Morphological and Phylogenetic Evidences Reveal Lasiodiplodia chonburiensis and L. theobromae Associated with Leaf Blight in Hevea brasiliensis in Southern Thailand
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Fungal Isolation
2.2. Pathogenicity Test
2.3. Morphology Study
2.4. Molecular Study
3. Results
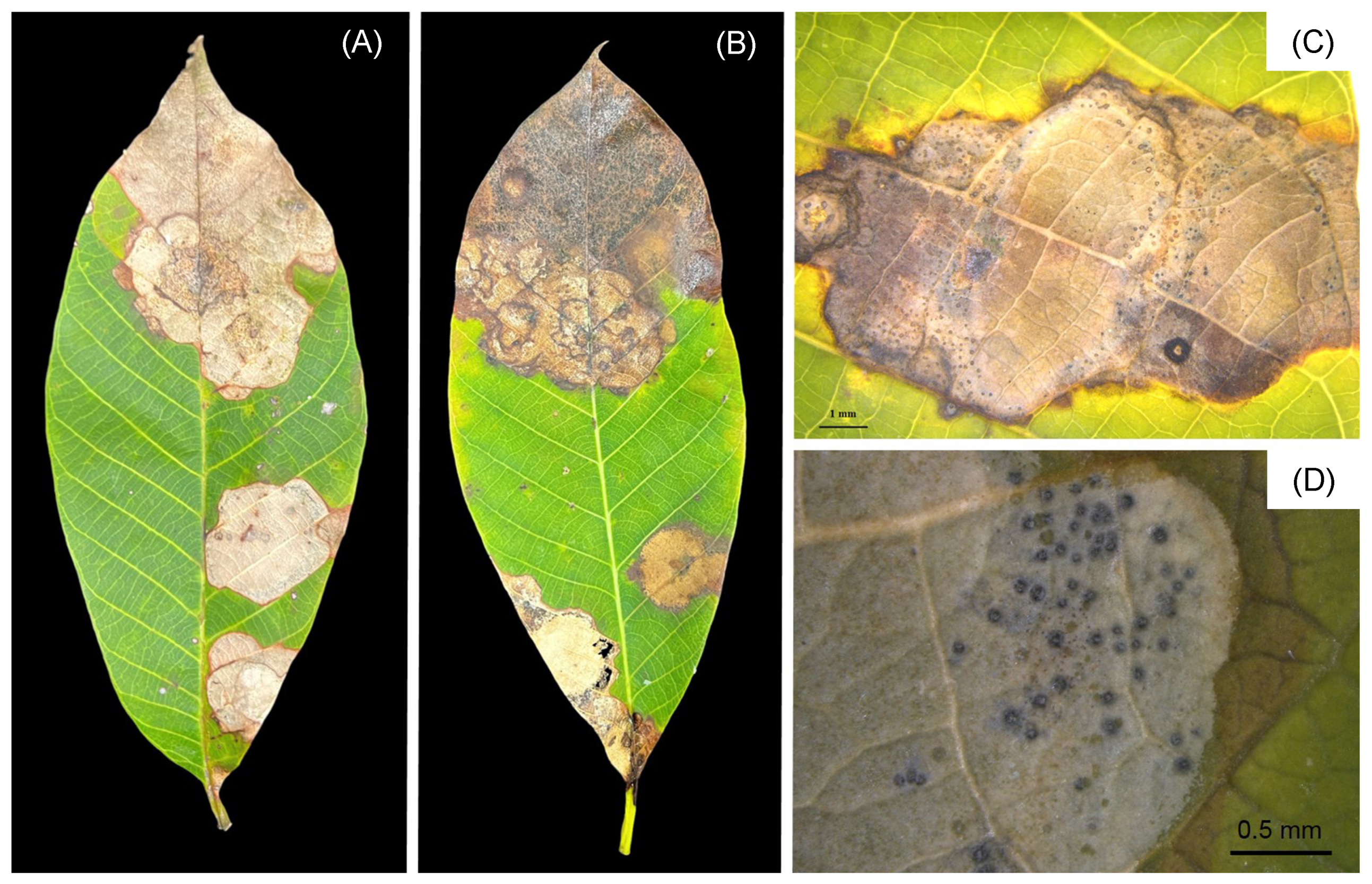
