Karyotypes of 10 Anuran Species from the Qinghai–Tibetan Plateau
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. Chromosome Preparation and Measurement
3. Results
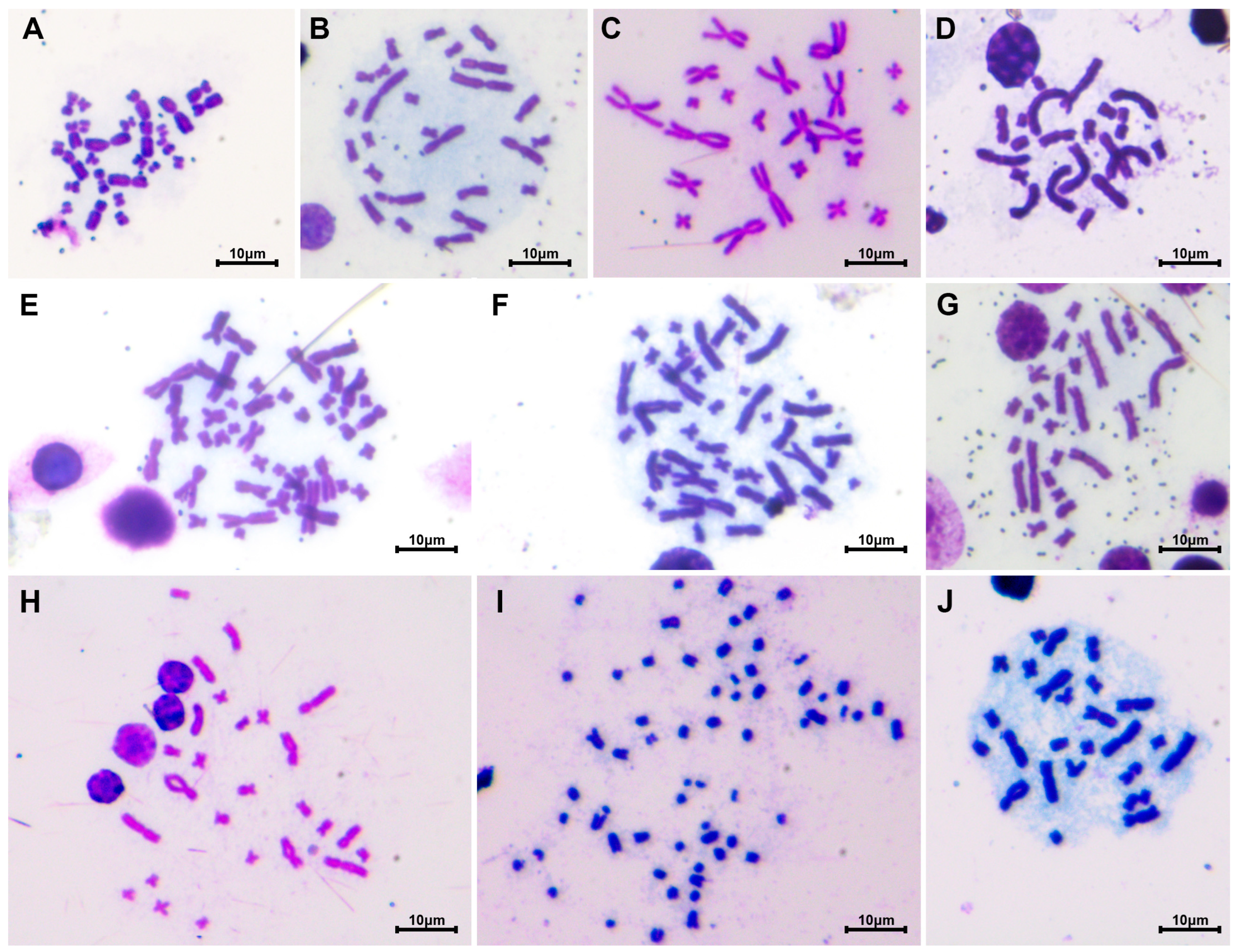
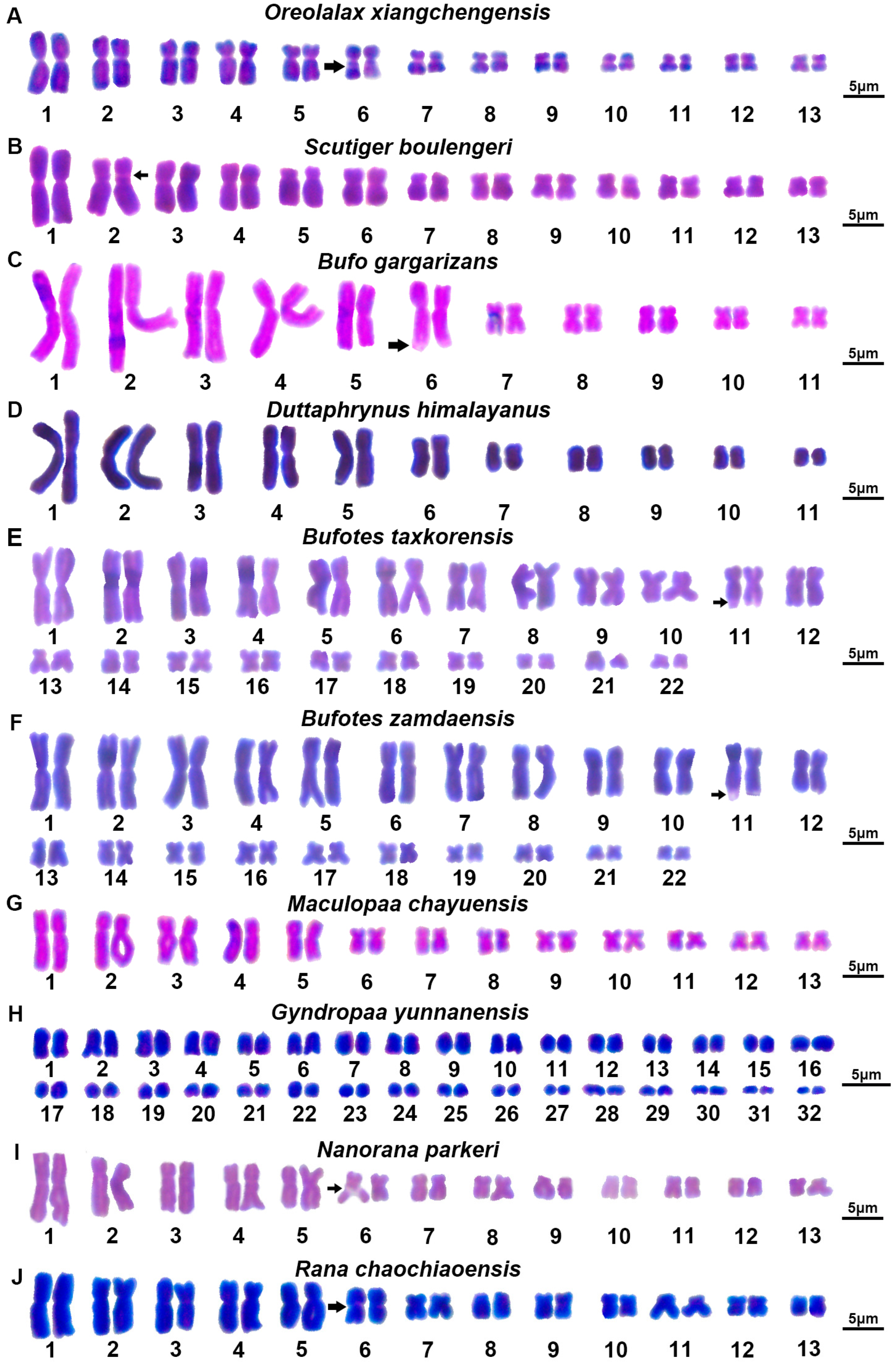
3.1. Karyotype of Megophryidae
3.2. Karyotype of Bufonidae
3.3. Karyotype of Dicroglossidae
3.4. Karyotype of Ranidae
4. Discussion
4.1. Karyotype Comparison
4.1.1. Karyotype of Megophryidae
4.1.2. Karyotype of Bufonidae
4.1.3. Karyotype of Dicroglossidae
4.1.4. Karyotype of Ranidae
4.2. Karyotypic Differences and Phylogenetic Differentiation
| Species | Chr. Number | Karyotype Composition | SM | ST | T | SC | Location | Ref. |
|---|---|---|---|---|---|---|---|---|
| Oreolalax xiangchengensis | 26 | 24M + 2SM | 3 | 6qinter | Weixi Lisu Autonomous County, Yunnan, China | -- | ||
| 26 | 20M + 6SM | 3, 4, 5 | 6qper | Zhongdian County, Yunnan Province, China | [25] | |||
| Scutiger boulengeri | 26 | 20M + 6SM | 5, 7, 8 | 2pinter | Dingjie County, Xizang, China | -- | ||
| 26 | 20M + 6SM | 5, 7, 8 | 2pinter | Dingqing, County, Xizang, China | -- | |||
| 26 | 20M + 6SM | 5, 7, 8 | 2pinter | Zhongba County, Xizang, China | -- | |||
| 26 | 22M + 2SM + 2ST | 7 | 5 | 2pinter | Kangding City, Sichuan Province, China | [26] | ||
| Bufo gargarizans | 22 | 22M | 6qinter | Lijiang Naxi autonomous county, China | -- | |||
| 22 | 18M + 4SM | 4, 9 | 6qinter | Heilongjiang Province, China | [22] | |||
| 22 | 18M + 4SM | 4, 9 | 6qinter | Beijing City, China | [22] | |||
| 22 | 18M + 4SM | 4, 9 | 6qinter | Shanghai City, China | [22] | |||
| 22 | 18M + 4SM | 4, 9 | 6qinter | Sichuan Province, China | [22] | |||
| 22 | 18M + 4SM | 4, 9 | 6qinter | Fujian Province, China | [22] | |||
| Duttaphrynus himalayanus | 22 | 22M | / | Dingjie County, Xizang, China | -- | |||
| Bufotes taxkorensi | 44 | 38M + 2ST + 4SM | 9, 13 | 21 | 11qter | Taxkorgan Tajik Autonomous County, Xinjiang, China | -- | |
| Bufotes zamdaensis | 44 | 36M + 8SM | 7, 8, 13, 14 | 11qter | Zanda County, Xizang, China | -- | ||
| 33 | / | / | / | / | / | Spiti River, India | [40] | |
| Bufotes pewzowi | 44 | 36M + 8SM | 7, 8, 13, 14 | 12qter | Hotan Prefecture, Xizang Province, China | [29] | ||
| Maculopaa chayuensi | 26 | 16M + 10SM | 2–4, 6, 13 | 6pper | Derung-Nu Autonomous County, Yunnan Province, China | -- | ||
| 26 | 16M + 10SM | 2–4, 6, 8 | 6pper | Lushui County, Yunnan Province, China | [30] | |||
| Gynandropaa yunnanensis | 64 | 64T | 1–32 | / | Binchuan County, Yunnan, China | -- | ||
| 64 | 64T | 1–32 | 4qper | Jingdong County, Yunnan Province, China | [31] | |||
| 64 | 64T | 1–32 | 2qinter | Jinping County, Yunnan Province, China | [22] | |||
| 64 | 64T | 1–32 | 15qinter | Tengchong City, Yunnan Province, China | [30] | |||
| Gynandropaa phrynoides | 64 | 64T | 1–32 | 18qinter | Yimen County, Yunnan Province, China | [22] | ||
| 64 | 64T | 1–32 | 20qinter | Qujing County, Yunnan Province, China | [32] | |||
