Artificial Seaweed Substrates Complement ARMS in DNA Metabarcoding-Based Monitoring of Temperate Coastal Macrozoobenthos
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sampling Design
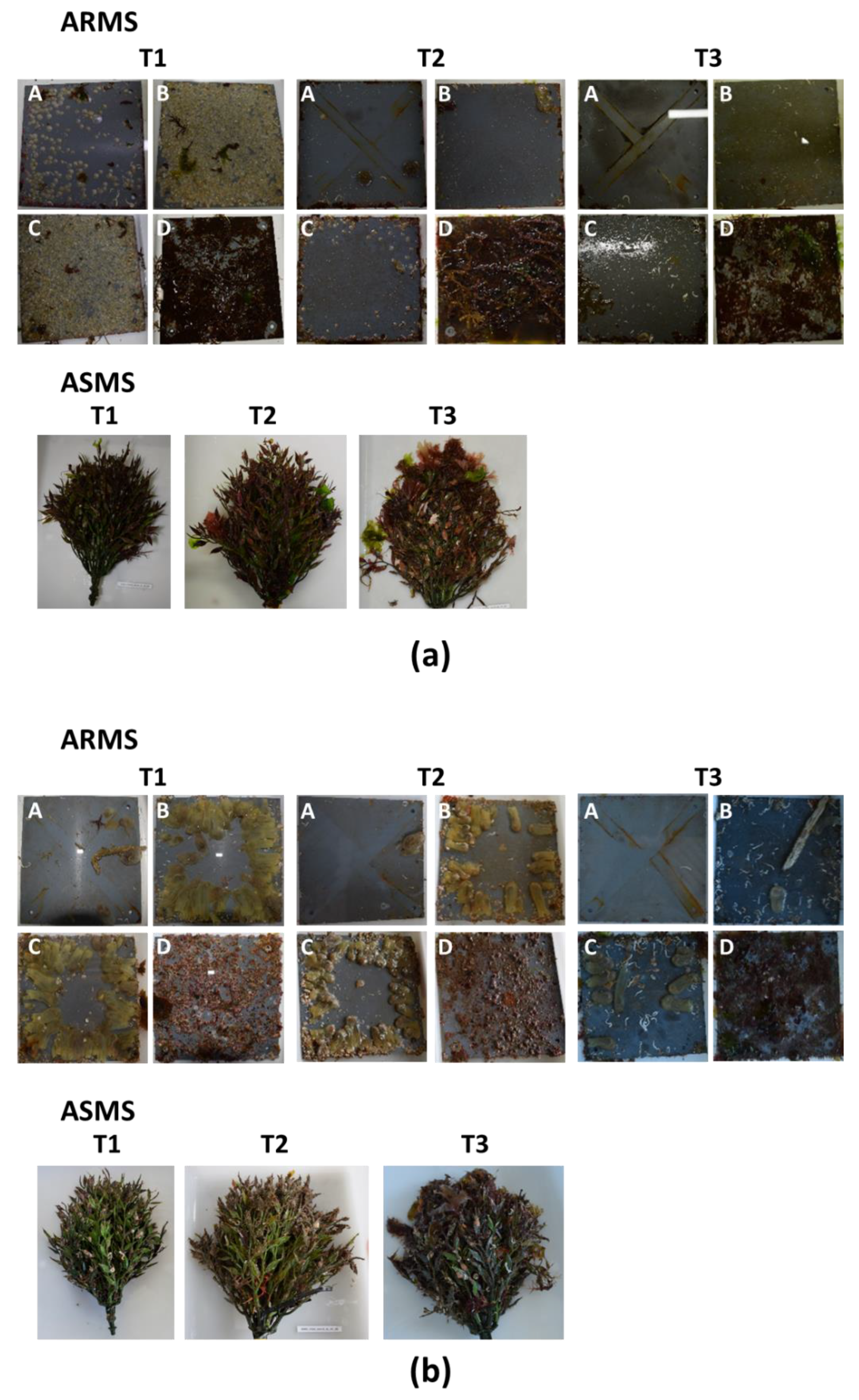
2.3. ARMS and ASMS Collection and Processing
2.4. DNA Metabarcoding
2.5. Data Processing
2.6. Statistical Analyses
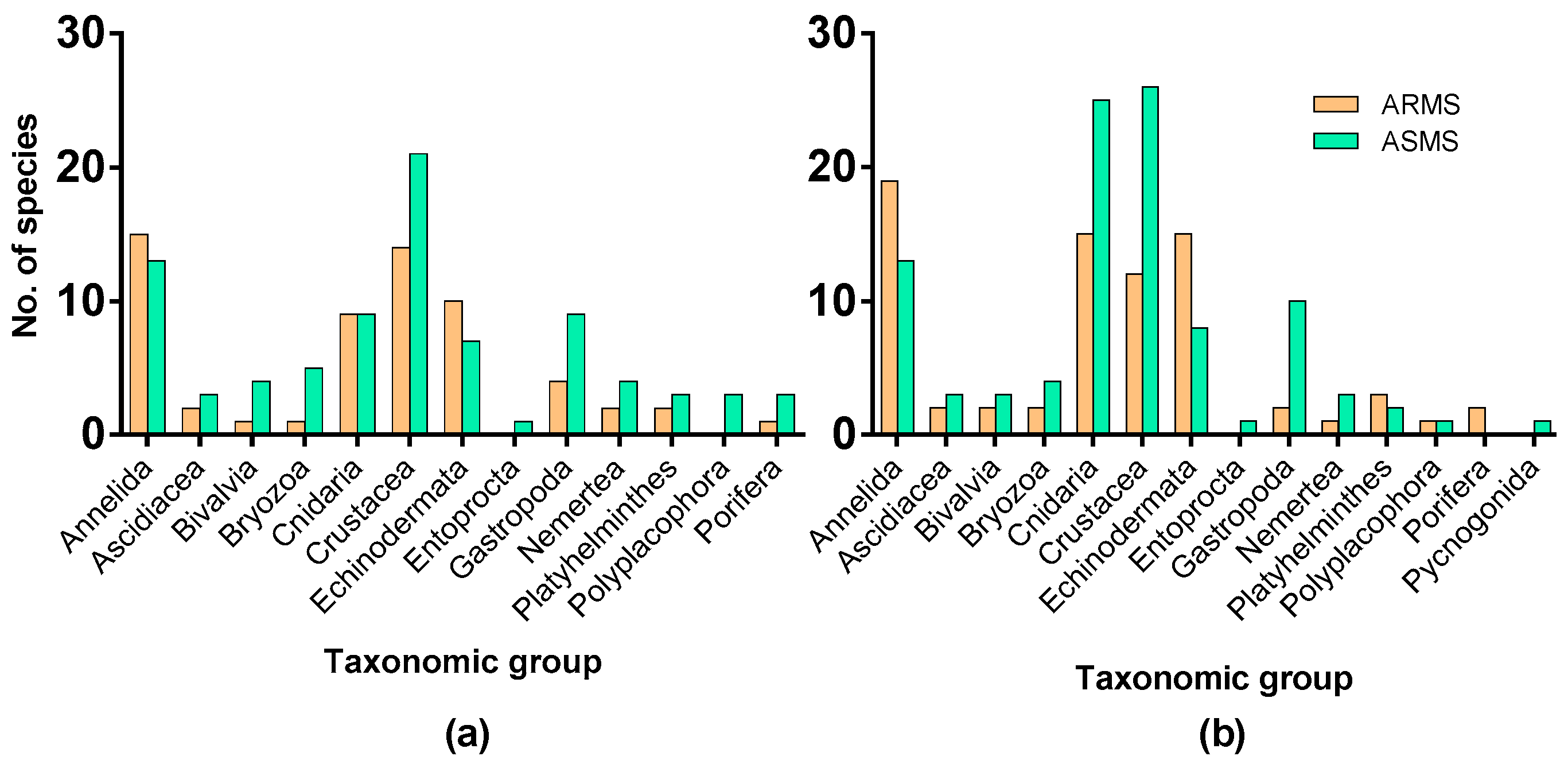
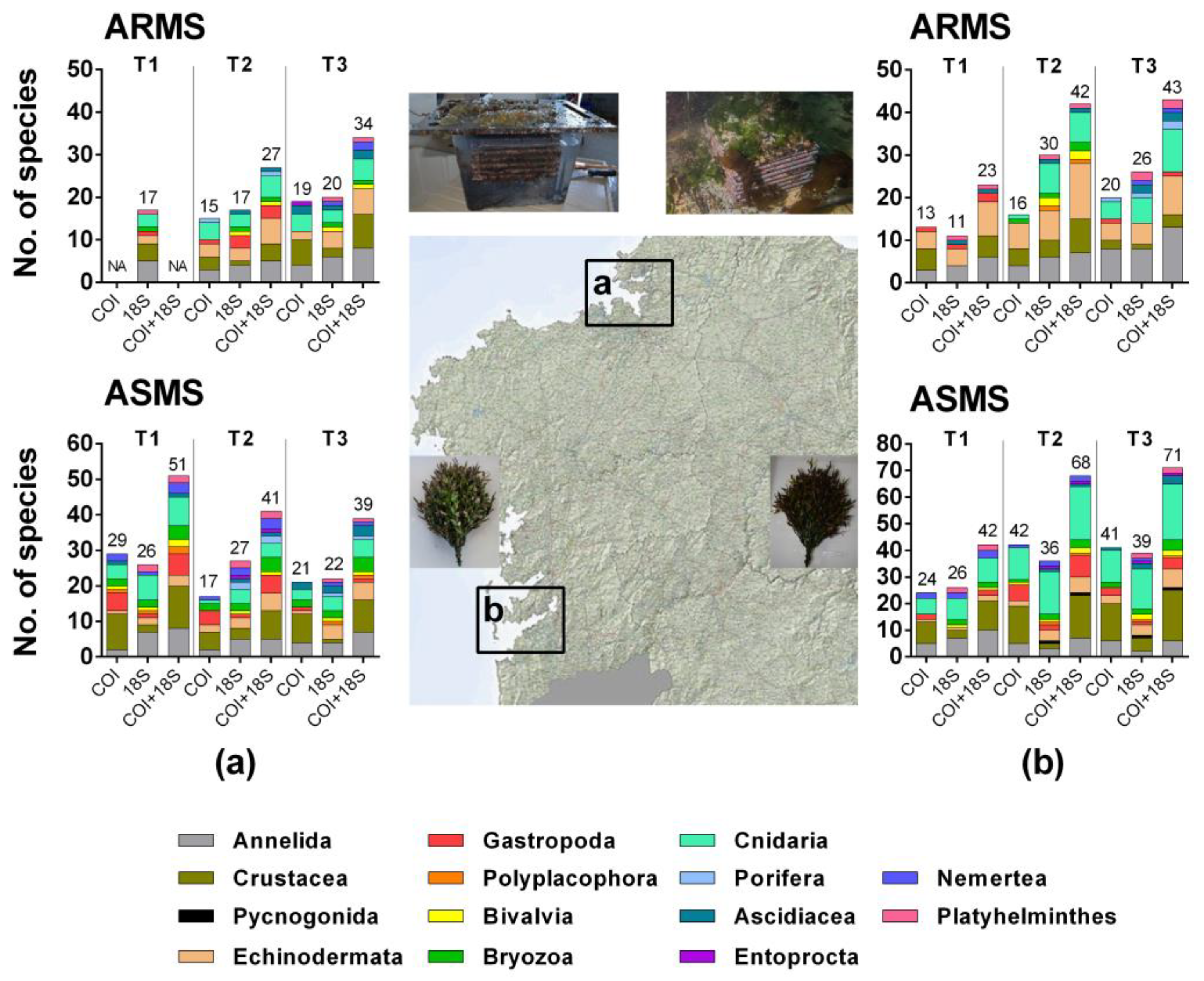
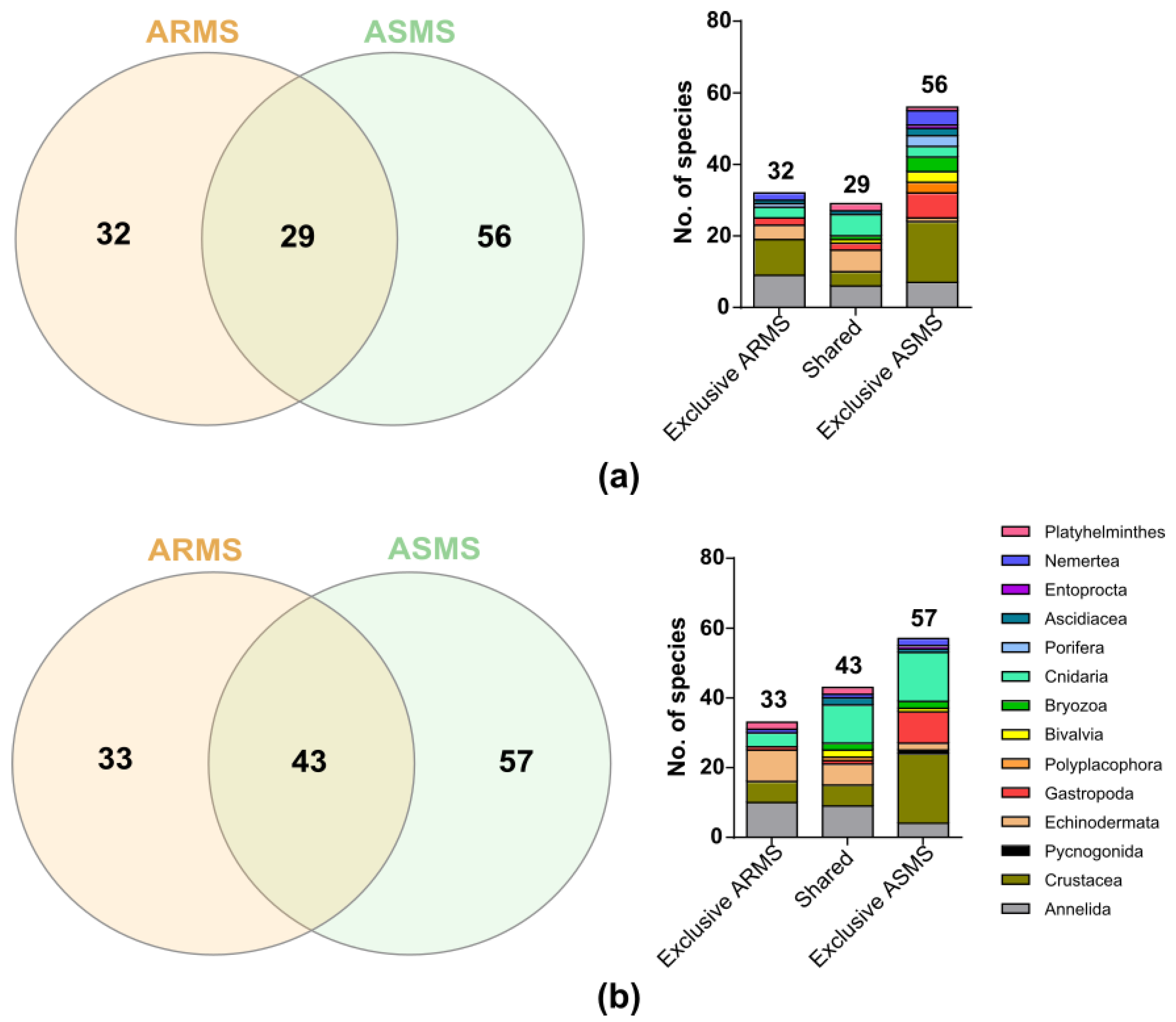
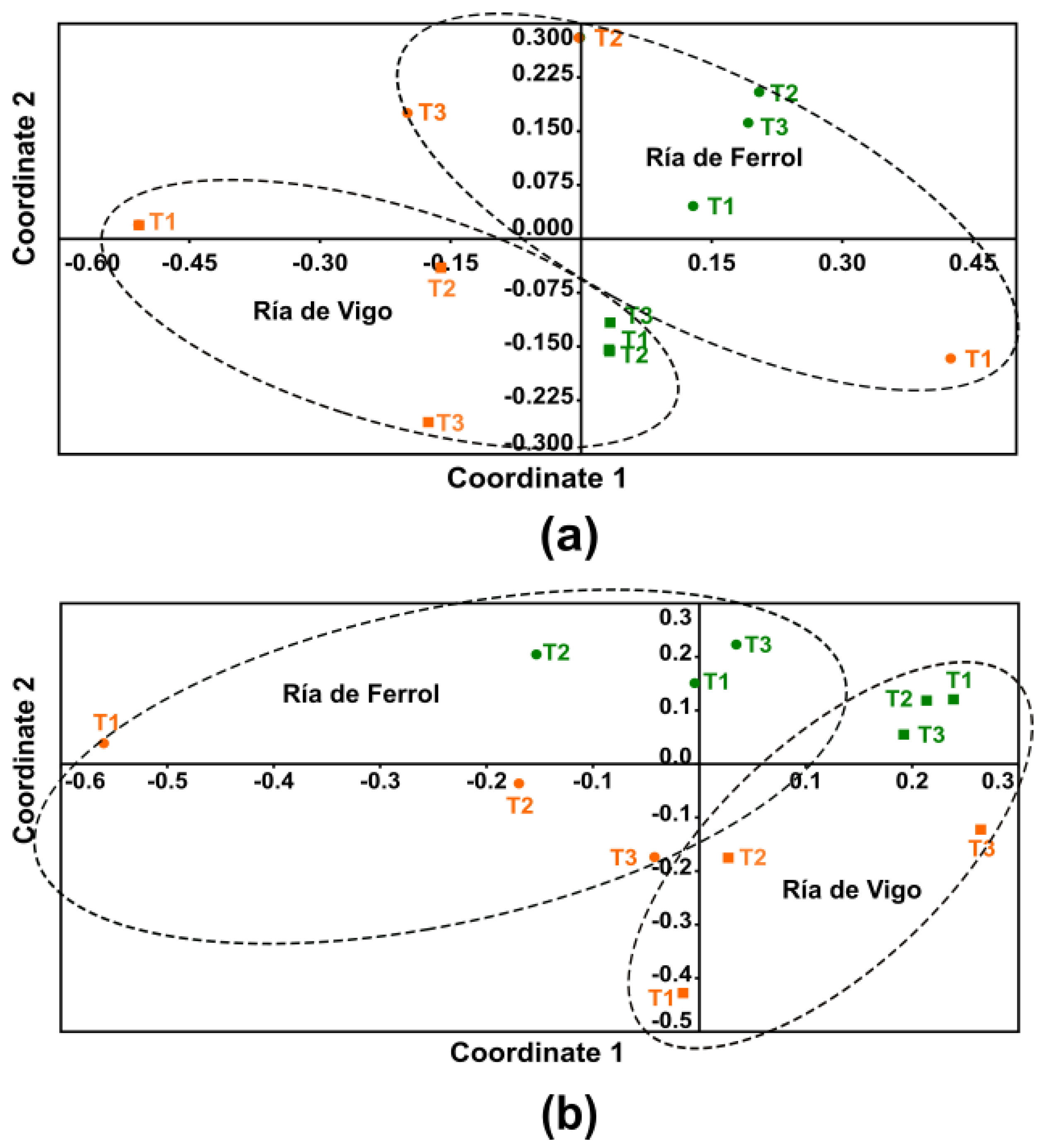
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doney, S.C.; Ruckelshaus, M.; Emmett Duffy, J.; Barry, J.P.; Chan, F.; English, C.A.; Galindo, H.M.; Grebmeier, J.M.; Hollowed, A.B.; Knowlton, N.; et al. Climate Change Impacts on Marine Ecosystems. Annu. Rev. Mar. Sci. 2012, 4, 11–37. [Google Scholar] [CrossRef]
- Niiranen, S.; Yletyinen, J.; Tomczak, M.T.; Blenckner, T.; Hjerne, O.; MacKenzie, B.R.; Müller-Karulis, B.; Neumann, T.; Meier, H.E.M. Combined Effects of Global Climate Change and Regional Ecosystem Drivers on an Exploited Marine Food Web. Glob. Chang. Biol. 2013, 19, 3327–3342. [Google Scholar] [CrossRef] [PubMed]
- Burrows, M.T.; Schoeman, D.S.; Buckley, L.B.; Moore, P.; Poloczanska, E.S.; Brander, K.M.; Brown, C.; Bruno, J.F.; Duarte, C.M.; Halpern, B.S.; et al. The Pace of Shifting Climate in Marine and Terrestrial Ecosystems. Science 2011, 334, 652–655. [Google Scholar] [CrossRef] [PubMed]
- Duarte, C.M. Global Change and the Future Ocean: A Grand Challenge for Marine Sciences. Front. Mar. Sci. 2014, 1, 63. [Google Scholar] [CrossRef]
- Spalding, M.D.; Fox, H.E.; Allen, G.R.; Davidson, N.; Ferdaña, Z.A.; Finlayson, M.; Halpern, B.S.; Jorge, M.A.; Lombana, A.; Lourie, S.A.; et al. Marine Ecoregions of the World: A Bioregionalization of Coastal and Shelf Areas. BioScience 2007, 57, 573–583. [Google Scholar] [CrossRef]
- Pereira, S.G.; Lima, F.P.; Queiroz, N.C.; Ribeiro, P.A.; Santos, A.M. Biogeographic Patterns of Intertidal Macroinvertebrates and Their Association with Macroalgae Distribution along the Portuguese Coast. Hydrobiologia 2006, 555, 185–192. [Google Scholar] [CrossRef]
