Abstract
The symbiotic relationship between macroorganisms, such as plants and animals, and the microorganisms in their environment plays a crucial role in shaping their physiology and ecology. Thus, many studies have examined microbial symbiosis in relation to plants, humans, and insects. However, little is known about the microbial diversity associated with isopods. Hence, in this study, we investigated the fungal diversity associated with two species of terrestrial isopods, Armadillidium nasatum and A. vulgare. In this study, we used a metabarcoding approach to compare fungal diversity between the two species for the first time. Our results indicated that A. nasatum had significantly greater mycobiome alpha diversity than A. vulgare. In contrast, fungal communities (beta diversity) did not differ significantly between hosts, except in beta dispersion of relative abundance. The majority of fungi identified belonged to Ascomycota and Basidiomycota, phyla that are dominated by saprotrophs. In conclusion, our findings shed light on the fungal communities associated with Armadillidium species, providing valuable insight into the biology of terrestrial isopods.
1. Introduction
Isopods, such as sea slaters, sow bugs, and pill bugs are a diverse group of crustaceans found in both aquatic and terrestrial environments [1,2,3]. The Oniscidea, a taxon of the Isopoda, comprises mostly terrestrial species, and its monophyly is well supported by numerous morphological apomorphies. These terrestrial isopods, commonly known as pill bugs or woodlice, are small crustaceans that play an important ecological role in many terrestrial ecosystems [4]. There are approximately 4000 known species of terrestrial isopod worldwide, inhabiting environments ranging from deserts to rainforests [2,3], and primarily found under fallen leaves and rocks [5,6]. Despite their small size and inconspicuous appearance, terrestrial isopods are incredibly important decomposers and nutrient recyclers in many ecosystems. As detritivores, terrestrial isopods break down organic materials from plants and fungal hyphae, facilitating the nutrient cycle in soil ecosystems [7,8,9]. In addition, isopods can be used in traditional medicines for their pharmacological and physiological benefits (e.g., anti-inflammation and antioxidation) [10,11,12]. Furthermore, they have great potential in food industry and agriculture as valuable food source for humans and animals [13,14]. Thus, in addition to their role in ecosystem functioning, isopods may also have valuable applications in various industries.
Since microorganisms influence the metabolism and evolution of their hosts [15,16,17], describing microbial diversity is key to enabling an efficient understanding of this relationship [18,19,20,21,22]. Although it has been investigated frequently in insect environments [23,24,25,26], research on isopods has been relatively neglected, with only a few studies focusing on bacteria [27,28,29]. Fungi are a major component of the soil microbial community and play important roles in nutrient cycling and organic matter decomposition. They form mutualistic associations with many terrestrial organisms, including plants, animals, and other fungi. Fungal communities associated with terrestrial isopods have received less attention compared to those associated with other soil organisms, such as plants and insects. However, given the major role fungi play as symbionts and parasites, they are an important microbial taxon to examine [30,31,32]. Several fungal species have been identified in isopods [33,34,35,36], and some produce compounds which possess pharmacological or cosmetic applications [37,38,39]. Fungi associated with isopods can be categorized as endosymbionts and foods. As endosymbionts, the isopods can have obligate gut symbionts or parasites [33,34], whereas terrestrial isopods feed on leaves colonized by decaying fungi or fungal mycelia [40,41], indicating that fungi detected in isopods is derived from food sources.
In recent years, the development of high-throughput sequencing technologies has revolutionized the study of microbial diversity and community composition at an unprecedented scale [42,43,44]. One such approach, metabarcoding, allows for the simultaneous sequencing of multiple barcoded sequences to identify and quantify the microbial communities present in a given sample. This method has been widely used to investigate microbial diversity in various ecosystems, including soil, water, and the gut microbiota of animals [43,45,46]. Compared to traditional culture-based methods, metabarcoding offers several advantages, including the ability to detect a wide range of microorganisms and identify rare or uncultivable species. In the context of studying microbial diversity associated with terrestrial isopods, the metabarcoding approach has the potential to provide a more comprehensive and accurate picture of community composition than traditional methods, and may reveal previously unknown interactions between isopods and microorganisms.
Armadillidium (family Armadillidiidae), a terrestrial isopod genus, is commonly known as a pill bug or roly poly [4]. It is characterized by its unique ability to roll into a ball as a defense mechanism. Armadillidium species are common and widespread and found in many habitats, including urban and suburban areas, forests, and agricultural fields. The genus Armadillidium includes over 100 species which have been the subject of extensive ecological and evolutionary research due to their unique traits, such as their ability to survive in diverse habitats, their role as decomposers, and their interactions with other organisms in the soil ecosystem [2,3]. South Korea is home to three Armadillidium species, among which Armadillidium nasatum and Armadillidium vulgare are the most common [47]. Previous research on Korean Armadillidium species has focused mainly on taxonomic and population genetics [47,48,49], with no study on microbial diversity associated with a terrestrial isopods.