| Gynandropaa sichuanensis | 64 | 64T | 32qter | Zhaojue County, Sichuan Province, China | [33] | |||
| Nanorana parkeri | 26 | 18M + 8SM | 2, 3, 6, 9 | 6qter | Dingjie and Nanmulin Counties, Xizang Province, China | -- | ||
| 26 | 16M + 10SM | 2–4, 6, 8, 9 | 6qter | Lasa City, Xizang Province, China | [26] | |||
| Rana chaochiaoensis | 26 | 16M + 8SM + 2ST | 2, 4, 11, 13 | 8 | 6qper | Weixi Lisu Autonomous County, Yunnan, China | -- | |
| 26 | 18M + 6SM + 2ST | 3, 9, 13 | 8 | 6qper | Kunming City, Yunnan Province, China | [34] | ||
| 26 | 16M + 8SM + 2ST | 2, 3, 9, 13 | 8 | 6qper | Zhongdian County, Yunnan Province, China | [22] | ||
| 26 | 14M + 10SM + 2ST | 2, 3, 7, 9, 13 | 8 | 6qper | Yanyuan City, Sichuan Province, China | [35] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, D.; Yang, Q.Y.; Wu, S.H. Physical Geography Pandect in China; Scinece Press: Beijing, China, 2015. [Google Scholar]
- Favre, A.; Päckert, M.; Pauls, S.U.; Jähnig, S.C.; Uhl, D.; Michalak, I.; Muellner-Riehl, A.N. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. 2015, 90, 236–253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.Z.; Deng, D.; Yang, Y.Q.; Liu, Y.H. Physical Geography of Hengduan Mountains; Science Press: Beijing, China, 1997. [Google Scholar]
- Fei, L.; Ye, C.Y.; Huang, Y.Z.; Liu, Y.M. Atlas of Amphibians of China; Henan Science and Technology Press: Zhengzhou, China, 1999. [Google Scholar]
- Zhao, E.M.; Yang, D. Amphibians and Reptiles of the Hengduan Mountains Region; Science Press: Beijing, China, 1997. [Google Scholar]
- Fei, L.; Ye, C.Y.; Jiang, J.P. Colored Atlas of Chinese Amphibians and Their Distributions; Sichuan Publishing House of Science and Technology: Chengdu, China, 2012. [Google Scholar]
- Frost, D.R. Amphibian Species of the World: An Online Reference. Version 6.1. Available online: https://amphibiansoftheworld.amnh.org/index.php (accessed on 7 March 2023).
- Hofmann, S.; Baniya, C.B.; Litvinchuk, S.N.; Miehe, G.; Li, J.T.; Schmidt, J. Phylogeny of spiny frogs Nanorana (Anura: Dicroglossidae) supports a Tibetan origin of a Himalayan species group. Ecol. Evol. 2019, 9, 14498–14511. [Google Scholar] [CrossRef]
- Zhou, W.W.; Yan, F.; Fu, J.Z.; Wu, S.F.; Murphy, R.W.; Che, J.; Zhang, Y.P. River islands, refugia and genetic structuring in the endemic brown frog Rana kukunoris (Anura, Ranidae) of the Qinghai-Tibetan Plateau. Mol. Ecol. 2013, 22, 130–142. [Google Scholar] [CrossRef]
- Hu, J.H.; Huang, Y.; Jiang, J.P.; Guisan, A.; Marske, K. Genetic diversity in frogs linked to past and future climate changes on the roof of the world. J. Anim. Ecol. 2019, 88, 953–963. [Google Scholar] [CrossRef]
- Hu, J.H.; Broennimann, O.; Guisan, A.; Wang, B.; Huang, Y.; Jiang, J.P. Niche conservatism in Gynandropaa frogs on the southeastern Qinghai-Tibetan Plateau. Sci. Rep. 2016, 6, 32624. [Google Scholar] [CrossRef]