3.1. Symptom Recognition and Fungal Isolates
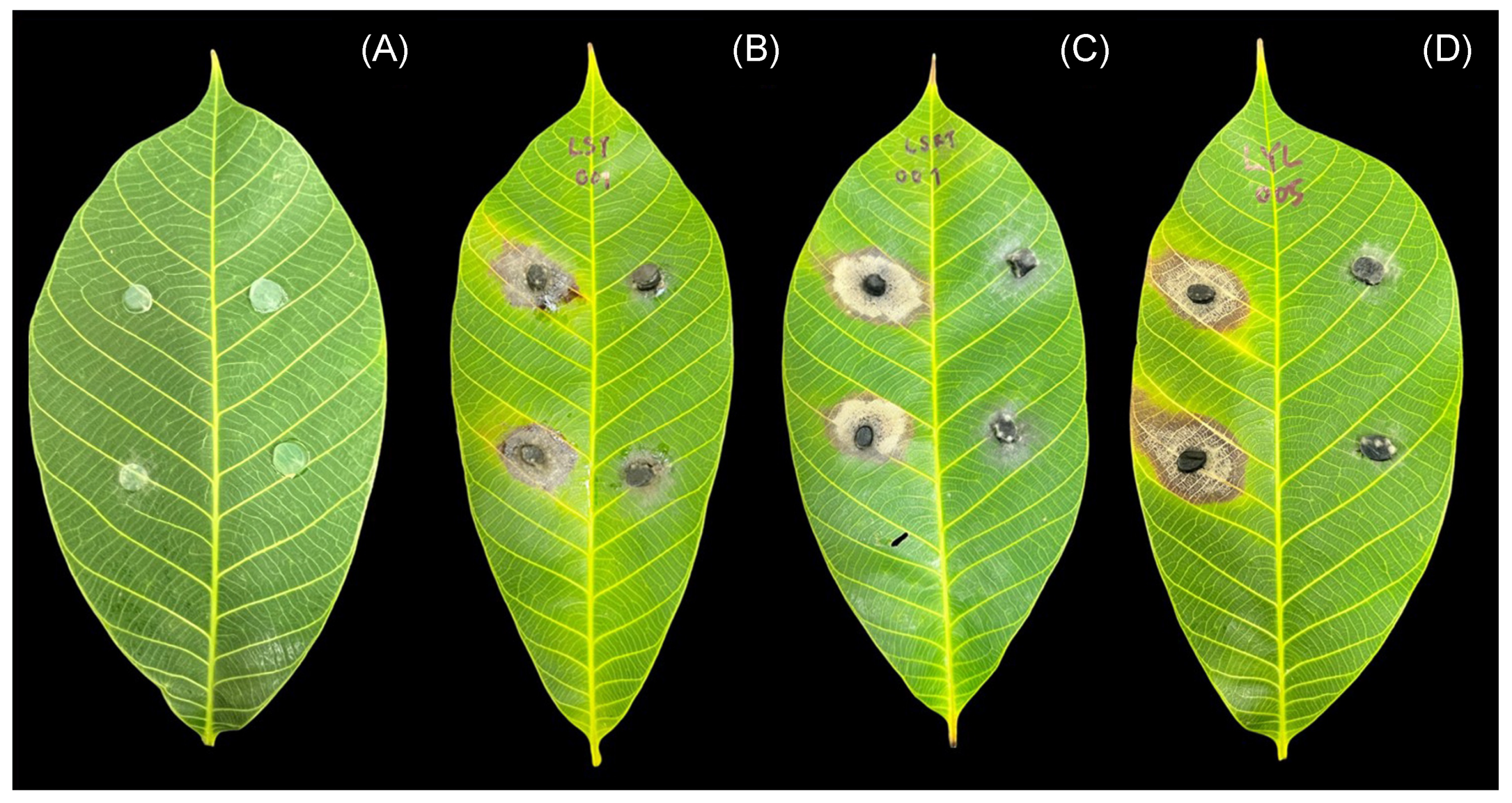
3.2. Pathogenicity of Lasiodiplodia
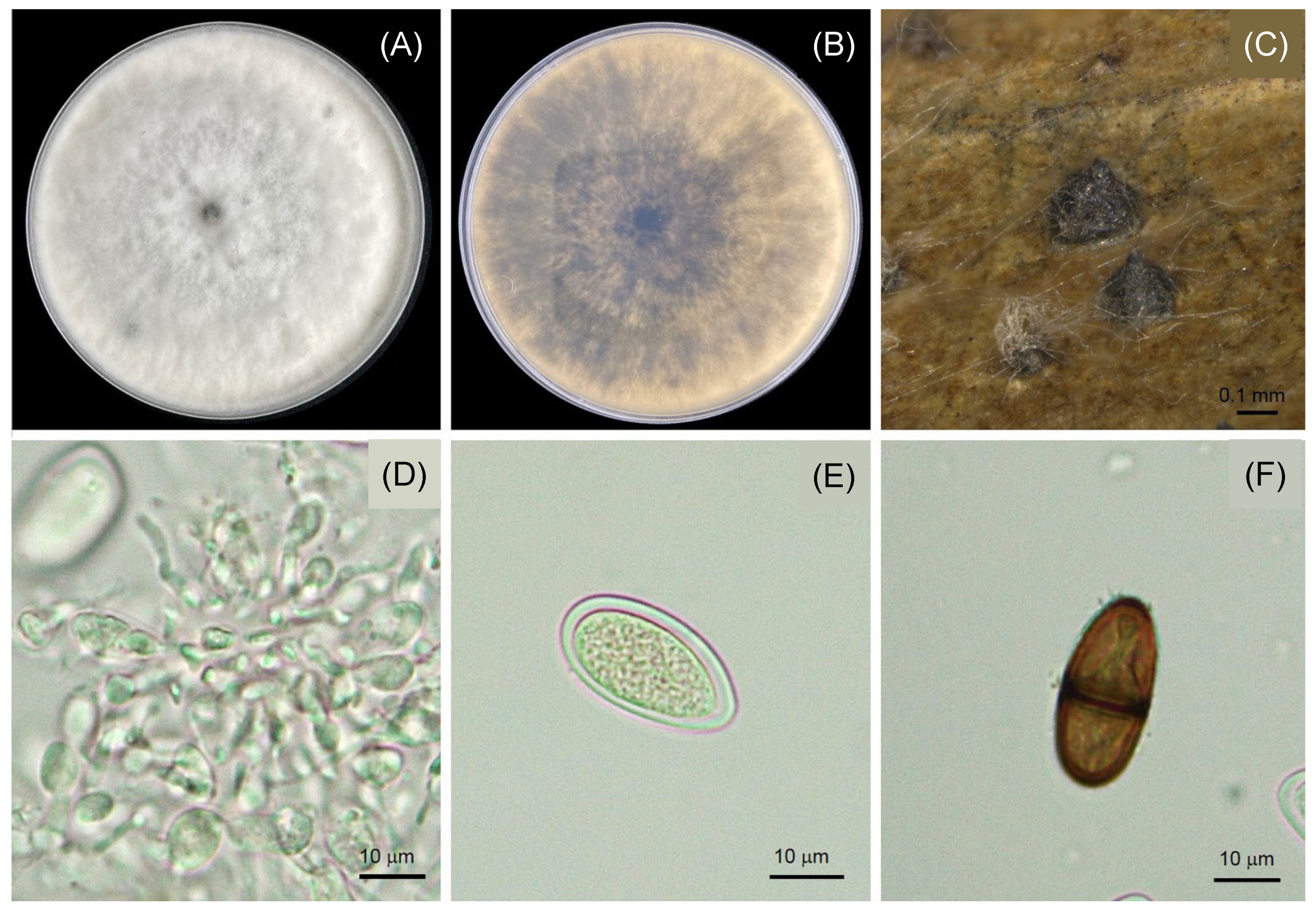
3.3. Morphological Characteristics of Lasiodiplodia
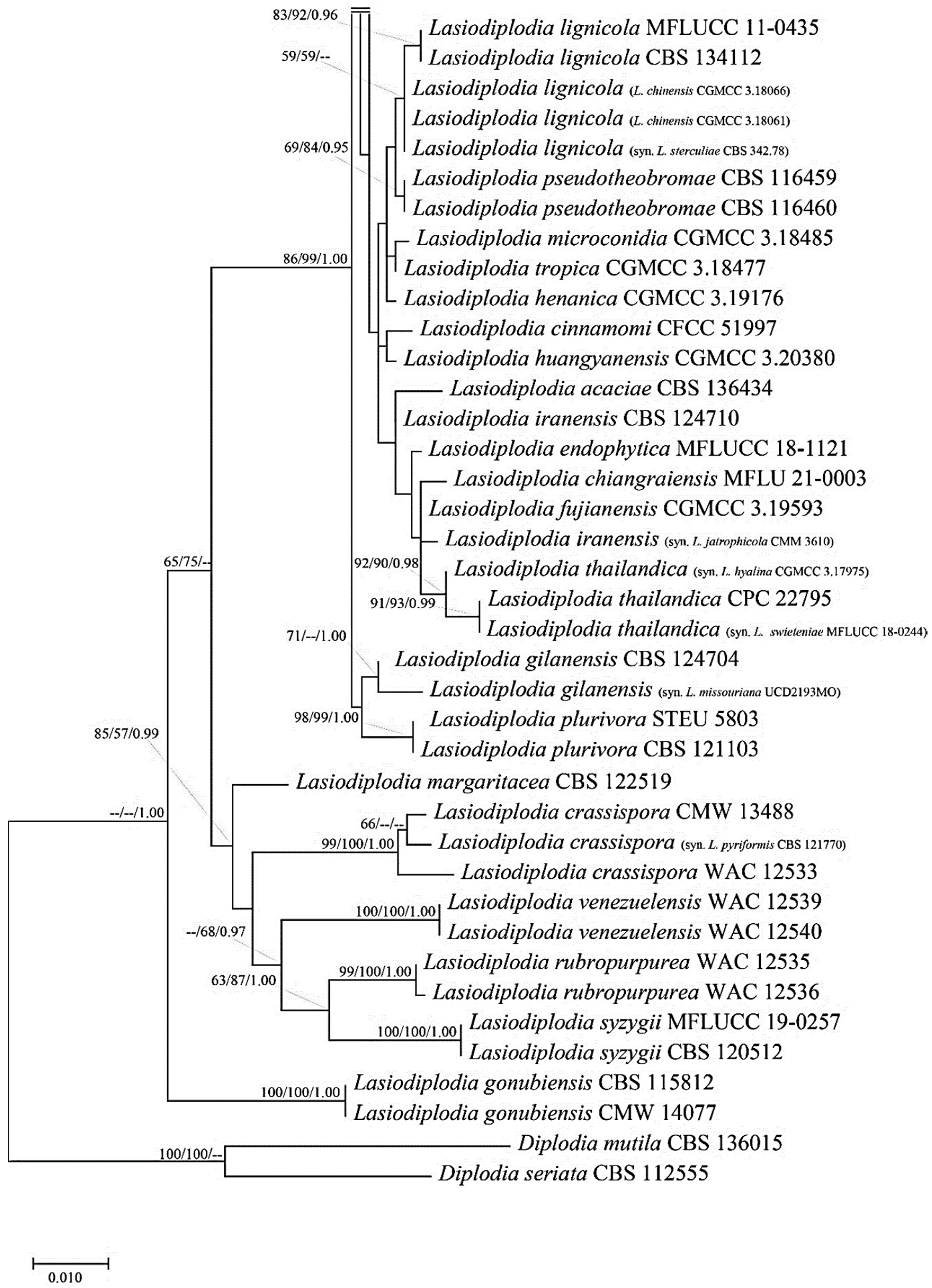
3.4. Molecular Properties of Lasiodiplodia
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nyaka Ngobisa, A.I.C.; Owona, N.P.A.; Doungous, O.; Godswill, N.N.; Njonje, S.W.; Ehabe, E.E. Characterization of Pestalotiopsis microspora, the causal agent of rubber leaf blight disease in Cameroon. Rubber Sci. 2018, 31, 112–120. [Google Scholar]
- Gazis, R.; Chaverri, P. Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea brasiliensis) in Peru. Fungal Ecol. 2010, 3, 240–254. [Google Scholar] [CrossRef]
- Fernando, S.; Silva, K.; Tennakoon, I.; Siriwardena, D.; Wijayaratne, C.; Nishantha, N. Re-Emergence of the Abnormal Leaf Fall Disease Condition in Sri Lankan Rubber; Plantations Rubber Research Institute of Sri Lanka (RRISL): Matugama, Sri Lanka, 2020.