Armadillidium vulgare is known to feed on decaying plant material and the microorganisms growing on it, thus its gut microbiota is likely to include a higher diversity of fungi. Some studies have investigated the fungal diversity associated with Armadillidium and related species, these studies have mainly focused on culturable fungi. To the best of our knowledge, studies investigating the fungal communities associated with Armadillidium using a metabarcoding approach have not yet been performed. Hence, this study is the first to investigate the diversity of fungi associated with A. nasatum and A. vulgare using a metabarcoding approach. The objective of this study was to provide information on the fungal diversity associated with the terrestrial isopod Armadillidium species. Specifically, by using metabarcoding technology, we expected to discover more diversity of fungi in isopods than previously known. We also hypothesized that fungal diversity and community structure would differ among Armadillidium species, and compared the characteristics of fungal communities between two Armadillidium species; A. nasatum and A. vulgare.
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
All isopod samples were collected from Central Park, Gwacheon, South Korea (37.431 N, 126.995 E) in September, 2022. The sampling sites in the park contained a variety of vegetation, including broad-leaved trees (e.g., Platanus occidentalis and Prunus serrulate), coniferous trees (e.g., Pinus spp.), and shrubs (e.g., Rhododendron spp.). Armadillidium species inhabit areas behind fallen leaves and rocks, and isopod samples were collected from four sites per species. The isopod samples were identified morphologically based on the description from previous studies [50,51]. One representative individual was randomly chosen from each site, and a total of eight representative individuals were selected for DNA extraction and amplicon sequencing. The surface of the samples was sacrificed and sterilized with 70% ethanol for 1 min and washed with sterilized distilled water for 1 min to remove soil and plant debris. After sterilization, the distilled water that was used for washing (100 µL × 3 replicates) was inoculated via a 90 mm PDA medium (BD Difco, Franklin Lakes, NJ, USA) to confirm the surface sterilization status of the isopod samples. After culturing at 25 °C for 2 weeks, it was confirmed that none of the microorganisms grew on the medium. We confirmed the death of the isopod samples by placing them in the freezer (−20 °C) for 5 min. One representative individual (c.a. 0.05 g) from each site was placed in a 2 mL tube containing a steel bead and homogenized (Taco™ Prep Bead Beater, GeneReach, Taichung, Taiwan). Genomic DNA was obtained from crushed samples using an AccuPrep DNA extraction kit following the manufacturer’s protocol (Bioneer, Daejeon, South Korea). DNA was stored in a freezer at −20 °C until further use.
2.2. Preparation for Illumina MiSeq
The fungal ribosomal internal transcribed spacer (ITS) region was amplified in two steps using an AccuPower PCR PreMix kit (Bioneer, South Korea). First, the full ITS region was amplified with ITS1Fngs and ITS4 primers [52,53] under the following conditions: 95 °C for 5 min; 35 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 40 s; and 72 °C for 5 min. Amplicons were confirmed using 1% gel electrophoresis and used for secondary PCR amplification of the ITS1 region. The primers ITS1Fngs and ITS2ngs [54] were attached to the MiSeq adapter, and the thermocycling conditions were as follows: 95 °C for 5 min; 15 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 40 s; and 72 °C for 7 min. Amplicons were again confirmed with 1% gel electrophoresis and then purified using an Expin™ PCR SV kit (GeneALL, Seoul, South Korea). For each sample, PCR was performed in triplicate using SimpliAmp™ Thermal Cycler (Applied Biosystems, Waltham, MA, USA) and the results were pooled together after measuring the DNA quantity using a Multiskan SkyHigh Microplate Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Sequencing was performed using Illumina MiSeq at Macrogen (Seoul, South Korea).
2.3. Bioinformatics and Statistics
Raw sequencing data were processed using the QIIME2 platform [55]. After demultiplexing and adapter trimming, sequence pairs were denoised and merged using DADA2 [56]. VSearch was used to cluster the operational taxonomic units (OTUs) based on 97% sequence similarity, and chimeric sequences were filtered [57]. Representative sequences of each OTU were grouped into taxa using the Naïve Bayesian classifier against NCBI RefSeq [58,59]. Before further analysis, all samples were normalized based on the lowest number of sequences (34,000 reads). As alpha diversity indices, richness (number of OTUs), diversity (Shannon diversity index), evenness (Pileu evenness index), and coverage (Good’s coverage index) were calculated in QIIME2. Alpha diversity and community structures were analyzed and visualized using the statistical packages ggplot2 [60], phyloseq [61], and vegan [62] in R version 4.1.2 [63]. After checking for normality using the Shapiro–Wilk test, alpha diversity indices were compared using t-tests and visualized with boxplots using ggplot2 package in R. A principal coordinates analysis (PCoA) was performed on binary Jaccard dissimilarities for presence/absence data, Bray–Curtis dissimilarities for abundance data, and a permutational multivariate analysis of variance using adonis in the vegan package. Multivariate homogeneity of group dispersions was calculated and tested using betadisper in the vegan package. Fungal trophic modes and traits were analyzed in FUNGuild [64]. All sequences generated from this study were deposited in the NCBI Sequence Read Archive under BioProject ID PRJNA905914.