- Fu, T.T.; Sun, Y.B.; Gao, W.; Long, C.B.; Yang, C.H.; Yang, X.W.; Zhang, Y.; Lan, X.Q.; Huang, S.; Jin, J.Q.; et al. The highest-elevation frog provides insights into mechanisms and evolution of defenses against high UV radiation. Proc. Natl. Acad. Sci. USA 2022, 119, e2212406119. [Google Scholar] [CrossRef]
- Xu, L.L.; Chen, H.; Zhang, M.J.; Zhu, W.; Chang, Q.; Lu, G.Q.; Chen, Y.H.; Jiang, J.P.; Zhu, L.F. Changes in the community structure of the symbiotic microbes of wild amphibians from the eastern edge of the Tibetan Plateau. Microbiologyopen 2020, 9, e1004. [Google Scholar] [CrossRef]
- Perkins, R.D.; Gamboa, J.R.; Jonika, M.M.; Lo, J.; Shum, A.; Adams, R.H.; Blackmon, H. A database of amphibian karyotypes. Chromosome Res. 2019, 27, 313–319. [Google Scholar] [CrossRef]
- Wellenreuther, M.; Bernatchez, L. Eco-Evolutionary Genomics of Chromosomal Inversions. Trends Ecol. Evol. 2018, 33, 427–440. [Google Scholar] [CrossRef]
- Noor, M.A.F.; Grams, K.L.; Bertucci, L.A.; Reiland, J. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 2001, 98, 12084–12088. [Google Scholar] [CrossRef]
- Tuttle, E.M.; Bergland, A.O.; Korody, M.L.; Brewer, M.S.; Newhouse, D.J.; Minx, P.; Stager, M.; Betuel, A.; Cheviron, Z.A.; Warren, W.C.; et al. Divergence and functional degradation of a sex chromosome-like supergene. Curr. Biol. 2016, 26, 344–350. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Ashman, T.L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26. [Google Scholar] [CrossRef]
- Yuan, X.Y.; Xia, Y.; Zeng, X.M. Suppressed recombination of sex chromosomes is not caused by chromosomal reciprocal translocation in spiny frog (Quasipaa boulengeri). Front. Genet. 2018, 9, 288. [Google Scholar] [CrossRef]
- Flemming, W. Zellsubstanz, Kern und Zelltheilung; FCW Vogel: Leipzig, Germany, 1882. [Google Scholar]
- Mohlhenrich, E.R.; Mueller, R.L. Genetic drift and mutational hazard in the evolution of salamander genomic gigantism. Evolution 2016, 70, 2865–2878. [Google Scholar] [CrossRef]
- Li, S.S. Cytotaxonomy of Amphibian in China; Science Press: Beijing, China, 2007. [Google Scholar]
- Schmid, M. Chromosome banding in Amphibia I. Constitutive heterochromatin and nucleolus organizer regions in Bufo and Hyla. Chromosoma 1978, 66, 361–388. [Google Scholar] [CrossRef]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Li, S.S. Cytogenetic study on three oreolalax pelobatoides from Yunnan. Acta Zool. Sin. 1991, 37, 216–223. [Google Scholar]
- Wu, G.F. Karyotypes of Scutiger boulengeri (Pelobatidae) of Sichuan and Altirana parkeri (Ranidae) of Xizang. Acta Herpetol. Sin. 1984, 3, 33–36. [Google Scholar]
- Stock, M.; Steinlein, C.; Lamatsch, D.K.; Schartl, M.; Schmid, M. Multiple origins of tetraploid taxa in the Eurasian Bufo viridis subgroup. Genetica 2005, 124, 255–272. [Google Scholar] [CrossRef]
- Dufresnes, C.; Mazepa, G.; Jablonski, D.; Oliveira, R.C.; Wenseleers, T.; Shabanov, D.A.; Auer, M.; Ernst, R.; Koch, C.; Ramirez-Chaves, H.E.; et al. Fifteen shades of green: The evolution of Bufotes toads revisited. Mol. Phylogenetics Evol. 2019, 141, 106615. [Google Scholar] [CrossRef]