- Henriques, S.; Pais, M.P.; Batista, M.I.; Teixeira, C.M.; Costa, M.J.; Cabral, H. Can Different Biological Indicators Detect Similar Trends of Marine Ecosystem Degradation? Ecol. Indic. 2014, 37, 105–118. [Google Scholar] [CrossRef]
- Bianchi, C.; Pronzato, R.; Cattaneo-Vietti, R.; Benedetti-Cecchi, L.; Morri, C.; Pansini, M.; Chemello, R.; Milazzo, M.; Fraschetti, S.; Terlizzi, A.; et al. Hard Bottoms. In Mediterranean Marine Benthos: A Manual of Methods for Its Sampling and Study. Biologia Marina Mediterranea; Gambi, M.C., Dappiano, M., Eds.; SIBM: Genova, Italy, 2004; Volume 11. [Google Scholar]
- Borja, A.; Elliott, M.; Andersen, J.H.; Berg, T.; Carstensen, J.; Halpern, B.S.; Heiskanen, A.-S.; Korpinen, S.; Lowndes, J.S.S.; Martin, G.; et al. Overview of Integrative Assessment of Marine Systems: The Ecosystem Approach in Practice. Front. Mar. Sci. 2016, 3, 20. [Google Scholar] [CrossRef]
- Danovaro, R.; Carugati, L.; Berzano, M.; Cahill, A.E.; Carvalho, S.; Chenuil, A.; Corinaldesi, C.; Cristina, S.; David, R.; Dell’Anno, A.; et al. Implementing and Innovating Marine Monitoring Approaches for Assessing Marine Environmental Status. Front. Mar. Sci. 2016, 3, 213. [Google Scholar] [CrossRef]
- Duarte, S.; Leite, B.; Feio, M.; Costa, F.; Filipe, A. Integration of DNA-Based Approaches in Aquatic Ecological Assessment Using Benthic Macroinvertebrates. Water 2021, 13, 331. [Google Scholar] [CrossRef]
- Tanhua, T.; Pouliquen, S.; Hausman, J.; O’Brien, K.; Bricher, P.; de Bruin, T.; Buck, J.J.H.; Burger, E.F.; Carval, T.; Casey, K.S.; et al. Ocean FAIR Data Services. Front. Mar. Sci. 2019, 6, 440. [Google Scholar] [CrossRef]
- Steyaert, M.; Priestley, V.; Osborne, O.; Herraiz, A.; Arnold, R.; Savolainen, V. Advances in Metabarcoding Techniques Bring Us Closer to Reliable Monitoring of the Marine Benthos. J. Appl. Ecol. 2020, 57, 2234–2245. [Google Scholar] [CrossRef]
- Bourlat, S.J.; Borja, A.; Gilbert, J.; Taylor, M.I.; Davies, N.; Weisberg, S.B.; Griffith, J.F.; Lettieri, T.; Field, D.; Benzie, J.; et al. Genomics in Marine Monitoring: New Opportunities for Assessing Marine Health Status. Mar. Pollut. Bull. 2013, 74, 19–31. [Google Scholar] [CrossRef]
