Genetic Variability and Connectivity in the Western Mediterranean Populations of the Bathyal Crab Geryon longipes
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Sequencing
2.2. Data Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caley, M.J.; Carr, M.H.; Hixon, M.A.; Hughes, T.P.; Jones, G.; Menge, B.A. Recruitment and the local dynamics of open marine populations. Annu. Rev. Ecol. Syst. 1996, 27, 477–500. [Google Scholar] [CrossRef]
- Pérez-Ruzafa, A.; González-Wängüemert, M.; Lenfant, P.; Marco, C.; García-Charton, J.A. Effects of fishing protection on the genetic structure of fish populations. Biol. Conserv. 2006, 129, 244–255. [Google Scholar] [CrossRef]
- Calderón, I.; Ortega, N.; Duran, S.; Becerro, M.; Pascual, M.; Turon, X. Finding the relevant scale: Clonality and genetic structure in a marine invertebrate (Crambe crambe, Porifera). Mol. Ecol. 2007, 16, 1799–1810. [Google Scholar] [CrossRef]
- Palero, F.; Abelló, P.; Macpherson, E.; Gristina, M.; Pascual, M. Phylogeography of the European spiny lobster (Palinurus elephas): Influence of current oceanographical features and historical processes. Mol. Phylogenet. Evol. 2008, 48, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L.J.; Fulton, C.J.; Hogg, A.M.; Joyce, K.E.; Radford, B.T.M.; Fraser, C.I. Climate-driven changes to ocean circulation and their inferred impacts on marine dispersal patterns. Glob. Ecol. Biogeogr. 2016, 25, 923–939. [Google Scholar] [CrossRef]
- Pascual, M.; Rives, B.; Schunter, C.; Macpherson, E. Impact of life history traits on gene flow: A multispecies systematic review across oceanographic barriers in the Mediterranean Sea. PLoS ONE 2017, 12, e0176419. [Google Scholar] [CrossRef] [PubMed]
- Canals, M.; Puig, P.; Durrieu de Madron, X.; Heussner, S.; Palanques, A.; Fabres, J. Flushing submarine canyons. Nature 2006, 444, 354–357. [Google Scholar] [CrossRef]
- Kelly, P.; Sulkin, S.D.; van Heukelem, W.F. A dispersal model for larvae of the deep sea red crab Geryon quinquedens based on behavioral regulation of vertical migration in the hatching stage. Mar. Biol. 1982, 72, 35–43. [Google Scholar] [CrossRef]
- Torres, A.P.; Reglero, P.; Hidalgo, M.; Abelló, P.; Simao, D.S.; Alemany, F.; Massutí, E.; Dos Santos, A. Contrasting patterns in the vertical distribution of decapod crustaceans throughout ontogeny. Hydrobiologia 2018, 808, 137–152. [Google Scholar] [CrossRef]
- Fernández, V.; Dietrich, D.E.; Haney, R.L.; Tintoré, J. Mesoscale, seasonal and interannual variability in the Mediterranean Sea using a numerical ocean model. Prog. Oceanogr. 2005, 66, 321–340. [Google Scholar] [CrossRef]
- Rio, M.H.; Poulain, P.M.; Pascual, A.; Mauri, E.; Larnicol, G.; Santoleri, R. A mean dynamic topography of the Mediterranean Sea computed from altimetric data, in-situ measurements and a general circulation model. J. Mar. Syst. 2007, 65, 484–508. [Google Scholar] [CrossRef]
- Coll, M.; Piroddi, C.; Steenbeek, J.; Kaschner, K.; Ben Rais Lasram, F.; Aguzzi, J.; Ballesteros, E.; Bianchi, C.N.; Corbera, J.; Dailianis, T.; et al. The Biodiversity of the Mediterranean Sea: Estimates, Patterns, and Threats. PLoS ONE 2010, 5, e11842. [Google Scholar] [CrossRef] [PubMed]
- Vasilakopoulos, P.; Raitsos, D.E.; Tzanatos, E.; Maravelias, C.D. Resilience and regime shifts in a marine biodiversity hotspot. Sci. Rep. 2017, 7, 13647. [Google Scholar] [CrossRef]
- Claudet, J.; Loiseau, C.; Sostres, M.; Zupan, M. Underprotected marine protected areas in a global biodiversity hotspot. One Earth 2020, 2, 380–384. [Google Scholar] [CrossRef]