3. Results
The eight samples yielded 1,044,406 reads (average: 130,551 reads/sample), and after filtering out low-quality and non-fungal sequences, a total of 484,999 reads (average: 60,625 reads/sample) remained for further analysis. Good’s coverage values indicated a sufficient number of sequences (0.999–1.000). Fungal diversity in the samples comprised 4 phyla, 15 classes, 38 orders, 65 families, 76 genera, and 153 OTUs. At the OTU level, a total of 116 and 78 fungal OTUs were detected from A. nasatum and A. vulgare, respectively. Among these, 41 OTUs were detected commonly in both Armadillidium species (Figure 1A). Among all the OTUs, Ascomycota was the phylum with the highest number of OTUs (100 OTUs), followed by Basidiomycota (50 OTUs). The top five classes with the most OTUs included three classes in Ascomycota (Dothideomycetes, Eurotiomycetes, and Sordariomycetes) and two classes in Basidiomycota (Agaricomycetes and Tremellomycetes). Sordariomycetes had the largest number of OTUs (53 OTUs) followed by Agaricomycetes (30 OTUs), Dothideomycetes (21 OTUs), Eurotiomycetes (18 OTUs), and Tremellomycetes (11 OTUs) (Figure 1B). The alpha diversity indices calculated from the fungal communities in two Armadillidium species differed significantly in terms of OTU richness (p = 0.009) and diversity (p = 0.038), but not in evenness (p = 0.181) (Figure 2). The number of OTUs and the Shannon–Weaver diversity index were significantly higher in A. nasatum compared to A. vulgare.
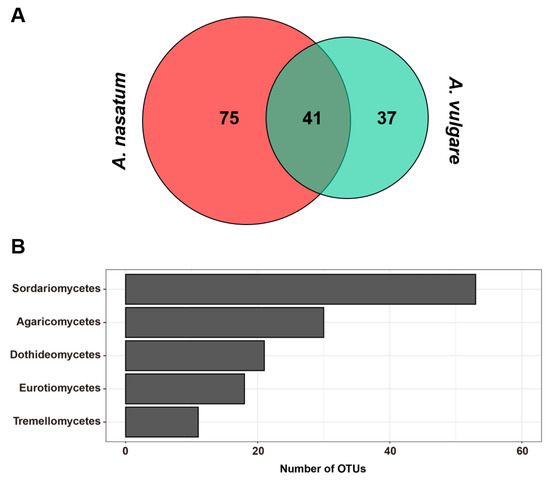
Figure 1.
(A) Venn diagrams showing number of OTUs in fungal communities associated with A. nasatum and A. vulgare. (B) The total number of OTUs for the Top 5 classes.
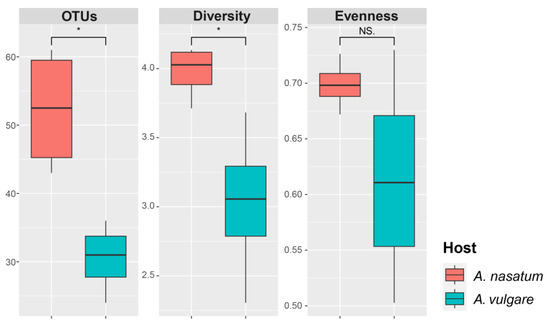
Figure 2.
Alpha diversity of fungal communities in A. nasatum and A. vulgare. OTUs: number of OTUs (richness measure), diversity: Shannon–Weaver diversity index, evenness: Pileu’s equitability index, asterisk (*): p < 0.05, NS.: p > 0.05.
Ordination analysis for the presence/absence dataset based on binary Jaccard dissimilarities showed that fungal communities did not differ significantly between hosts (p = 0.067) (Figure 3A). Additionally, beta dispersion of the fungal community in A. vulgare was relatively high, but there was no difference between hosts (p = 0.481) (Figure 3C). Bray–Curtis dissimilarities on abundance data yielded similar results: fungal communities did not differ significantly between hosts (p = 0.221) (Figure 3B), while beta dispersion was significantly higher in A. vulgare samples (p = 0.012) (Figure 3D).
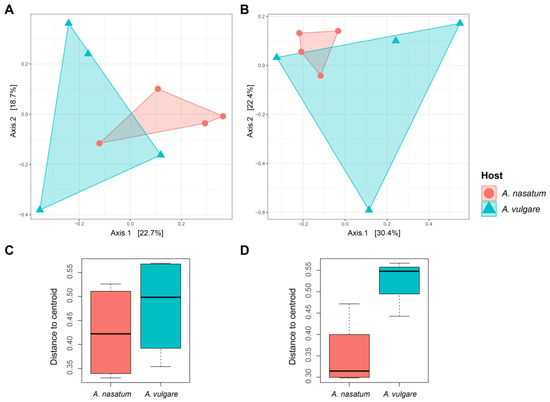
Figure 3.
Structures of fungal communities associated with A. nasatum and A. vulgare. PCoA plots using (A) binary Jaccard and (B) Bray–Curtis dissimilarities. Beta dispersion values based on (C) binary Jaccard and (D) Bray–Curtis dissimilarities.