- Pornsuriya, C.; Chairin, T.; Thaochan, N.; Sunpapao, A. Identification and characterization of Neopestalotiopsis fungi associated with a novel leaf fall disease of rubber trees (Hevea brasiliensis) in Thailand. J. Phytopathol. 2020, 168, 416–427. [Google Scholar] [CrossRef]
- Huda-Shakirah, A.R.; Mohamed Nor, N.M.I.; Zakaria, L.; Leong, Y.H.; Mohd, M.H. Lasiodiplodia theobromae as a causal pathogen of leaf blight, stem canker, and pod rot of Theobroma cacao in Malaysia. Sci. Rep. 2022, 12, 8966. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Slik, J.W.F.; Jeon, Y.S.; Tomlinson, K.W.; Yang, X.; Wang, J.; Kerfahi, D.; Porazinska, D.L.; Adams, J.M. Tropical forest conversion to rubber plantation affects soil micro- & mesofaunal community & diversity. Sci. Rep. 2019, 9, 5893. [Google Scholar] [PubMed]
- Thithuan, N.; Bunjonsiri, P.; Sunpapao, A. Morphology and behavior of gametes and zoospores from the plant-parasitic green algae, Cephaleuros (Chlorophyta, Ulvophyceae). Pac. Sci. 2019, 73, 403–410. [Google Scholar] [CrossRef]
- Jacob, J.; Gitz, V.; Gohet, E.; Kadir, A.B.; Nair, L.; Salvatore, P.; Nghia, N.A.; Sergey, B.; Michael, B.; Omar, C.P.; et al. Natural rubber contributions to adaptation to climate change. In Proceedings of the XV World Forestry Congress, Coex, Seoul, Republic of Korea, 2–6 May 2022. [Google Scholar]
- Thaochan, N.; Pornsuriya, C.; Chairin, T.; Chomnunti, P.; Sunpapao, A. Morphological and molecular characterization of Calonectria foliicola associated with leaf blight on rubber tree (Hevea brasiliensis) in Thailand. J. Fungi 2022, 8, 986. [Google Scholar] [CrossRef]
- Laohasakul, B.; Boonyapipat, P.; Plodpai, P. First Report of Phytophthora citrophthora Causing Leaf Fall of Para Rubber Tree (Hevea brasiliensis) in Thailand. Plant Dis. 2017, 101, 1057. [Google Scholar] [CrossRef]
- Nam, M.H.; Park, M.S.; Kim, H.S.; Kim, T.I.; Lee, E.M.; Park, J.D.; Kim, H.G. 2016. First report of dieback caused by Lasiodiplodia theobromae in strawberry plants in Korea. Mycobiology 2016, 44, 319–324. [Google Scholar] [CrossRef]
- Rosado, A.W.C.; Machado, A.R.; Freire, F.D.C.O.; Pereira, O.L. Phylogeny, identification, and pathogenicity of Lasiodiplodia associated with postharvest stem-end rot of coconut in Brazil. Plant Dis. 2016, 100, 561–568. [Google Scholar] [CrossRef]
- Saeed, E.E.; Sham, A.; AbuZarqa, A.; Al Shurafa, K.A.; Al Naqbi, T.S.; Iratni, R.; El-Tarabily, K.; AbuQamar, S.F. Detection and management of mango dieback disease in the United Arab Emirates. Int. J. Mol. Sci. 2017, 18, 2086. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Cruz, M.A.; Almaguer-Vargas, G.; Leyva-Mir, S.G.; Colinas-León, M.T.; Correia, K.C.; Camacho-Tapia, M.; Robles-Yerena, L.; Michereff, S.J.; Tovar-Pedraza, J.M. Phylogeny, distribution and pathogenicity of Lasiodiplodia species associated with cankers and dieback symptoms of Persian lime in Mexico. Plant Dis. 2019, 103, 1156–1165. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Li, H.; Zhou, L.; Chen, F. Lasiodiplodia pseudotheobromae causes stem canker of Chinese hackberry in China. J. For. Res. 2019, 31, 2571–2580. [Google Scholar] [CrossRef]