- Cahill, A.E.; Pearman, J.K.; Borja, A.; Carugati, L.; Carvalho, S.; Danovaro, R.; Dashfield, S.; David, R.; Féral, J.-P.; Olenin, S.; et al. A Comparative Analysis of Metabarcoding and Morphology-Based Identification of Benthic Communities across Different Regional Seas. Ecol. Evol. 2018, 8, 8908–8920. [Google Scholar] [CrossRef]
- Aylagas, E.; Borja, A.; Muxika, I.; Rodriguez-Ezpeleta, N. Adapting metabarcoding-based benthic biomonitoring into routine marine ecological status assessment networks. Ecol. Ind. 2018, 95, 194–202. [Google Scholar] [CrossRef]
- Ros, M.; Vázquez-Luis, M.; Guerra-García, J.M. The Role of Marinas and Recreational Boating in the Occurrence and Distribution of Exotic Caprellids (Crustacea: Amphipoda) in the Western Mediterranean: Mallorca Island as a Case Study. J. Sea Res. 2013, 83, 94–103. [Google Scholar] [CrossRef]
- Pearman, J.K.; Aylagas, E.; Voolstra, C.R.; Anlauf, H.; Villalobos, R.; Carvalho, S. Disentangling the Complex Microbial Community of Coral Reefs Using Standardized Autonomous Reef Monitoring Structures (ARMS). Mol. Ecol. 2019, 28, 3496–3507. [Google Scholar] [CrossRef]
- Sedano, F.; Tierno de Figueroa, J.M.; Navarro-Barranco, C.; Ortega, E.; Guerra-García, J.M.; Espinosa, F. Do Artificial Structures Cause Shifts in Epifaunal Communities and Trophic Guilds across Different Spatial Scales? Mar. Environ. Res. 2020, 158, 104998. [Google Scholar] [CrossRef]
- Pavloudi, C.; Yperifanou, E.; Kristoffersen, J.; Dailianis, T.; Gerovasileiou, V. Artificial Reef Monitoring Structures (ARMS) Providing Insights on Hard Substrate Biodiversity and Community Structure of the Eastern Mediterranean Sea. ACA 2021, 4, e64760. [Google Scholar] [CrossRef]
- Zimmerman, T.L.; Martin, J.W. Artificial Reef Matrix Structures (Arms): An Inexpensive and Effective Method for Collecting Coral Reef-Associated Invertebrates. GCR 2004, 16, 59–64. [Google Scholar] [CrossRef]
- Leray, M.; Knowlton, N. DNA Barcoding and Metabarcoding of Standardized Samples Reveal Patterns of Marine Benthic Diversity. Proc. Natl. Acad. Sci. USA 2015, 112, 2076–2081. [Google Scholar] [CrossRef] [PubMed]