Major fungal phyla associated with A. nasatum and A. vulgare included Ascomycota (33.8–65.1%) and Basidiomycota (34.9–66.2%) (Table 1; Figure 4A). The other phyla (Chytridiomycota and Mucoromycota) were detected with very low abundance: less than 0.2% in all samples except for the An3 sample (Chytridiomycota, 4.1%). At the class level, Agaricomycetes (28.9–66.0%) and Sordariomycetes (9.8–48.4%) were dominant in most samples, except for one A. vulgare sample (Av3), where Dothideomycetes (48.5%) was dominant. The two species differed in taxonomic-composition patterns (Figure 4B, Supplementary Figure S1). Across all A. nasatum samples, dominant taxa were Acanthophysium (Stereaceae, Russulales), Aspergillus, (Aspergillaceae, Eurotiales), Penicillium (Aspergillaceae, Eurotiales), Fusarium (Nectriaceae, Hypocreales), Trichoderma (Hypocreaceae, Hypocreales), and Purpureocillium (Ophiocordycipitaceae, Hypocreales). However, dominant taxa differed within A. vulgare samples (Figure 4B, Supplementary Figure S1). Penicillium and Phallus (Phallaceae, Phallales) were dominant on Av1; Ceratobasidium (Ceratobasidiaceae, Cantharellales), Fusarium, and Phallus on Av2; Leptospora (Dothideomycetes incertae sedis) on Av3; and Acanthophysium on Av4. Given the trophic mode of fungal communities, saprotrophs were dominant in all samples (55.8–78.9%) except for Av2, which was dominated by pathotroph-saprotroph-symbiotrophs (54.0%) (Figure 5A). Among saprotrophs, white rot (32.4–64.3%) and undefined saprotroph (34.0–45.2%) were the most abundant traits (Figure 5B).

Table 1.
Abundance percentage of fungal genera in A. nasatum and A. vulgare.
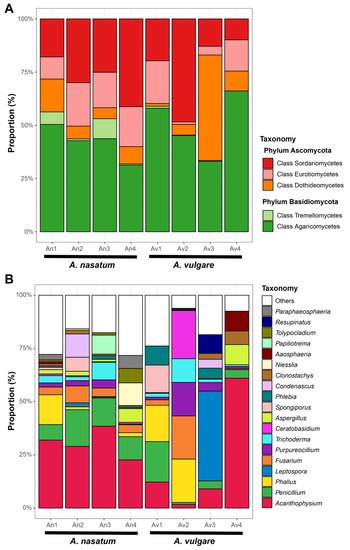
Figure 4.
Taxonomic composition of fungal communities in A. nasatum and A. vulgare. (A) Phyla and classes. (B) Genera.
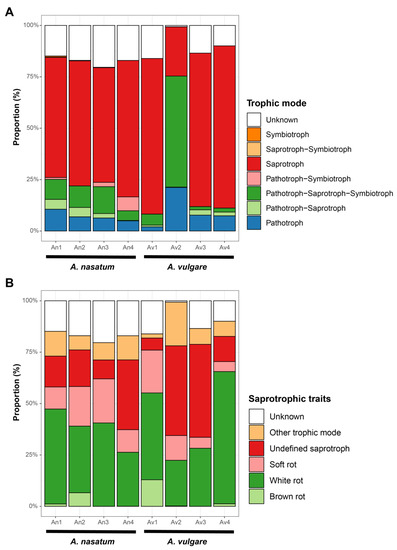
Figure 5.
Functional composition of fungal communities in A. nasatum and A. vulgare. (A) Trophic mode. (B) Saprotrophic traits.
4. Discussion
In this study, we used metabarcoding techniques to investigate for the first time fungal diversity associated with A. nasatum and A. vulgare. Previous culture-dependent approaches have uncovered only a few symbiotic fungi, but our method showed that many species have been overlooked. We detected a total of 153 fungal species in A. nasatum and A. vulgare. Given the higher richness and diversity of fungi associated with A. nasatum, most fungal OTUs from A. vulgare were also present in A. nasatum. Ordination analysis of both presence/absence and abundance data confirmed the similarity in fungal communities between the two species. One difference, however, was that the beta-diversity dispersion quantified based on Bray-Curtis dissimilarities was significantly larger in the fungal community of A. vulgare. This outcome indicates that dominant taxa varied considerably within A. vulgare-associated fungal communities. Further, our results suggest that A. vulgare may be more of a generalist in diet and microhabitat, while A. nasatum is more likely to be specialist. Hence, A. nasatum shares more dominant OTUs within fungal communities associated with conspecific Armadillidium.
Dominant Armadillidium-associated fungi belong to Ascomycota (Dothideomycetes, Eurotiomycetes, and Sordariomycetes) and Basidiomycota (Agaricomycetes). Fungi become associated with terrestrial isopods either via endosymbionts (e.g., obligate gut symbionts and parasites) or food [33,34,35]. While Asellariales and Harpellales (phylum Kickxellomycota) are known to be obligate gut-inhabiting fungi previously found in isopods [33,35], neither were detected in this study. Thus, the major source of fungi we detected likely came from food. Although Armadillidium species are omnivorous, their major sources of nutrients are wood debris and fallen leaves [65], with fungal hyphae in plant materials or soil providing a key nutrient source [41].