- Wu, M.; Zhao, Y.J. A preliminary study of the karyotype of Bufo viridis laurenti in Xinjang. Zool. Res. 1987, 8, 339–343. [Google Scholar]
- Li, S.S.; Hu, J.S. The study on the karyotypes, C-banding and Ag-NORs of four paa species in China. Zool. Res. 1996, 17, 84–88. [Google Scholar]
- Li, S.S. On the karyotypes and Ag-NORs of three sympatrically paa frogs in Yunnan Province. Acta Zool. Sin. 1994, 40, 317–323. [Google Scholar]
- Liu, W.G.; Jiu, R.G. A special karyotype in the genus Rana—An investigation of the karyotype, C-banding and Ag-stained NORs of Rana phrynoides Boulenger. Acta Herpetol. Sin. 1984, 11, 61–64. [Google Scholar]
- Wu, G.F.; Zhao, E.M. A rare karyotype of anurans, the karyotype of Rana Phrynoides. Acta Herpetol. Sin. 1984, 3, 29–32. [Google Scholar]
- Li, S.S.; Wang, Y.X.; Li, C.Y.; Wang, R.F.; Liu, G.Z. A comparative investigation of the karyotypes from four Amphibian species. Zool. Res. 1981, 2, 17–24. [Google Scholar]
- Zeng, X.M.; Ye, C.Y.; Fei, L.; Jiang, J.P.; Xie, F. The karyotype and NORs investigations of four brown frogs. Zool. Res. 1998, 19, 412–414. [Google Scholar]
- Yang, D.T. From Water to Land; Chinese Forestry Press: Beijing, China, 1983. [Google Scholar]
- Yang, D.T.; Rao, D.Q. Amphibia and Reptilia of Yunnan; Yunnan Science and Technology Press: Kunming, China, 2008. [Google Scholar]
- Hou, Y.M.; Shi, S.C.; Hu, D.M.; Deng, Y.; Jiang, J.P.; Xie, F.; Wang, B. A new species of the toothed toad Oreolalax (Anura, Megophryidae) from Sichuan Province, China. Zookeys 2020, 929, 93–115. [Google Scholar] [CrossRef]
- Fei, L.; Ye, C.Y.; Huang, Y.Z.; Chen, X.N. Taxonomic studies on Bufo viridis from west China. Zool. Res. 1999, 20, 294–300. [Google Scholar]
- Litivinchuk, S.N.; Borkin, L.J.; Skorinov, D.V.; Mazepa, G.A.; Pasynkova, R.A.; Dedukh, D.A. Unusual trripoid speciation in green toads of Bufo viridis complex in high Asia. In Proceedings of the Problems of Herpetology, Minsk, Republic of Belarus, 24–27 September 2012. [Google Scholar]
| Species | Locality | Longitude (°E) | Latitude (°N) | Gender | Voucher Number |
|---|---|---|---|---|---|
| Oreolalax xiangchengensis | Weixi Lisu Autonomous County, Yunnan, China | 98.94837582 | 27.68311651 | ♀ | CIB5334220116 |
| Oreolalax xiangchengensis | Weixi Lisu Autonomous County, Yunnan, China | 98.94837582 | 27.68311651 | ♀ | CIB5334220118 |
| Scutiger boulengeri | Dingjie County, Xizang, China | 87.081458 | 28.592544 | ♀ | CIBXJ2021121 |
| Scutiger boulengeri | Dingqing County, Xizang, China | 95.416961 | 31.053237 | ♂ | CIBXJ2021124 |
| Scutiger boulengeri | Dingqing County, Xizang, China | 95.416961 | 31.053237 | ♂ | CIBXJ2021125 |
| Scutiger boulengeri | Zhongba County, Xizang, China | 84.047854 | 29.775521 | ♂ | CIBXJ2021133 |
| Scutiger boulengeri | Zhongba County, Xizang, China | 84.047854 | 29.775521 | ♀ | CIBXJ2021134 |
| Bufo gargarizans | Lijiang Naxi autonomous county | 99.712708 | 27.06504 | ♂ | CIByN201909214 |
