura Științifică: Bucharest, Romania, 1969. [Google Scholar]
- Taberlet, P.; Fumagalli, L.; Wust-Saucy, A.-G.; Cosson, J.-F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998, 7, 453–464. [Google Scholar] [CrossRef]
- Provan, J.; Bennett, K.D. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 2008, 23, 564–571. [Google Scholar] [CrossRef]
- Bálint, M.; Ujvárosi, L.; Theissinger, K.; Lehrian, S.; Mészáros, N.; Pauls, S.U. The Carpathians as a major diversity hotspot in Europe. In Biodiversity Hotspots; Zachos, F.E., Habel, J.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 189–205. [Google Scholar] [CrossRef]
- Epure, E. Die Fauna der Hirudineen im Banat. Ann. Sci. Univ. Jassy 1944, 30, 29–37. [Google Scholar]
- Cristea, V. Contribuții la studiul hirudineelor din Balta Cătușa. Lucr. Șt. Instit. Ped. Galați 1969, 3, 149–158. [Google Scholar]
- Manoleli, D. Contributions to the knowledge of the Hirudinea from the Eastern Romanian Plain and from the Mounts of Vrancea. Trav. Mus. Hist. Nat. “Grigore Antipa” 1974, 14, 79–83. [Google Scholar]
- Cristea, V.; Manoleli, D. Conspectus des Sangsues (Hirudinea) de Roumanie, avec une clé de détermination. Trav. Mus. Hist. Nat. “Grigore Antipa” 1977, 18, 23–56. [Google Scholar]
- Neacșu, P.I. Contrubutions à la conaîssance de l’écologie des sangsues dans le Delta du Danube (etangs Matița et Merhei). An. Univ. Bucur. Biol. 1983, 32, 51–56. [Google Scholar]
- Neacșu, P.I.; Marinescu, T. Contributions to the ecology of the Hirudinea-Populations (Annelida-Hirudinea) from the pond Valea Oranei (Distr. Dolj). An. Univ. Bucur. Biol. 1984, 33, 67–73. [Google Scholar]
- Gagiu, A. The first recorded occurrence of Hirudo verbana Carena, 1820 (Hirudinea: Arhynchobdellida: Hirudinidae) in Romania. Trav. Mus. Hist. Nat. “Grigore Antipa” 2010, 53, 7–11. [Google Scholar] [CrossRef]
- Fermaș, T.A.; Cristofor, S. Contributions to knowledge of structural dynamics of the populations of medicinal leeches in the Brăila marshes complex (the Lower Danube wetland system). An. Șt. Univ. „A.I. Cuza” Iași Ser. Biol. Anim. 2013, S3, 29–36. [Google Scholar]
- Iorgu, I.Ș.; Surugiu, V.; Gheoca, V.; Popa, O.P.; Popa, L.O.; Sîrbu, I.; Pârvulescu, L.; Iorgu, E.I.; Manci, C.O.; Fusu, L.; et al. Ghid Sintetic Pentru Monitorizarea Speciilor de Nevertebrate de Interes Comunitar din România; S.C. CCAT S.R.L. & S.C. Integra Trading S.R.L.: Bucharest, Romania, 2015. [Google Scholar]
- Surugiu, V. On the presence of the European medicinal leech Hirudo medicinalis Linnaeus, 1758 (Annelida: Hirudinea) in Romania. Trav. Mus. Hist. Nat. “Grigore Antipa” 2018, 61, 7–11. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Bandelt, H.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Csaikl, U.M.; Bordács, S.; Burg, K.; Coart, E.; Cottrell, J.; van Dam, B.; Deans, J.D.; Dumolin-Lapègue, S.; Fineschi, S.; et al. Chloroplast DNA variation in European white oaks: Phylogeography and patterns of diversity based on data from over 2600 populations. For. Ecol. Manag. 2002, 156, 5–26. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1 km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carre, G.; García Marquéz, J.R.; Gruber, B.; Lafourcade, B.; Leitao, P.J.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- Hair, J.F.; Black, W.C.; Babin, B.J.; Anderson, R.E.; Tatham, R.L. Multivariate Data Analysis; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2006. [Google Scholar]
- Harding, T.; Tusell, F. Cat: Analysis and Imputation of Categorical-Variable Datasets with Missing Values. 2023. Available online: https://cran.r-project.org/web/packages/cat/index.html (accessed on 20 April 2024).