The dominant fungi detected in Armadillidium are saprotrophic, with the majority being white rot or undefined. Because their habitats are plant materials and organic layers, they are abundantly consumed by terrestrial isopods [40,66,67]. Saprotrophic fungi are known to have enzymes that degrade plant materials. For example, Aspergillus, Acanthophysium, Phallus, Penicillium, and Trichoderma are high in β-glucosidase, cellulase, chitinase, and proteases [68,69,70]. In addition, fungi (e.g., Aspergillus) associated with marine isopods Limnoria lignorum and Sphaeroma serratum have higher cellulase activity than bacteria from the same host [71]. Therefore, it is possible that fungal communities associated with Armadillidium species may provide enzymes to degrade organic materials and benefit host nutrition uptake. Some arthropods rely on microbial enzymes for digestion [26,72]. Generally, the gut microbiome can play this role, but sometimes the host ingests microorganisms for this purpose. However, the study did not test this hypothesis, so it is more relevant that the saprotrophic fungi were detected in high abundance in Armadillidium species because they decompose fallen leaves and isopods feed on the decaying plant materials. The noteworthy distribution pattern of saprotrophic traits in Armadillidium-associated fungal communities is the higher abundance of soft and white rots as compared to other traits (e.g., brown rot). Previous research on the wood-boring marine isopod, Limnoria lignorum, has revealed that the ingestion of wood fragments partially decomposed by fungi and bacteria aids in the isopod’s nutrient consumption [73]. This finding suggests that the decomposition of wood structures by fungi is crucial for effective nutrient digestion and consumption by isopods. Lignin is a heterogeneous polymer of aromatic residues and cellulose, which is a challenging material to decompose [74]. Since white rot and soft rot fungi are capable of degrading lignin [75], they assist isopods in extracting nutrients, such as cellulose, from wood, making it easier for them to consume. Consequently, Armadillidium likely prefer consuming wood degraded by white rot or soft rot fungi, leading to a higher abundance of these saprotrophic traits in Armadillidium-associated fungal communities.
While the results of this study provide important insights into the fungal diversity and community composition associated with Armadillidium species, there are a few limitations. First, it included a relatively small sample size and limited number of sampling sites. Only four individuals of each species were sampled from a single park in Gwacheon, South Korea, which may not fully represent the overall fungal community associated with Armadillidium. Second, the study was conducted within a specific geographic region, therefore, the results cannot be generalized to other regions without further investigation. Despite these limitations, this study provides valuable information on the fungal diversity associated with Armadillidium and highlights the importance of considering terrestrial isopod-associated fungi in biodiversity studies. Future studies with larger sample sizes and more extensive sampling across multiple regions will advance our understanding of the fungal communities associated with Armadillidium and other terrestrial isopods. Additionally, incorporating other environmental variables, such as soil properties and vegetation cover, will provide novel insights into the factors driving fungal community composition and diversity in these systems.
5. Conclusions
In conclusion, we found diverse fungi associated with A. nasatum and A. vulgare, most of which are saprotrophic. Fungal richness and diversity were significantly higher in A. nasatum than in A. vulgare, while community structure generally did not differ between the two species. One exception was in beta dispersion, suggesting that A. vulgare has a wide range of niche breadths. The saprotrophic fungi detected in Armadillidium species are thought to be either food for the isopods or a by-product of food consumption. However, the enzymes from these fungi may also contribute to host nutrient uptake and digestion. Further research on the diversity and functional traits of fungal communities will provide valuable insight into the biology and ecology of terrestrial isopods.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15040533/s1, Figure S1: Taxonomic composition of fungal communities in A. nasatum and A. vulgare.
Author Contributions