| Duttaphrynus himalayanus | Dingjie County, Xizang, China | 87.081458 | 28.592544 | ♀ | CIBXJ2021130 |
| Duttaphrynus himalayanus | Dingjie County, Xizang, China | 87.081458 | 28.592544 | ♂ | CIBXJ2021131 |
| Duttaphrynus himalayanus | Dingjie County, Xizang, China | 87.081458 | 28.592544 | ♂ | CIBXJ2021132 |
| Bufotes taxkorensi | Taxkorgan Tajik Autonomous County, Xinjiang, China | 75.21498056 | 37.83938056 | ♂ | CIBXJ2021119 |
| Bufotes taxkorensi | Taxkorgan Tajik Autonomous County, Xinjiang, China | 75.21498056 | 37.83938056 | ♂ | CIBXJ2021126 |
| Bufotes taxkorensi | Taxkorgan Tajik Autonomous County, Xinjiang, China | 75.21498056 | 37.83938056 | ♂ | CIBXJ2021127 |
| Bufotes zamdaensis | Zanda County, Xizang, China | 79.984408 | 31.534458 | ♂ | CIBXJ2021120 |
| Bufotes zamdaensis | Zanda County, Xizang, China | 79.984408 | 31.534458 | ♂ | CIBXJ2021135 |
| Bufotes zamdaensis | Zanda County, Xizang, China | 79.984408 | 31.534458 | ♂ | CIBXJ2021136 |
| Maculopaa chayuensis | Derung-Nu Autonomous County, Yunnan, China | 98.59665692 | 27.76807508 | ♂ | CIByN201909280 |
| Maculopaa chayuensis | Derung-Nu Autonomous County, Yunnan, China | 98.59665692 | 27.76807508 | ♀ | CIByN201909282 |
| Gynandropaa yunnanensis | Binchuan County, Yunnan, China | 100.331225 | 25.91264 | ♂ | CIB5334220131 |
| Nanorana parkeri | Dingjie County, Xizang, China | 87.081458 | 28.592544 | ♀ | CIBXJ2021122 |
| Nanorana parkeri | Dingjie County, Xizang, China | 87.081458 | 28.592544 | ♂ | CIBXJ2021123 |
| Nanorana parkeri | Nanmulin County, Xizang, China | 89.1059 | 29.34904722 | ♂ | CIBXJ2021128 |
| Rana chaochiaoensis | Weixi Lisu Autonomous County, Yunnan, China | 99.42701547 | 27.58048148 | ♂ | CIB5334220132 |
| Rana chaochiaoensis | Weixi Lisu Autonomous County, Yunnan, China | 99.42701547 | 27.58048148 | ♀ | CIB5334220104 |
| Chr. | Index (Mean ± SD) | Oreolalax xiangchengensis | Scutiger boulenger | Bufo gargarizans | Duttaphrynus himalayanus | Bufotes taxkorensi | Bufotes zamdaensis | Maculopaa chayuensi | Gynandropaa yunnanensis | Nanorana parkeri | Rana chaochiaoensis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | AR | 1.15 ± 0.04 | 1.23 ± 0.06 | 1.22 ± 0.06 | 1.29 ± 0.07 | 1.18 ± 0.07 | 1.19 ± 0.05 | 1.45 ± 0.05 | -- | 1.20 ± 0.05 | 1.16 ± 0.06 |
| CI | 46.48 ± 0.84 | 44.79 ± 0.90 | 45.08 ± 0.87 | 43.58 ± 0.87 | 45.79 ± 1.02 | 45.57 ± 0.95 | 40.88 ± 0.85 | -- | 45.47 ± 0.84 | 46.34 ± 1.02 | |
| RL | 15.00 ± 0.59 | 15.92 ± 0.62 | 16.06 ± 0.57 | 17.44 ± 0.76 | 8.23 ± 0.34 | 8.77 ± 0.33 | 14.09 ± 0.54 | 5.57 ± 0.23 | 15.27 ± 0.57 | 13.55 ± 0.53 | |
| LC | M | M | M | M | M | M | M | T | M | M | |
| 2 | AR | 1.49 ± 0.07 | 1.36 ± 0.06 | 1.26 ± 0.05 | 1.06 ± 0.05 | 1.16 ± 0.05 | 1.26 ± 0.05 | 1.99 ± 0.09 | -- | 2.32 ± 0.10 | 1.95 ± 0.09 |
| CI | 40.21 ± 0.78 | 42.45 ± 0.84 | 44.23 ± 0.82 | 48.55 ± 0.97 | 46.31 ± 1.01 | 44.21 ± 0.88 | 33.44 ± 0.67 | -- | 30.09 ± 0.59 | 33.95 ± 0.70 | |