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudik, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Sutton, L.J.; Puschendorf, R. Climatic niche of the Saker Falcon Falco cherrug: Predicted new areas to direct population surveys in Central Asia. Ibis 2020, 162, 27–41. [Google Scholar] [CrossRef]
- Fehérvári, P.; Solt, S.; Palatitz, P.; Barna, K.; Agoston, A.; Gergely, J.; Nagy, A.; Nagy, K.; Harnos, A. Allocating active conservation measures using species distribution models: A case study of Red-footed Falcon breeding site management in the Carpathian Basin. Anim. Conserv. 2012, 15, 648–657. [Google Scholar] [CrossRef]
- Brezeanu, G.; Popescu-Marinescu, V. Studiul hidrobiologic al bazinului inferior al Cernei (Cerna, Belareca și Mehadica). Hidrobiologia 1964, 5, 65–94. [Google Scholar]
- Popescu-Marinescu, V. Structura zoocenozelor bentonice din Dunăre, în sectorul românesc, în perioada 1971–1986. Hidrobiologia 1992, 20, 111–134. [Google Scholar]
- Slatkin, M.; Hudson, R.R. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 1991, 129, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar] [PubMed]
- Leon, N. Zoologia medicală a ţăranului român. Arh. Soc. Şt. Liter. Iaşi 1897, 8, 249–278. [Google Scholar]
- Ardeleanu, C. The Danubian leech trade in the 19th century. The global market of a tiny product. Rev. Istor. 2018, 29, 177–188. [Google Scholar]
- Scriban, I. Contribution à la Faune des Hirudinées d’eau douce de Roumanie. Ann. Sci. Univ. Jassy 1904, 3, 17–20. [Google Scholar]
- Scriban, I. Contributions à l’anatomie et à l’histologie des Hirudinées. Ann. Sci. Univ. Jassy 1910, 6, 147–286. [Google Scholar]
- Kutschera, U. The Hirudo medicinalis species complex. Sci. Nat. 2012, 99, 433–434. [Google Scholar] [CrossRef]
- Sağlam, N.; Saunders, R.; Lang, S.A.; Shain, D.H. A new species of Hirudo (Annelida: Hirudinidae): Historical biogeography of Eurasian medicinal leeches. BMC Zool. 2016, 1, 5. [Google Scholar] [CrossRef]
- Trontelj, P.; Utevsky, S.Y. Phylogeny and phylogeography of medicinal leeches (genus Hirudo): Fast dispersal and shallow genetic structure. Mol. Phylogenet. Evol. 2012, 63, 475–485. [Google Scholar] [CrossRef]
- Utevsky, S.; Zagmajster, M.; Atemasov, A.; Zinenko, O.; Utevska, O.; Utevsky, A.; Trontelj, P. Distribution and status of medicinal leeches (genus Hirudo) in the Western Palaearctic: Anthropogenic, ecological, or historical effects? Aquat. Conserv. Mar. Freshw. Ecosyst. 2010, 20, 198–210. [Google Scholar] [CrossRef]
- Arias, A.; Surugiu, V.; Carballeira, R.; Popa, O.P.; Popa, L.O.; Utevsky, S. Unravelling the extent of diversity within the Iberian medicinal leeches (Hirudinea: Hirudo) using molecules and morphology. Biology 2021, 10, 315. [Google Scholar] [CrossRef] [PubMed]
- Utevsky, S.; Zinenko, A.I.; Atemasov, A.A.; Huseynov, M.A.; Utevska, O.M.; Utevsky, A.Y. New information on the distribution of the medicinal leech (genus Hirudo) in the Iberian Peninsula, the Caucasus and Central Asia. Lauterbornia 2008, 65, 119–130. [Google Scholar]