- Chen, J.; Zhu, Z.; Fu, Y.; Cheng, J.; Xie, J.; Lin, Y. Identification of Lasiodiplodia pseudotheobromae causing fruit rot of citrus in China. Plants 2021, 10, 202. [Google Scholar] [CrossRef]
- Pečenka, J.; Tekielska, D.A.; Kocanová, M.; Peňázová, E.; Berraf-Tebbal, A.; Eichmeier, A. First report of Lasiodiplodia theobromae causing decline of blueberry (Vaccinium corymbosum L.) in the Czech Republic. Plant Dis. 2021, 105, 215. [Google Scholar] [CrossRef]
- Gnanesh, B.N.; Arunakumar, G.S.; Tejaswi, A.; Supriya, M.; Manojkumar, H.B.; Devi, S.S. Characterization and pathogenicity of Lasiodiplodia theobromae causing black root rot and identification of novel sources of resistance in mulberry collections. Plant Pathol. J. 2022, 38, 272–286. [Google Scholar] [CrossRef]
- Chantarasiri, A.; Boontanom, P. Fusarium solani and Lasiodiplodia pseudotheobromae, fungal pathogens causing stem rot disease on durian trees (Durio zibethinus) in Eastern Thailand. New Dis. Rep. 2021, 44, 1–3. [Google Scholar] [CrossRef]
- Dianda, O.Z.; Wonni, I.; Ouédraogo, L.; Sankara, P.; Tollenaere, C.; Del Ponte, E.M.; Fernandez, D. Lasiodiplodia species associated with mango (Mangifera indica L.) decline in Burkina Faso and influence of climatic factors on the disease distribution. Physiol. Mol. Plant Pathol. 2003, 126, 102041. [Google Scholar] [CrossRef]
- Burgess, T.I.; Barber, P.A.; Mohali, S.; Pegg, G.; de Beer, W.; Wingfield, M.J. Three new Lasiodiplodia spp. from the tropics, recognized based on DNA sequence comparisons and morphology. Mycologia 2006, 98, 423–435. [Google Scholar] [CrossRef]
- Meng, C.R.; Zhang, Q.; Yang, Z.F.; Geng, K.; Zeng, X.Y.; Chethana, K.T.; Wang, Y. Lasiodiplodia syzygii sp. nov. (Botryosphaeriaceae) causing post-harvest water-soaked brown lesions on Syzygium samarangense in Chiang Rai, Thailand. Biodivers. Data J. 2021, 9, e60604. [Google Scholar] [CrossRef]
- Saitoh, K.; Togashi, K.; Arie, T.; Teraoka, T. A simple method for a mini-preparation of fungal DNA. J. Gen. Plant Pathol. 2006, 72, 348–350. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetic. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Rongquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Phillips, A.J.; Alves, A.; Abdollahzadeh, J.; Slippers, B.; Wingfield, M.J.; Groenewald, J.Z.; Crous, P.W. The Botryosphaeriaceae: Genera and species known from culture. Stud Mycol. 2013, 76, 51–167. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; McKenzie, E.H.; Bhat, D.J.; Phillips, A.J.; Wanasinghe, D.N.; Samarakoon, M.C.; Jayawardena, R.S.; Dissanayake, A.J.; Tennakoon, D.S.; et al. Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Divers. 2018, 93, 1–160. [Google Scholar] [CrossRef]
- Alves, A.; Crous, P.W.; Correia, A.; Phillips, A.J.L. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Divers. 2008, 28, 1–13. [Google Scholar]
- Munirah, M.S.; Azmi, A.R.; Yong, S.Y.C.; Nur, A.I.M.Z. Characterization of Lasiodiplodia theobromae and L. pseudotheobromae causing fruit rot on pre-harvest mango in Malaysia. Plant Pathol. Quar. 2017, 7, 202–213. [Google Scholar] [CrossRef]