| RL | 12.28 ± 0.48 | 11.92 ± 0.47 | 15.86 ± 0.68 | 14.47 ± 0.55 | 7.84 ± 0.30 | 8.61 ± 0.33 | 12.86 ± 0.50 | 4.99 ± 0.21 | 12.21 ± 0.47 | 11.72 ± 0.47 | |
| LC | M | M | M | M | M | M | SM | T | SM | SM | |
| 3 | AR | 1.71 ± 0.06 | 1.38 ± 0.05 | 1.52 ± 0.06 | 1.48 ± 0.07 | 1.11 ± 0.06 | 1.20 ± 0.06 | 1.81 ± 0.07 | -- | 2.35 ± 0.11 | 1.36 ± 0.05 |
| CI | 36.85 ± 0.70 | 42.10 ± 0.83 | 39.75 ± 0.74 | 40.32 ± 0.79 | 47.29 ± 1.00 | 45.55 ± 0.92 | 35.59 ± 0.68 | -- | 29.83 ± 0.55 | 42.37 ± 0.85 | |
| RL | 10.71 ± 0.40 | 10.39 ± 0.38 | 14.04 ± 0.57 | 13.59 ± 0.54 | 7.77 ± 0.31 | 7.41 ± 0.29 | 10.91 ± 0.44 | 4.98 ± 0.19 | 11.37 ± 0.47 | 10.93 ± 0.42 | |
| LC | SM | M | M | M | M | M | SM | T | SM | M | |
| 4 | AR | 1.61 ± 0.08 | 1.29 ± 0.07 | 1.63 ± 0.10 | 1.52 ± 0.07 | 1.16 ± 0.07 | 1.07 ± 0.04 | 1.97 ± 0.09 | -- | 1.50 ± 0.10 | 1.73 ± 0.07 |
| CI | 38.35 ± 0.70 | 43.68 ± 0.83 | 37.98 ± 0.78 | 39.71 ± 0.77 | 46.32 ± 0.85 | 48.20 ± 0.94 | 33.65 ± 0.69 | -- | 40.03 ± 0.81 | 36.63 ± 0.69 | |
| RL | 10.59 ± 0.42 | 9.76 ± 0.37 | 11.98 ± 0.48 | 11.92 ± 0.48 | 7.21 ± 0.29 | 7.15 ± 0.28 | 10.31 ± 0.40 | 4.40 ± 0.17 | 10.32 ± 0.40 | 10.73 ± 0.46 | |
| LC | M | M | M | M | M | M | SM | T | M | SM | |
| 5 | AR | 1.59 ± 0.08 | 2.37 ± 0.13 | 1.07 ± 0.06 | 1.28 ± 0.07 | 1.49 ± 0.08 | 1.30 ± 0.07 | 1.33 ± 0.06 | -- | 1.35 ± 0.07 | 1.53 ± 0.09 |
| CI | 38.56 ± 0.78 | 29.70 ± 0.60 | 48.39 ± 0.97 | 43.88 ± 0.86 | 40.14 ± 0.90 | 43.41 ± 0.87 | 42.85 ± 0.87 | -- | 42.56 ± 0.83 | 39.52 ± 0.80 | |
| RL | 8.84 ± 0.34 | 8.08 ± 0.34 | 10.18 ± 0.42 | 11.15 ± 0.46 | 7.21 ± 0.30 | 6.85 ± 0.27 | 9.39 ± 0.37 | 4.14 ± 0.17 | 9.69 ± 0.38 | 10.12 ± 0.39 | |
| LC | M | SM | M | M | M | M | M | T | M | M | |
| 6 | AR | 1.30 ± 0.06 | 1.29 ± 0.07 | 1.54 ± 0.06 | 1.43 ± 0.08 | 1.56 ± 0.05 | 1.33 ± 0.07 | 1.91 ± 0.10 | -- | 1.76 ± 0.09 | 1.21 ± 0.05 |
| CI | 43.54 ± 0.89 | 43.76 ± 0.75 | 39.39 ± 0.82 | 41.23 ± 0.80 | 39.11 ± 0.75 | 42.84 ± 0.85 | 34.38 ± 0.69 | -- | 36.25 ± 0.75 | 45.22 ± 0.98 | |
| RL | 8.60 ± 0.35 | 7.98 ± 0.30 | 10.10 ± 0.43 | 8.04 ± 0.31 | 6.76 ± 0.25 | 6.73 ± 0.26 | 6.11 ± 0.24 | 3.93 ± 0.16 | 6.47 ± 0.28 | 7.66 ± 0.32 | |
| LC | M | M | M | M | M | M | SM | T | SM | M | |
| 7 | AR | 1.17 ± 0.05 | 1.76 ± 0.08 | 1.34 ± 0.07 | 1.27 ± 0.05 | 1.76 ± 0.08 | 1.10 ± 0.07 | 1.58 ± 0.09 | -- | 1.57 ± 0.08 | 1.42 ± 0.06 |
| CI | 46.19 ± 0.85 | 36.19 ± 0.72 | 42.66 ± 0.87 | 44.09 ± 0.97 | 36.24 ± 0.75 | 47.67 ± 0.90 | 38.75 ± 0.80 | -- | 38.85 ± 0.81 | 41.40 ± 0.78 | |
| RL | 5.73 ± 0.23 | 5.94 ± 0.25 | 5.15 ± 0.20 | 5.42 ± 0.19 | 6.10 ± 0.25 | 5.59 ± 0.23 | 6.31 ± 0.27 | 4.30 ± 0.17 | 6.20 ± 0.26 | 5.63 ± 0.23 | |
| LC | M | SM | M | M | SM | M | M | T | M | M | |
| 8 | AR | 1.33 ± 0.06 | 1.84 ± 0.09 | 1.11 ± 0.05 | 1.23 ± 0.05 | 1.89 ± 0.12 | 1.89 ± 0.08 | 1.52 ± 0.07 | -- | 1.23 ± 0.06 | 3.33 ± 0.16 |
| CI | 42.88 ± 0.81 | 35.22 ± 0.69 | 47.41 ± 1.01 | 44.84 ± 0.83 | 34.63 ± 0.71 | 34.66 ± 0.64 | 39.63 ± 0.72 | -- | 44.81 ± 0.97 | 23.09 ± 0.46 | |