- Ransome, E.; Geller, J.B.; Timmers, M.; Leray, M.; Mahardini, A.; Sembiring, A.; Collins, A.G.; Meyer, C.P. The Importance of Standardization for Biodiversity Comparisons: A Case Study Using Autonomous Reef Monitoring Structures (ARMS) and Metabarcoding to Measure Cryptic Diversity on Mo’orea Coral Reefs, French Polynesia. PLoS ONE 2017, 12, e0175066. [Google Scholar] [CrossRef] [PubMed]
- Templado, J.; Paulay, G.; Gittenberger, A.; Meyer, C. Chapter 11. Sampling the Marine Realm. ABC Taxa 2010, 8, 273–307. [Google Scholar]
- Plaisance, L.; Caley, M.J.; Brainard, R.E.; Knowlton, N. The Diversity of Coral Reefs: What Are We Missing? PLoS ONE 2011, 6, e25026. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.J.M.; Ip, Y.C.A.; Bauman, A.G.; Huang, D. MinION-in-ARMS: Nanopore Sequencing to Expedite Barcoding of Specimen-Rich Macrofaunal Samples From Autonomous Reef Monitoring Structures. Front. Mar. Sci. 2020, 7, 448. [Google Scholar] [CrossRef]
- Obst, M.; Exter, K.; Allcock, A.L.; Arvanitidis, C.; Axberg, A.; Bustamante, M.; Cancio, I.; Carreira-Flores, D.; Chatzinikolaou, E.; Chatzigeorgiou, G.; et al. A Marine Biodiversity Observation Network for Genetic Monitoring of Hard-Bottom Communities (ARMS-MBON). Front. Mar. Sci. 2020, 7, 572680. [Google Scholar] [CrossRef]
- David, R.; Uyarra, M.C.; Carvalho, S.; Anlauf, H.; Borja, A.; Cahill, A.E.; Carugati, L.; Danovaro, R.; De Jode, A.; Feral, J.-P.; et al. Lessons from Photo Analyses of Autonomous Reef Monitoring Structures as Tools to Detect (Bio-)Geographical, Spatial, and Environmental Effects. Mar. Pollut. Bull. 2019, 141, 420–429. [Google Scholar] [CrossRef]
- Carreira-Flores, D.; Neto, R.; Ferreira, H.; Cabecinha, E.; Díaz-Agras, G.; Gomes, P.T. Artificial Substrates as Sampling Devices for Marine Epibenthic Fauna: A Quest for Standardization. Reg. Stud. Mar. Sci. 2020, 37, 101331. [Google Scholar] [CrossRef]