- Pipattanapuckdee, A.; Boonyakait, D.; Tiyayon, C.; Seehanam, P.; Ruangwong, O.-U. Lasiodiplodia pseudotheobromae causes postharvest fruit rot of longan in Thailand. Australas. Plant Dis. Notes 2019, 14, 21. [Google Scholar] [CrossRef]
- Coutinho, I.B.L.; Freire, F.C.O.; Lima, C.S.; Lima, J.S.; Goncalves, F.J.T.; Machado, A.R.; Silva, A.M.S.; Cardoso, J.E. Diversity of genus Lasiodiplodia associated with perennial tropical fruit plants in north-eastern Brazil. Plant Pathol. 2017, 66, 90–104. [Google Scholar] [CrossRef]
- Slippers, B.; Wingfield, M.J. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: Diversity, ecology, and impact. Fungal Biol. Rev. 2007, 21, 90–106. [Google Scholar] [CrossRef]
- Mehmood, N.; Riaz, A.; Hassan, I.; Ghuffar, S.; Sattar, A.; Aurangzeb, W.; Azhar, S. First report of Lasiodiplodia theobromae as causal agent of strawberry dieback in Punjab, Pakistan. J. Plant Pathol. 2021, 103, 1355–1356. [Google Scholar] [CrossRef]
- Aguilera-Cogley, V.; Atencio, R.; Jaén, M. First report of Lasiodiplodia theobromae causing branch canker and dieback of cashew in Panama. Plant Dis. 2022, 45, e12063. [Google Scholar] [CrossRef]
- Trakunyingcharoen, T.; Cheewangkoon, R.; To-anun, C. Phylogenetic study of the Botryosphaeriaceae species associated with avocado and Pará rubber in Thailand. Chiang Mai J. Sci. 2015, 42, 104–116. [Google Scholar]
- Daengsuwan, W.; Wonglom, P.; Sunpapao, A. First report of Lasiodiplodia theobromae causing spadix rot in Anthurium andraeanum. J. Phytopathol. 2019, 168, 129–133. [Google Scholar] [CrossRef]
- Slippers, B.; Roux, J.; Wingfield, M.J.; van der Walt, F.J.; Jami, F.; Mehl, J.W.; Marais, G.J. Confronting the constraints of morphological taxonomy in the Botryosphaeriales. Persoonia 2014, 33, 155–168. [Google Scholar] [CrossRef]
- de Silva, N.I.; Phillips, A.J.L.; Liu, J.K.; Lumyong, S.; Hyde, K.D. Phylogeny and morphology of Lasiodiplodia species associated with Magnolia forest plants. Sci. Rep. 2019, 9, 14355. [Google Scholar] [CrossRef]
- Xia, G.; Manawasinghe, I.S.; Phillips, A.J.L.; You, C.; Jayawardena, R.S.; Luo, M.; Hyde, K.D. Lasiodiplodia fici sp. nov., Causing Leaf Spot on Ficus altissima in China. Pathogens 2022, 11. [Google Scholar] [CrossRef]
- Fukada, F.; Kodama, S.; Nishiuchi, T.; Kajikawa, N.; Kubo, Y. Plant pathogenic fungi Colletotrichum and Magnaporthe share a common G1 phase monitoring strategy for proper appressorium development. New Phytol. 2019, 222, 1909–1923. [Google Scholar] [CrossRef]
- de Jong, J.C.; McCormack, B.J.; Smirnoff, N.; Talbot, N.J. Glycerol generates turgor in rice blast. Nature 1997, 389, 244–245. [Google Scholar] [CrossRef]
- Perfect, S.E.; Hughes, H.B.; O’Connell, R.J.; Green, J.R. Colletotrichum—A model genus for studies on pathology and fungal–plant interactions. Fungal Genet. Biol. 1999, 27, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Sakalidis, M.L.; Ray, J.D.; Lanoiselet, V.; Hardy, G.E.S.; Burgess, T.I. Pathogenic Botryosphaeriaceae associated with Mangifera indica in the Kimberley region of Western Australia. Eur. J. Plant Pathol. 2011, 130, 379–391. [Google Scholar] [CrossRef]