| RL | 5.14 ± 0.19 | 5.76 ± 0.25 | 4.62 ± 0.18 | 5.07 ± 0.19 | 5.97 ± 0.25 | 5.49 ± 0.22 | 5.83 ± 0.23 | 3.92 ± 0.15 | 5.24 ± 0.22 | 5.59 ± 0.25 | |
| LC | M | SM | M | M | SM | SM | M | T | M | ST | |
| 9 | AR | 1.40 ± 0.06 | 1.39 ± 0.06 | 1.50 ± 0.08 | 1.16 ± 0.05 | 1.50 ± 0.07 | 1.48 ± 0.04 | 1.42 ± 0.06 | -- | 2.56 ± 0.16 | 1.55 ± 0.06 |
| CI | 41.61 ± 0.80 | 41.75 ± 0.91 | 40.01 ± 0.80 | 46.40 ± 0.93 | 40.00 ± 0.78 | 40.40 ± 0.89 | 41.35 ± 0.79 | -- | 28.07 ± 0.55 | 39.15 ± 0.75 | |
| RL | 4.94 ± 0.21 | 5.48 ± 0.22 | 4.52 ± 0.18 | 4.79 ± 0.19 | 5.55 ± 0.23 | 5.33 ± 0.21 | 5.08 ± 0.22 | 4.19 ± 0.18 | 5.16 ± 0.20 | 5.46 ± 0.22 | |
| LC | M | M | M | M | M | M | M | T | SM | M | |
| 10 | AR | 1.03 ± 0.04 | 1.30 ± 0.08 | 1.13 ± 0.06 | 1.05 ± 0.06 | 1.14 ± 0.05 | 1.67 ± 0.08 | 1.27 ± 0.07 | -- | 1.38 ± 0.07 | 1.38 ± 0.07 |
| CI | 49.23 ± 1.05 | 43.52 ± 0.87 | 46.99 ± 0.95 | 48.78 ± 0.97 | 46.66 ± 0.88 | 37.48 ± 0.81 | 43.99 ± 0.92 | -- | 42.05 ± 0.84 | 42.09 ± 0.79 | |
| RL | 4.79 ± 0.16 | 5.37 ± 0.24 | 3.86 ± 0.16 | 4.60 ± 0.21 | 5.20 ± 0.21 | 5.27 ± 0.20 | 5.08 ± 0.19 | 3.61 ± 0.14 | 4.93 ± 0.20 | 4.93 ± 0.21 | |
| LC | M | M | M | M | M | M | M | T | M | M | |
| 11 | AR | 1.04 ± 0.04 | 1.31 ± 0.08 | 1.06 ± 0.06 | 1.07 ± 0.06 | 1.06 ± 0.04 | 1.28 ± 0.04 | 1.50 ± 0.08 | -- | 1.45 ± 0.05 | 2.23 ± 0.11 |
| CI | 49.13 ± 0.96 | 43.24 ± 0.87 | 48.64 ± 0.89 | 48.35 ± 1.01 | 48.43 ± 1.00 | 43.78 ± 0.88 | 40.05 ± 0.76 | -- | 40.82 ± 0.81 | 31.00 ± 0.66 | |
| RL | 4.64 ± 0.18 | 4.68 ± 0.19 | 3.62 ± 0.13 | 3.50 ± 0.12 | 5.06 ± 0.20 | 4.43 ± 0.17 | 4.74 ± 0.19 | 3.45 ± 0.14 | 4.89 ± 0.21 | 4.87 ± 0.20 | |
| LC | M | M | M | M | M | M | M | T | M | SM | |
| 12 | AR | 1.18 ± 0.07 | 1.15 ± 0.05 | 1.27 ± 0.06 | 1.42 ± 0.08 | 1.23 ± 0.05 | -- | 1.33 ± 0.06 | 1.30 ± 0.06 | ||
| CI | 45.86 ± 0.86 | 46.44 ± 0.99 | 44.14 ± 1.01 | 41.32 ± 0.79 | 44.92 ± 0.89 | -- | 42.85 ± 0.82 | 43.40 ± 0.90 | |||
| RL | 4.49 ± 0.17 | 4.49 ± 0.17 | 4.61 ± 0.18 | 4.11 ± 0.18 | 4.69 ± 0.18 | 3.55 ± 0.15 | 4.33 ± 0.17 | 4.35 ± 0.18 | |||
| LC | M | M | M | M | M | T | M | M | |||
| 13 | AR | 1.23 ± 0.05 | 1.09 ± 0.06 | 1.74 ± 0.07 | 1.90 ± 0.11 | 1.96 ± 0.12 | -- | 1.37 ± 0.07 | 1.96 ± 0.10 | ||
| CI | 44.84 ± 1.03 | 47.75 ± 0.99 | 36.54 ± 0.75 | 34.48 ± 0.70 | 33.80 ± 0.63 | -- | 42.15 ± 0.83 | 33.78 ± 0.73 | |||
| RL | 4.26 ± 0.17 | 4.23 ± 0.18 | 2.89 ± 0.13 | 3.27 ± 0.13 | 4.60 ± 0.19 | 3.66 ± 0.14 | 3.92 ± 0.17 | 4.47 ± 0.17 | |||
| LC | M | M | SM | SM | SM | T | M | SM | |||
| 14 | AR | 1.80 ± 0.07 | 1.37 ± 0.08 | -- | |||||||
| CI | 35.73 ± 0.71 | 42.26 ± 0.79 | -- | ||||||||
| RL | 2.92 ± 0.11 | 2.89 ± 0.11 | 3.02 ± 0.13 | ||||||||
| LC | SM | M | T | ||||||||
| 15 | AR | 1.14 ± 0.05 | 1.30 ± 0.06 | -- | |||||||
| CI | 46.72 ± 0.87 | 43.45 ± 0.89 | -- | ||||||||
| RL | 2.52 ± 0.11 | 2.73 ± 0.11 | 3.02 ± 0.12 | ||||||||
| LC | M | M | T | ||||||||
| 16 | AR | 1.20 ± 0.06 | 1.03 ± 0.04 | -- | |||||||
| CI | 45.54 ± 0.91 | 49.22 ± 10.00 | -- | ||||||||
| RL | 2.46 ± 0.11 | 2.63 ± 0.10 | 2.70 ± 0.10 | ||||||||
| LC | M | M | T | ||||||||