Conceptualization, S.-Y.O.; Methodology, S.-Y.O.; Validation, Y.C. and S.-Y.O.; Formal Analysis, S.-Y.O.; Investigation, Y.C. and S.-Y.O.; Resources, S.-Y.O.; Data Curation, Y.C. and S.-Y.O.; Writing—Original Draft Preparation, Y.C. and S.-Y.O.; Writing—Review and Editing, Y.C. and S.-Y.O.; Visualization, S.-Y.O.; Supervision, S.-Y.O.; Project Administration, Y.C. and S.-Y.O.; Funding Acquisition, Y.C. and S.-Y.O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Financial Program for Customized Research Capabilities in 2022 from Changwon National University.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Raw sequences were deposited in NCBI SRA database under accession number PRJNA905914.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Zimmer, M. Nutrition in Terrestrial Isopods (Isopoda: Oniscidea): An Evolutionary-Ecological Approach. Biol. Rev. 2002, 77, 455–493. [Google Scholar] [CrossRef] [PubMed]
- Schmalfuss, H. World Catalog of Terrestrial Isopods (Isopoda: Oniscidea). Stuttgarter Beiträge zur Naturkunde, Serie A 2003, 654, 1–341. [Google Scholar]
- Sfenthourakis, S.; Taiti, S. Patterns of Taxonomic Diversity among Terrestrial Isopods. ZooKeys 2015, 515, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C. Phylogeny of the Terrestrial Isopoda (Oniscidea): A Review. Arthropod Syst. Phylogeny 2008, 66, 191–226. [Google Scholar]
- Faberi, A.J.; López, A.N.; Clemente, N.L.; Manetti, P.L. Importance of Diet in the Growth, Survivorship and Reproduction of the No-Tillage Pest Armadillidium vulgare (Crustacea: Isopoda). Rev. Chil. Hist. Nat. 2011, 84, 407–417. [Google Scholar] [CrossRef]
- Špaldoňová, A.; Frouz, J. The Role of Armadillidium vulgare (Isopoda: Oniscidea) in Litter Decomposition and Soil Organic Matter Stabilization. Appl. Soil Ecol. 2014, 83, 186–192. [Google Scholar] [CrossRef]
- Zimmer, M.; Pennings, S.C.; Buck, T.L.; Carefoot, T.H. Species-Specific Patterns of Litter Processing by Terrestrial Isopods (Isopoda: Oniscidea) in High Intertidal Salt Marshes and Coastal Forests. Funct. Ecol. 2002, 16, 596–607. [Google Scholar] [CrossRef]
- Zimmer, M.; Pennings, S.C.; Buck, T.L.; Carefoot, T.H. Salt Marsh Litter and Detritivores: A Closer Look at Redundancy. Estuaries 2004, 27, 753–769. [Google Scholar] [CrossRef]
- Elisabeth, H. Evolutionary Adaptation of Oniscidean Isopods to Terrestrial Life: Structure, Physiology and Behavior. Terr. Arthropod Rev. 2011, 4, 95–130. [Google Scholar] [CrossRef]
- Guo, S.; Ren, M.; Song, S.; Wei, P.; Luo, J. Evaluation of Antinociceptive and Anti-Inflammatory Effects of Aqueous Extract of Armadillidium Vulgare Latreille. Chin. J. Integr. Med. 2017, 23, 138–145. [Google Scholar] [CrossRef]
- Alves, R.R.; Alves, H.N. The Faunal Drugstore: Animal-Based Remedies Used in Traditional Medicines in Latin America. J. Ethnobiol. Ethnomedicine 2011, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Liu, X.; Mikaye, M.S.; Zhao, H.; Zhang, Y. Traditional Chinese medicine in the treatment of high incidence diseases in cold areas: The thrombotic diseases. Frigid Zone Med. 2021, 1, 23–44. [Google Scholar] [CrossRef]
- Xu, L.; Li, Y.; Liu, Y.; Mi, H.; Jiang, X.; Sun, Y.; Zhao, H.; Chen, D.; Wang, L. A Comprehensive Evaluation of the Potential of Semiterrestrial Isopods, Ligia Exotica, as a New Animal Food. Sci. Rep. 2021, 11, 7213. [Google Scholar] [CrossRef] [PubMed]
- McMonigle, O. Pillbugs and Other Isopods: Cultivating Vivarium Clean-Up Crews and Feeders for Dart Frogs, Arachnids, and Insects; Coachwhip Publications: Greenville, OH, USA, 2013. [Google Scholar]
- Cordovez, V.; Dini-Andreote, F.; Carrion, V.; Raaijmakers, J. Ecology and Evolution of Plant Microbiomes. Annu. Rev. Microbiol. 2019, 73, 69–88. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.M.; Sun, E.W.; Rogers, G.B.; Keating, D.J. The Influence of the Gut Microbiome on Host Metabolism Through the Regulation of Gut Hormone Release. Front. Physiol. 2019, 10, 428. [Google Scholar] [CrossRef] [PubMed]
- Trevelline, B.K.; Kohl, K.D. The Gut Microbiome Influences Host Diet Selection Behavior. Proc. Natl. Acad. Sci. USA 2022, 119, e2117537119. [Google Scholar] [CrossRef]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of Microorganisms in the Evolution of Animals and Plants: The Hologenome Theory of Evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef]
- Gilbert, S.F.; McDonald, E.; Boyle, N.; Buttino, N.; Gyi, L.; Mai, M.; Prakash, N.; Robinson, J. Symbiosis as a Source of Selectable Epigenetic Variation: Taking the Heat for the Big Guy. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 671–678. [Google Scholar] [CrossRef]
- Feldhaar, H. Bacterial Symbionts as Mediators of Ecologically Important Traits of Insect Hosts. Ecol. Entomol. 2011, 36, 533–543. [Google Scholar] [CrossRef]
- Brucker, R.M.; Bordenstein, S.R. Speciation by Symbiosis. Trends Ecol. Evol. 2012, 27, 443–451. [Google Scholar] [CrossRef]
- Brucker, R.M.; Bordenstein, S.R. The Hologenomic Basis of Speciation: Gut Bacteria Cause Hybrid Lethality in the Genus Nasonia. Science 2013, 341, 667–669. [Google Scholar] [CrossRef] [PubMed]