- Carreira-Flores, D.; Neto, R.; Ferreira, H.; Cabecinha, E.; Díaz-Agras, G.; Gomes, P.T. Two Better than One: The Complementary of Different Types of Artificial Substrates on Benthic Marine Macrofauna Studies. Mar. Environ. Res. 2021, 171, 105449. [Google Scholar] [CrossRef]
- Lobo, J.; Shokralla, S.; Costa, M.H.; Hajibabaei, M.; Costa, F.O. DNA Metabarcoding for High-Throughput Monitoring of Estuarine Macrobenthic Communities. Sci. Rep. 2017, 7, 15618. [Google Scholar] [CrossRef]
- Carvalho, S.; Aylagas, E.; Villalobos, R.; Kattan, Y.; Berumen, M.; Pearman, J.K. Beyond the Visual: Using Metabarcoding to Characterize the Hidden Reef Cryptobiome. Proc. R. Soc. B 2019, 286, 20182697. [Google Scholar] [CrossRef]
- Duarte, S.; Vieira, P.E.; Leite, B.R.; Teixeira, M.A.L.; Neto, J.M.; Costa, F.O. Macrozoobenthos Monitoring in Portuguese Transitional Waters in the Scope of the Water Framework Directive Using Morphology and DNA Metabarcoding. Estuar. Coast. Shelf Sci. 2023, 281, 108207. [Google Scholar] [CrossRef]
- Pearman, J.K.; Chust, G.; Aylagas, E.; Villarino, E.; Watson, J.R.; Chenuil, A.; Borja, A.; Cahill, A.E.; Carugati, L.; Danovaro, R.; et al. Pan-Regional Marine Benthic Cryptobiome Biodiversity Patterns Revealed by Metabarcoding Autonomous Reef Monitoring Structures. Mol. Ecol. 2020, 29, 4882–4897. [Google Scholar] [CrossRef]
- Moreira, J.; Díaz-Agras, G.; Candás, M.; Señarís, M.P.; Urgorri, V. Leptostracans (Crustacea: Phyllocarida) from the Ría de Ferrol (Galicia, NW Iberian Peninsula), with Description of a New Species of Nebalia Leach, 1814. Sci. Mar. 2009, 73, 269–285. [Google Scholar] [CrossRef]
- Urgorri, V.; Troncoso, J.; Dobarro, J. Malacofauna asociada a una biocenosis de maerl en la ría de Ferrol (Galicia, NO España). An. Biol. 1992, 18, 161–174. [Google Scholar]
- Prego, R.; Fraga, F. A Simple Model to Calculate the Residual Flows in a Spanish Ria. Hydrographic Consequences in the Ria of Vigo. Estuar. Coast. Shelf Sci. 1992, 34, 603–615. [Google Scholar] [CrossRef]
- Barroso, C.; Moreira, M.; Gibbs, P. Comparison of Imposex and Intersex Development in Four Prosobranch Species for TBT Monitoring of a Southern European Estuarine System (Ria de Aveiro, NW Portugal). Mar. Ecol. Prog. Ser. 2000, 201, 221–232. [Google Scholar] [CrossRef]