- Khanzada, M.A.; Lodhi, A.M.; Shahzad, S. Mango dieback and gummosis in Sindh, Pakistan caused by Lasiodiplodia theobromae. Plant Health Prog. 2004, 15, 13. [Google Scholar] [CrossRef]
- Endes, A.; Kayim, M.; Eskalen, A. First report of Lasiodiplodia theobromae, L. pseudotheobromae, and Diplodia seriata causing bot canker and gummosis of nectarines in Turkey. Plant Dis. 2016, 100, 2321. [Google Scholar] [CrossRef]
- Mahadevakumar, S.; Joy, J.; Mamatha Bhanu, L.S.; Niranjan Raj, S.; Sharvani, K.A.; Sowmya, R.; Chandranayaka, S. First report of Lasiodiplodia theobromae associated with panicle blight of grapes (Vitis vinifera L.) in India. Plant Dis. 2022, 107, 225. [Google Scholar] [CrossRef]
- Pinizzotto, S.; Kadir, A.A.S.A.; Gitz, V.; Sainte-Beuve, J.; Naire, L.; Gohet, E.; Penot, E.; Meybeck, A. Natural Rubber and Climate Change: A Policy Paper; FTA Brief; CIFOR: Bogor, Indonesia, 2021; Volume 6, pp. 1–24. [Google Scholar]
- Ismail, T.; Gohet, E. Impact of climate change on latex harvesting Tajuddin. Nat. Rubber Syst. Clim. Change (FTA Work. Rep.) 2020, 150, 01008. [Google Scholar]


| Species | Conidia | Paraphyses | Sources | |
|---|---|---|---|---|
| Size | Septation | |||
| Lasiodiplodia chonburiensis | 15.0–30.0 × 10.0–15.0 | - | - | [30] |
| 19.4–25.2 × 10.3–13.4 | 1 | Septate | This study (LSrt001) | |
| 20.7–25.4 × 10.5–13.6 | 1 | Septate | This study (LSrt002) | |
| 17.7–24.9 × 11.2–12.4 | 1 | Septate | This study (LST001) | |
| 18.2–24.6 × 10.0–11.4 | 1 | Septate | This study (LST002) | |
| 19.3–26.9 × 11.1–14.6 | 1 | Septate | This study (LST003) | |
| L. pseudotheobromae | 25.5–30.5 × 14.8–17.2 | 1 | Septate | [31] |
| 25.5–27.3 × 12.7–14.6 | 1 | Septate | [32] | |
| 23.7–28.2 × 12.4–14.9 | 1 | Septate | [33] | |
| L. theobromae | 26.2–27.0 × 14.0–14.4 | 1 | Septate | [31] |
| 19.7–26.7 × 10.9–15.3 | 1 | Septate | [34] | |
| 23.0–27.4 × 12.2–14.7 | 1 | Septate | This study (LYL005) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pornsuriya, C.; Thaochan, N.; Chairin, T.; Sunpapao, A. Morphological and Phylogenetic Evidences Reveal Lasiodiplodia chonburiensis and L. theobromae Associated with Leaf Blight in Hevea brasiliensis in Southern Thailand. Diversity 2023, 15, 961. https://doi.org/10.3390/d15090961
Pornsuriya C, Thaochan N, Chairin T, Sunpapao A. Morphological and Phylogenetic Evidences Reveal Lasiodiplodia chonburiensis and L. theobromae Associated with Leaf Blight in Hevea brasiliensis in Southern Thailand. Diversity. 2023; 15(9):961. https://doi.org/10.3390/d15090961
Chicago/Turabian StylePornsuriya, Chaninun, Narit Thaochan, Thanunchanok Chairin, and Anurag Sunpapao. 2023. "Morphological and Phylogenetic Evidences Reveal Lasiodiplodia chonburiensis and L. theobromae Associated with Leaf Blight in Hevea brasiliensis in Southern Thailand" Diversity 15, no. 9: 961. https://doi.org/10.3390/d15090961
APA StylePornsuriya, C., Thaochan, N., Chairin, T., & Sunpapao, A. (2023). Morphological and Phylogenetic Evidences Reveal Lasiodiplodia chonburiensis and L. theobromae Associated with Leaf Blight in Hevea brasiliensis in Southern Thailand. Diversity, 15(9), 961. https://doi.org/10.3390/d15090961