- Utevsky, S.; Atemasov, A.A.; Mazepa, G.O.; Utevsky, A.Y.; Utevska, O.M.; Zinenko, O.I. New information on the distribution of the medicinal leech, Hirudo (Hirudinea) in Ukraine, Central Asia, Azerbaijan and the Northern Caucasus. Vestn. Zool. 2008, 42, 56–75. (In Russian) [Google Scholar]
- Utevsky, S.; Trontelj, P. Phylogeography of the southern medicinal leech, Hirudo verbana: A response to Živić et al. (2015). Aquat. Ecol. 2016, 50, 97–100. [Google Scholar] [CrossRef]
- Lukin, E.I. Piyavki [Leeches]. Fauna SSSR; Academy of Sciences of the USSR: Nauka, Leningrad, Soviet Union, 1976; Volume 1. (In Russian) [Google Scholar]
- Utevsky, S.Y.; Utevsky, A.Y.; Utevskaya, O.M. A finding of the officinal medicinal leech Hirudo medicinalis f. offcinalis in Ukraine. Vestn. Zool. 1998, 32, 37. (In Russian) [Google Scholar]
- Todorov, M.; Grozeva, S.; Hubenov, Z.; Kenderov, L.; Trichkova, T. Taxonomic status and distribution of medicinal leeches of the genus Hirudo L. (Hirudinea) in Bulgaria. Acta Zool. Bulg. 2016, 68, 171–182. [Google Scholar]
- Živić, I.; Radosavljević, T.; Stojanović, K.; Petrović, A. The first molecular characterization of the genus Hirudo on the territory of Serbia: Estimation of endangerment. Aquat. Ecol. 2015, 49, 81–90. [Google Scholar] [CrossRef]
- Bănărescu, P. Distribution pattern of the aquatic fauna of the Balkan Peninsula. In Balkan Biodiversity—Pattern and Process in the European Hotspot; Griffiths, H.I., Kryštufek, B., Reed, J.M., Eds.; Kluwer: Dordrecht, Germany, 2004; pp. 203–217. [Google Scholar] [CrossRef]
- Hewitt, G. Mediterranean peninsulas: The evolution of hotspots. In Biodiversity Hotspots; Zachos, F.E., Habel, J.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 123–147. [Google Scholar] [CrossRef]
- Wielstra, B.; Crnobrnja-Isailović, J.; Litvinchuk, S.N.; Reijnen, B.T.; Skidmore, A.K.; Sotiropoulos, K.; Toxopeus, A.G.; Tzankov, N.; Vukov, T.; Arntzen, J.W. Tracing glacial refugia of Triturus newts based on mitochondrial DNA phylogeography and species distribution modeling. Front. Zool. 2013, 10, 13. [Google Scholar] [CrossRef]
- Lyubas, A.A.; Kondakov, A.V.; Tomilova, A.A.; Gofarov, M.Y.; Eliseeva, T.A.; Konopleva, E.S.; Vikhrev, I.V.; Yunitsyna, O.A.; Pešić, V.; Bolotov, I.N. Taxonomic reassessment of freshwater mussels from the Western Balkans reveals an overlooked but critical refugium and defines conservation priorities. Diversity 2022, 14, 935. [Google Scholar] [CrossRef]
- Gómez, A.; Lunt, D.H. Refugia within refugia: Patterns of phylogeographic concordance in the Iberian Peninsula. In Phylogeography of Southern European Refugia. Evolutionary Perspectives on the Origins and Conservation of European Biodiversity; Weiss, S., Ferrand, N., Eds.; Springer: Dordrecht, Germany, 2007; pp. 155–188. [Google Scholar] [CrossRef]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef]
- Lică, A. (Hirudinaria S.R.L., Murighiol, TL, Romania). Personal communication, 13 July 2024.