- Prego, R.; Filgueiras, A.V.; Santos-Echeandía, J. Temporal and Spatial Changes of Total and Labile Metal Concentration in the Surface Sediments of the Vigo Ria (NW Iberian Peninsula): Influence of Anthropogenic Sources. Mar. Pollut. Bull. 2008, 56, 1031–1042. [Google Scholar] [CrossRef]
- Leite, B.R.; Vieira, P.E.; Troncoso, J.S.; Costa, F.O. Comparing species detection success between molecular markers in DNA metabarcoding of coastal macroinvertebrates. Metabarcoding Metagenomics 2021, 5, e70063. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Dewaard, J.R.; Hebert, P.D.N. An Inexpensive, Automation-Friendly Protocol for Recovering High-Quality DNA. Mol. Ecol. Notes 2006, 6, 998–1002. [Google Scholar] [CrossRef]
- Steinke, D.; deWaard, S.L.; Sones, J.E.; Ivanova, N.V.; Prosser, S.W.J.; Perez, K.; Braukmann, T.W.A.; Milton, M.; Zakharov, E.V.; deWaard, J.R.; et al. Message in a Bottle—Metabarcoding Enables Biodiversity Comparisons across Ecoregions. GigaScience 2022, 11, giac040. [Google Scholar] [CrossRef] [PubMed]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A New Versatile Primer Set Targeting a Short Fragment of the Mitochondrial COI Region for Metabarcoding Metazoan Diversity: Application for Characterizing Coral Reef Fish Gut Contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Lobo, J.; Costa, P.M.; Teixeira, M.A.; Ferreira, M.S.; Costa, M.H.; Costa, F.O. Enhanced Primers for Amplification of DNA Barcodes from a Broad Range of Marine Metazoans. BMC Ecol. 2013, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Stoeck, T.; Bass, D.; Nebel, M.; Christen, R.; Jones, M.D.M.; Breiner, H.-W.; Richards, T.A. Multiple Marker Parallel Tag Environmental DNA Sequencing Reveals a Highly Complex Eukaryotic Community in Marine Anoxic Water. Mol. Ecol. 2010, 19, 21–31. [Google Scholar] [CrossRef]
- Lejzerowicz, F.; Esling, P.; Pillet, L.; Wilding, T.A.; Black, K.D.; Pawlowski, J. High-Throughput Sequencing and Morphology Perform Equally Well for Benthic Monitoring of Marine Ecosystems. Sci. Rep. 2015, 5, 13932. [Google Scholar] [CrossRef]