- Esposti, M.D.; Romero, E.M. The Functional Microbiome of Arthropods. PLoS ONE 2017, 12, e0176573. [Google Scholar] [CrossRef] [PubMed]
- Jing, T.-Z.; Qi, F.-H.; Wang, Z.-Y. Most Dominant Roles of Insect Gut Bacteria: Digestion, Detoxification, or Essential Nutrient Provision? Microbiome 2020, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Malassigné, S.; Minard, G.; Vallon, L.; Martin, E.; Valiente Moro, C.; Luis, P. Diversity and Functions of Yeast Communities Associated with Insects. Microorganisms 2021, 9, 1552. [Google Scholar] [CrossRef] [PubMed]
- Schapheer, C.; Pellens, R.; Scherson, R. Arthropod-Microbiota Integration: Its Importance for Ecosystem Conservation. Front. Microbiol. 2021, 12, 702763. [Google Scholar] [CrossRef]
- Dittmer, J.; Lesobre, J.; Moumen, B.; Bouchon, D. Host Origin and Tissue Microhabitat Shaping the Microbiota of the Terrestrial Isopod Armadillidium vulgare. FEMS Microbiol. Ecol. 2016, 92, fiw063. [Google Scholar] [CrossRef]
- Wenzel, M.A.; Douglas, A.; Piertney, S.B. Microbiome Composition within a Sympatric Species Complex of Intertidal Isopods (Jaera albifrons). PLoS ONE 2018, 13, e0202212. [Google Scholar] [CrossRef]
- Delhoumi, M.; Catania, V.; Zaabar, W.; Tolone, M.; Quatrini, P.; Achouri, M.S. The Gut Microbiota Structure of the Terrestrial Isopod Porcellionides pruinosus (Isopoda: Oniscidea). Eur. Zool. J. 2020, 87, 357–368. [Google Scholar] [CrossRef]
- Newbound, M.; Mccarthy, M.A.; Lebel, T. Fungi and the Urban Environment: A Review. Landsc. Urban Plan. 2010, 96, 138–145. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in Aquatic Ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354. [Google Scholar] [CrossRef]
- Bahram, M.; Netherway, T. Fungi as Mediators Linking Organisms and Ecosystems. FEMS Microbiol. Rev. 2022, 46, fuab058. [Google Scholar] [CrossRef] [PubMed]
- Cafaro, M.J. Baltomyces, a New Genus of Gut-Inhabiting Fungus in an Isopod. Mycologia 1999, 91, 517–519. [Google Scholar] [CrossRef]
- Cafaro, M.J. Gut Fungi of Isopods: The Genus Palavascia. Mycologia 2000, 92, 361–369. [Google Scholar] [CrossRef]
- White, M.M. Legerioides, a New Genus of Harpellales in Isopods and Other Trichomycetes from New England, USA. Mycologia 1999, 91, 1021–1030. [Google Scholar] [CrossRef]
- Jaber, S.; Mercier, A.; Knio, K.; Brun, S.; Kambris, Z. Isolation of Fungi from Dead Arthropods and Identification of a New Mosquito Natural Pathogen. Parasit. Vectors 2016, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, J.; Liu, J.; Zhu, H.; Sun, B.; Wang, J.; Zhang, J.; Luo, Z.; Yao, G.; Xue, Y.; et al. Armochaetoglobins A–J: Cytochalasan Alkaloids from Chaetomium globosum TW1-1, a Fungus Derived from the Terrestrial Arthropod Armadillidium vulgare. J. Nat. Prod. 2015, 78, 1193–1201. [Google Scholar] [CrossRef]
- Han, W.-B.; Wang, G.-Y.; Tang, J.-J.; Wang, W.-J.; Liu, H.; Gil, R.R.; Navarro-Vázquez, A.; Lei, X.; Gao, J.-M. Herpotrichones A and B, Two Intermolecular [4 + 2] Adducts with Anti-Neuroinflammatory Activity from a Herpotrichia Species. Org. Lett. 2020, 22, 405–409. [Google Scholar] [CrossRef]
- Zhai, Y.-J.; Huo, G.-M.; Wei, J.; Lin, L.-B.; Zhang, Q.; Li, J.-N.; Chen, X.; Han, W.-B.; Gao, J.-M. Structures and Absolute Configurations of Butenolide Derivatives from the Isopod-Associated Fungus Pidoplitchkoviella terricola. Phytochemistry 2022, 193, 112981. [Google Scholar] [CrossRef]
- Crowther, T.W.; Stanton, D.W.G.; Thomas, S.M.; A’Bear, A.D.; Hiscox, J.; Jones, T.H.; Voříšková, J.; Baldrian, P.; Boddy, L. Top-down Control of Soil Fungal Community Composition by a Globally Distributed Keystone Consumer. Ecology 2013, 94, 2518–2528. [Google Scholar] [CrossRef]
- Póss, A.M.; Bogdányi, F.T.; Tóth, F. Consumption of Fungi-Infected Fallen Pear Leaves by the Common Woodlouse. Acta Phytopathol. Entomol. Hung. 2022, 57, 79–91. [Google Scholar] [CrossRef]
- Nayfach, S.; Roux, S.; Seshadri, R.; Udwary, D.; Varghese, N.; Schulz, F.; Wu, D.; Paez-Espino, D.; Chen, I.-M.; Huntemann, M.; et al. A Genomic Catalog of Earth’s Microbiomes. Nat. Biotechnol. 2021, 39, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshizawa, S. The OceanDNA MAG Catalog Contains over 50,000 Prokaryotic Genomes Originated from Various Marine Environments. Sci. Data 2022, 9, 305. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global Diversity and Geography of Soil Fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Anslan, S.; Nilsson, R.H.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L.; Bahram, M. Great Differences in Performance and Outcome of High-Throughput Sequencing Data Analysis Platforms for Fungal Metabarcoding. MycoKeys 2018, 39, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Phukhamsakda, C.; Nilsson, R.H.; Bhunjun, C.S.; de Farias, A.R.G.; Sun, Y.-R.; Wijesinghe, S.N.; Raza, M.; Bao, D.-F.; Lu, L.; Tibpromma, S.; et al. The Numbers of Fungi: Contributions from Traditional Taxonomic Studies and Challenges of Metabarcoding. Fungal Divers. 2022, 114, 327–386. [Google Scholar] [CrossRef]