- IUCN. IUCN Red List Categories and Criteria: Version 3.1, 2nd ed.; International Union for Conservation of Nature and Natural Resources: Gland, Switzerland; Cambridge, UK, 2012. [Google Scholar]
- Surugiu, V. Hirudo verbana Carena, 1820. In The Red Book of Invertebrates from Romania; Murariu, D., Maican, S., Eds.; Romanian Academy Publishing House: Bucharest, Romania, 2021; p. 39. [Google Scholar]



| Population | N | h | Hd | π | S | Tajima’s D | Fu’s Fs | R2 Statistics |
|---|---|---|---|---|---|---|---|---|
| Dorohoi | 3 | 2 | 0.667 | 0.00081 | 1 | – | – | – |
| Buciumeni | 5 | 1 | 0.000 | 0.00000 | 0 | – | – | – |
| Bujoru | 8 | 2 | 0.429 | 0.00081 | 1 | 0.33350 (p > 0.10) | 0.536 | 0.2041 |
| Băclănești | 7 | 4 | 0.714 | 0.00162 | 3 | −1.35841 (p > 0.10) | −1.798 | 0.1650 |
| Căptălan | 3 | 2 | 0.667 | 0.00126 | 2 | – | – | – |
| Cristești-Jijia | 15 | 3 | 0.448 | 0.00220 | 4 | −0.17792 (p > 0.10) | 1.263 | 0.1429 |
| Ilia | 3 | 1 | 0.000 | 0.00000 | 0 | – | – | – |
| Ipoteşti | 6 | 2 | 0.533 | 0.00101 | 1 | 0.85057 (p > 0.10) | 0.625 | 0.2667 |
| Chiriloaia Lake | 3 | 3 | 1.000 | 0.00252 | 2 | – | – | – |
| Harapu Island | 9 | 3 | 0.417 | 0.00084 | 2 | −1.36240 (p > 0.10) | −1.081 | 0.2079 |
| Letea | 10 | 4 | 0.800 | 0.00202 | 3 | −0.02107 (p > 0.10) | −0.742 | 0.0450 |
| Murighiol | 5 | 1 | 0.000 | 0.00000 | 0 | – | – | – |
| Pietroşani | 3 | 2 | 0.667 | 0.00126 | 1 | – | – | – |
| Cârja | 7 | 3 | 0.667 | 0.00198 | 3 | −0.65405 (p > 0.10) | 0.110 | 0.2464 |
| Probota | 3 | 1 | 0.000 | 0.00000 | 0 | – | – | – |
| Sabangia | 4 | 2 | 0.500 | 0.00095 | 3 | −0.61237 (p > 0.10) | 0.172 | 0.4330 |
| Satchinez | 10 | 5 | 0.844 | 0.00227 | 3 | 0.47343 (p > 0.10) | −1.803 | 0.1944 |
| Vadu | 4 | 4 | 1.000 | 0.00315 | 3 | 0.16766 (p > 0.10) | −2.181 | 0.2003 |
| Viișoara | 3 | 3 | 1.000 | 0.00162 | 2 | – | – | – |
| Romanian H. verbana pop. | 133 | 23 | 0.757 | 0.00245 | 25 | −2.06935 (p < 0.05) | −19.652 | 0.0275 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popa, O.P.; Ștefan, A.; Baltag, E.Ș.; Stratan, A.A.; Popa, L.O.; Surugiu, V. High Genetic Diversity of Hirudo verbana Carena, 1820 (Annelida: Hirudinea: Hirudinidae) in Romania Confirms That the Balkans Are Refugia Within Refugium. Diversity 2024, 16, 726. https://doi.org/10.3390/d16120726
Popa OP, Ștefan A, Baltag EȘ, Stratan AA, Popa LO, Surugiu V. High Genetic Diversity of Hirudo verbana Carena, 1820 (Annelida: Hirudinea: Hirudinidae) in Romania Confirms That the Balkans Are Refugia Within Refugium. Diversity. 2024; 16(12):726. https://doi.org/10.3390/d16120726
Chicago/Turabian StylePopa, Oana Paula, Andrei Ștefan, Emanuel Ștefan Baltag, Ana Alexandra Stratan, Luis Ovidiu Popa, and Victor Surugiu. 2024. "High Genetic Diversity of Hirudo verbana Carena, 1820 (Annelida: Hirudinea: Hirudinidae) in Romania Confirms That the Balkans Are Refugia Within Refugium" Diversity 16, no. 12: 726. https://doi.org/10.3390/d16120726
APA StylePopa, O. P., Ștefan, A., Baltag, E. Ș., Stratan, A. A., Popa, L. O., & Surugiu, V. (2024). High Genetic Diversity of Hirudo verbana Carena, 1820 (Annelida: Hirudinea: Hirudinidae) in Romania Confirms That the Balkans Are Refugia Within Refugium. Diversity, 16(12), 726. https://doi.org/10.3390/d16120726