- Illumina 16S. Metagenomic Sequencing Library Preparation. In Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System; Illumina: San Diego, CA, USA, 2013. [Google Scholar]
- Comeau, A.M.; Douglas, G.M.; Langille, M.G.I. Microbiome Helper: A Custom and Streamlined Workflow for Microbiome Research. MSystems 2017, 2, e00127-16. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality Control and Preprocessing of Metagenomic Datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Ratnasingham, S. MBRAVE: The Multiplex Barcode Research And Visualization Environment. BISS 2019, 3, e37986. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Leite, B.R.; Vieira, P.E.; Teixeira, M.A.L.; Lobo-Arteaga, J.; Hollatz, C.; Borges, L.M.S.; Duarte, S.; Troncoso, J.S.; Costa, F.O. Gap-analysis and annotated reference library for supporting macroinvertebrate metabarcoding in Atlantic Iberia. Reg. Stud. Mar. Sci. 2020, 36, 101307. [Google Scholar] [CrossRef]
- Lavrador, A.S.; Fontes, J.T.; Vieira, P.E.; Costa, F.O.; Duarte, S. Compilation, Revision, and Annotation of DNA Barcodes of Marine Invertebrate Non-Indigenous Species (NIS) Occurring in European Coastal Regions. Diversity 2023, 15, 174. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate High-Throughput Multiple Sequence Alignment of Ribosomal RNA Genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A Web-Based Tool for the Analysis of Sets through Venn Diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Hammer, O.; Harper, D.; Ryan, P. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Al-Rshaidat, M.M.D.; Snider, A.; Rosebraugh, S.; Devine, A.M.; Devine, T.D.; Plaisance, L.; Knowlton, N.; Leray, M. Deep COI Sequencing of Standardized Benthic Samples Unveils Overlooked Diversity of Jordanian Coral Reefs in the Northern Red Sea. Genome 2016, 59, 724–737. [Google Scholar] [CrossRef]
- Gaither, M.R.; DiBattista, J.D.; Leray, M.; Heyden, S. Metabarcoding the Marine Environment: From Single Species to Biogeographic Patterns. Environ. DNA 2022, 4, 3–8. [Google Scholar] [CrossRef]