- Kwon, D.H. Terrestrial Isopoda (Crustacea) from Korea. Korean J. Zool. 1993, 36, 133–158. [Google Scholar]
- Kwon, D.H. Terrestrial Isopoda (Crustacea) from Cheju Island, Korea. Anim. Syst. Evol. Divers. 1995, 11, 509–538. [Google Scholar]
- Song, J.-H. A New Record of Porcellio scaber (Isopoda: Oniscidea: Porcellionidae) from South Korea, with Notes on Its Variation. Anim. Syst. Evol. Divers. 2020, 36, 309–315. [Google Scholar] [CrossRef]
- Shultz, J.W. A Guide to the Identification of the Terrestrial Isopoda of Maryland, U.S.A. (Crustacea). ZooKeys 2018, 801, 207–228. [Google Scholar] [CrossRef]
- Kim, E.J.; Moon, D.H.; Jo, U.B. Taxonomic Study on the Four Species of Terrestrial Isopods, Oniscoidae, from the Pusan Area in Korea. J. Sci. 1990, 49, 225–251. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.L. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: San Diego, CA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Põlme, S.; Riit, T.; Liiv, I.; Kõljalg, U.; Kisand, V.; Nilsson, H.; Hildebrand, F.; et al. Shotgun Metagenomes and Multiple Primer Pair-Barcode Combinations of Amplicons Reveal Biases in Metabarcoding Analyses of Fungi. MycoKeys 2015, 10, 1–43. [Google Scholar] [CrossRef]
- Tedersoo, L.; Tooming-Klunderud, A.; Anslan, S. PacBio Metabarcoding of Fungi and Other Eukaryotes: Errors, Biases and Perspectives. New Phytol. 2018, 217, 1370–1385. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2′s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Wickham, H. Package ‘Ggplot2′: Elegant Graphics for Data Analysis, Version 3.2.1.; Springer: New York, NY, USA, 2016. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, Version 2.6-4. 2020, 1–263. Available online: https://CRAN.R-project.org/package=vegan (accessed on 20 November 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An Open Annotation Tool for Parsing Fungal Community Datasets by Ecological Guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Paris, O.H. The Ecology of Armadillidium vulgare (Isopoda: Oniscoidea) in California Grassland: Food, Enemies, and Weather. Ecol. Monogr. 1963, 33, 1–22. [Google Scholar] [CrossRef]
- Crowther, T.W.; Boddy, L.; Hefin Jones, T. Functional and Ecological Consequences of Saprotrophic Fungus–Grazer Interactions. ISME J. 2012, 6, 1992–2001. [Google Scholar] [CrossRef] [PubMed]
- Crowther, T.W.; Thomas, S.M.; Maynard, D.S.; Baldrian, P.; Covey, K.; Frey, S.D.; van Diepen, L.T.A.; Bradford, M.A. Biotic Interactions Mediate Soil Microbial Feedbacks to Climate Change. Proc. Natl. Acad. Sci. USA 2015, 112, 7033–7038. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, Y.; Wang, H.; Zu, Y. Differences in the Activities of Eight Enzymes from Ten Soil Fungi and Their Possible Influences on the Surface Structure, Functional Groups, and Element Composition of Soil Colloids. PLoS ONE 2014, 9, e111740. [Google Scholar] [CrossRef] [PubMed]
- de Passos, D.F.; Pereira, N.; Castro, A.M. de A Comparative Review of Recent Advances in Cellulases Production by Aspergillus, Penicillium and Trichoderma Strains and Their Use for Lignocellulose Deconstruction. Curr. Opin. Green Sustain. Chem. 2018, 14, 60–66. [Google Scholar] [CrossRef]
- Yoon, S.-M.; Kim, Y.-S.; Kim, Y.-K.; Kim, T.-J. A Novel Endo-β-1,4-Xylanase from Acanthophysium sp. KMF001, a Wood Rotting Fungus. J. Korean Wood Sci. Technol. 2018, 46, 670–680. [Google Scholar] [CrossRef]
- El-Shanshoury, A.R.; Mona, M.H.; Shoukr, F.A.; El-Bossery, A.M. The Enumeration and Characterization of Bacteria and Fungi Associated with Marine Wood-Boring Isopods, and the Ability of These Microorganisms to Digest Cellulose and Wood. Mar. Biol. 1994, 119, 321–326. [Google Scholar] [CrossRef]
- Banerjee, S.; Maiti, T.K.; Roy, R.N. Enzyme Producing Insect Gut Microbes: An Unexplored Biotechnological Aspect. Crit. Rev. Biotechnol. 2022, 42, 384–402. [Google Scholar] [CrossRef]
- Daniel, G.; Nilsson, T.; Cragg, S. Limnoria lignorum Ingest Bacterial and Fungal Degraded Wood. Holz Als Roh-Werkst. 1991, 49, 488–490. [Google Scholar] [CrossRef]
- Wong, D.W.S. Structure and Action Mechanism of Ligninolytic Enzymes. Appl. Biochem. Biotechnol. 2009, 157, 174–209. [Google Scholar] [CrossRef]
- Cragg, S.M.; Beckham, G.T.; Bruce, N.C.; Bugg, T.D.; Distel, D.L.; Dupree, P.; Etxabe, A.G.; Goodell, B.S.; Jellison, J.; McGeehan, J.E.; et al. Lignocellulose Degradation Mechanisms across the Tree of Life. Curr. Opin. Chem. Biol. 2015, 29, 108–119. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).