| 17 | AR | 1.17 ± 0.06 | 1.18 ± 0.07 | -- | |||||||
| CI | 46.04 ± 0.93 | 45.79 ± 1.03 | -- | ||||||||
| RL | 2.36 ± 0.10 | 2.45 ± 0.09 | 3.08 ± 0.13 | ||||||||
| LC | M | M | T | ||||||||
| 18 | AR | 1.61 ± 0.09 | 1.22 ± 0.06 | -- | |||||||
| CI | 38.30 ± 0.79 | 45.02 ± 0.93 | -- | ||||||||
| RL | 2.22 ± 0.09 | 2.39 ± 0.10 | 2.70 ± 0.10 | ||||||||
| LC | M | M | T | ||||||||
| 19 | AR | 1.10 ± 0.06 | 1.27 ± 0.07 | -- | |||||||
| CI | 47.67 ± 1.02 | 44.08 ± 0.82 | -- | ||||||||
| RL | 2.00 ± 0.08 | 2.18 ± 0.08 | 2.76 ± 0.11 | ||||||||
| LC | M | M | T | ||||||||
| 20 | AR | 1.04 ± 0.05 | 1.16 ± 0.06 | -- | |||||||
| CI | 49.09 ± 0.99 | 46.30 ± 0.90 | -- | ||||||||
| RL | 1.86 ± 0.08 | 2.00 ± 0.08 | 2.60 ± 0.10 | ||||||||
| LC | M | M | T | ||||||||
| 21 | AR | 1.43 ± 0.06 | 3.76 ± 0.18 | -- | |||||||
| CI | 41.13 ± 0.83 | 20.99 ± 0.42 | -- | ||||||||
| RL | 1.67 ± 0.07 | 1.94 ± 0.08 | 2.44 ± 0.10 | ||||||||
| LC | M | ST | T | ||||||||
| 22 | AR | 1.23 ± 0.05 | 1.51 ± 0.06 | -- | |||||||
| CI | 44.92 ± 0.96 | 39.86 ± 0.78 | -- | ||||||||
| RL | 1.60 ± 0.06 | 1.79 ± 0.07 | 2.81 ± 0.10 | ||||||||
| LC | M | M | T | ||||||||
| 23 | AR | -- | |||||||||
| CI | -- | ||||||||||
| RL | 2.60 ± 0.11 | ||||||||||
| LC | T | ||||||||||
| 24 | AR | -- | |||||||||
| CI | -- | ||||||||||
| RL | 2.60 ± 0.10 | ||||||||||
| LC | T | ||||||||||
| 25 | AR | -- | |||||||||
| CI | -- | ||||||||||
| RL | 2.54 ± 0.09 | ||||||||||
| LC | T | ||||||||||
| 26 | AR | -- | |||||||||
| CI | -- | ||||||||||
| RL | 2.23 ± 0.09 | ||||||||||
| LC | T | ||||||||||
| 27 | AR | -- | |||||||||
| CI | -- | ||||||||||
| RL | 1.91 ± 0.08 | ||||||||||
| LC | T | ||||||||||
| 28 | AR | -- | |||||||||
| CI | -- | ||||||||||
| RL | 1.91 ± 0.07 | ||||||||||
| LC | T | ||||||||||
| 29 | AR | -- | |||||||||
| CI | -- | ||||||||||
| RL | 1.91 ± 0.08 | ||||||||||
| LC | T | ||||||||||
| 30 | AR | -- | |||||||||
| CI | -- | ||||||||||
| RL | 1.75 ± 0.07 | ||||||||||
| LC | T | ||||||||||
| 31 | AR | -- | |||||||||
| CI | -- | ||||||||||
| RL | 1.43 ± 0.06 | ||||||||||
| LC | T | ||||||||||
| 32 | AR | -- | |||||||||
| CI | -- | ||||||||||
| RL | 1.32 ± 0.05 | ||||||||||
| LC | T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Shi, S.; Lu, N.; Shen, C.; Jiang, J. Karyotypes of 10 Anuran Species from the Qinghai–Tibetan Plateau. Diversity 2023, 15, 947. https://doi.org/10.3390/d15090947
Chen Q, Shi S, Lu N, Shen C, Jiang J. Karyotypes of 10 Anuran Species from the Qinghai–Tibetan Plateau. Diversity. 2023; 15(9):947. https://doi.org/10.3390/d15090947
Chicago/Turabian StyleChen, Qiheng, Shengchao Shi, Ningning Lu, Cheng Shen, and Jianping Jiang. 2023. "Karyotypes of 10 Anuran Species from the Qinghai–Tibetan Plateau" Diversity 15, no. 9: 947. https://doi.org/10.3390/d15090947
APA StyleChen, Q., Shi, S., Lu, N., Shen, C., & Jiang, J. (2023). Karyotypes of 10 Anuran Species from the Qinghai–Tibetan Plateau. Diversity, 15(9), 947. https://doi.org/10.3390/d15090947