- Ip, Y.C.A.; Chang, J.J.M.; Oh, R.M.; Quek, Z.B.R.; Chan, Y.K.S.; Bauman, A.G.; Huang, D. Seq’ and ARMS Shall Find: DNA (Meta)Barcoding of Autonomous Reef Monitoring Structures across the Tree of Life Uncovers Hidden Cryptobiome of Tropical Urban Coral Reefs. Mol. Ecol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Pearman, J.K.; Anlauf, H.; Irigoien, X.; Carvalho, S. Please Mind the Gap—Visual Census and Cryptic Biodiversity Assessment at Central Red Sea Coral Reefs. Mar. Environ. Res. 2016, 118, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.C.M. Macroalgas Formadoras de Habitat: Espaço Neutro para Macrofauna Marinha? Master’s Thesis, Universidade do Minho, Braga, Portugal, 2017. [Google Scholar]
- Plaisance, L.; Knowlton, N.; Paulay, G.; Meyer, C. Reef-Associated Crustacean Fauna: Biodiversity Estimates Using Semi-Quantitative Sampling and DNA Barcoding. Coral Reefs 2009, 28, 977–986. [Google Scholar] [CrossRef]
- Wolfe, K.; Mumby, P.J. RUbble Biodiversity Samplers: 3D-printed Coral Models to Standardize Biodiversity Censuses. Methods Ecol. Evol. 2020, 11, 1395–1400. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leite, B.R.; Duarte, S.; Troncoso, J.S.; Costa, F.O. Artificial Seaweed Substrates Complement ARMS in DNA Metabarcoding-Based Monitoring of Temperate Coastal Macrozoobenthos. Diversity 2023, 15, 657. https://doi.org/10.3390/d15050657
Leite BR, Duarte S, Troncoso JS, Costa FO. Artificial Seaweed Substrates Complement ARMS in DNA Metabarcoding-Based Monitoring of Temperate Coastal Macrozoobenthos. Diversity. 2023; 15(5):657. https://doi.org/10.3390/d15050657
Chicago/Turabian StyleLeite, Barbara R., Sofia Duarte, Jesús S. Troncoso, and Filipe O. Costa. 2023. "Artificial Seaweed Substrates Complement ARMS in DNA Metabarcoding-Based Monitoring of Temperate Coastal Macrozoobenthos" Diversity 15, no. 5: 657. https://doi.org/10.3390/d15050657
APA StyleLeite, B. R., Duarte, S., Troncoso, J. S., & Costa, F. O. (2023). Artificial Seaweed Substrates Complement ARMS in DNA Metabarcoding-Based Monitoring of Temperate Coastal Macrozoobenthos. Diversity, 15(5), 657. https://doi.org/10.3390/d15050657