- García-Merchán, V.H.; Robainas-Barcia, A.; Abelló, P.; Macpherson, E.; Palero, F.; García-Rodríguez, M.; Gil de Sola, L.; Pascual, M. Phylogeographic patterns of decapod crustaceans at the Atlantic-Mediterranean transition. Mol. Phylogen. Evol. 2012, 62, 664–672. [Google Scholar] [CrossRef]
- Pascual, M.; Palero, F.; García-Merchán, V.H.; Macpherson, E.; Robainas-Barcia, A.; Mestres, F.; Roda, T.; Abelló, P. Temporal and spatial genetic differentiation in the crab Liocarcinus depurator across the Atlantic-Mediterranean transition. Sci. Rep. 2016, 6, 29892. [Google Scholar] [CrossRef] [PubMed]
- Mestres, F.; Sellés, M.; Rojo, E.; Lagares, C.; Serra, B.; Ojeda, V.; Abelló, P. La conectividad entre poblaciones del cangrejo marino Liocarcinus depurator en la transición Atlanto-mediterránea. In X Foro Iberoamericano De Los Recursos Marinos Y La Acuicultura; AFRIMAR-AFIRMA: Las Palmas de Gran Canarias, Spain, 2021; pp. 495–511. [Google Scholar]
- Ojeda, V.; Serra, B.; Lagares, C.; Rojo-Francàs, E.; Sellés, M.; Marco-Herrero, E.; García, E.; Farré, M.; Arenas, C.; Abelló, P.; et al. Interannual fluctuations in connectivity among crab populations (Liocarcinus depurator) along the Atlantic-Mediterranean transition. Sci. Rep. 2022, 12, 9797. [Google Scholar] [CrossRef] [PubMed]
- Abelló, P.; Carbonell, A.; Torres, P. Biogeography of epibenthic crustaceans on the shelf and upper slope off the Iberian Peninsula Mediterranean coasts: Implications for the establishment of natural management areas. Sci. Mar. 2002, 66 (Suppl. 2), 183–198. [Google Scholar] [CrossRef]
- Rufino, M.M.; Abelló, P.; Yule, A.B.; Torres, P. Geographic, bathymetric and inter-annual variability in the distribution of Liocarcinus depurator (Brachyura: Portunidae) along the Mediterranean coast of the Iberian Peninsula. Sci. Mar. 2005, 69, 503–518. [Google Scholar] [CrossRef]
- Clark, P.F. A comparative study of zoeal morphology in the genus Liocarcinus (Crustacea: Braclhyura: Portunidae). Zool. J. Linn. Soc. 1984, 82, 273–290. [Google Scholar] [CrossRef]
- Ingle, R.W. Larval Stages of Northeastern Atlantic Crabs. An Illustrated Key; Chapman & Hall: London, UK, 1992. [Google Scholar]
- Abelló, P.; Guerao, G. Temporal variability in the vertical and mesoscale spatial distribution of crab megalopae (Crustacea: Decapoda) in the northwestern Mediterranean. Estuar. Coast. Shelf Sci. 1999, 49, 129–139. [Google Scholar] [CrossRef]
- Abelló, P.; Valladares, F.J. Bathyal decapod crustaceans of the Catalan Sea (northwestern Mediterranean). Mésogée 1988, 48, 97–102. [Google Scholar]
- Cartes, J.E. Deep-sea decapod fauna of the western Mediterranean: Bathymetric distribution and biogeographic aspects. Crustaceana 1993, 65, 29–40. [Google Scholar] [CrossRef]
- Company, J.B.; Maiorano, P.; Tselepides, A.; Politou, C.Y.; Plaity, W.; Rotllant, G.; Sardá, F. Deep-sea decapod crustaceans in the western and central Mediterranean Sea: Preliminary aspects of species distribution, biomass and population structure. Sci. Mar. 2004, 68 (Suppl. 3), 73–86. [Google Scholar] [CrossRef]
- Attrill, M.J.; Hartnoll, R.G.; Rice, A.L.; Thurston, M.H. A depth-related distribution of the red crab, Geryon trispinosus (Herbst) [=G. tridens Kroyer]: Indications of vertical migration. Progr. Oceanogr. 1990, 24, 197–206. [Google Scholar] [CrossRef]
- Attrill, M.; Hartnoll, R.G.; Rice, A.L. Aspects of the biology of the deep-sea crab Geryon trispinosus from the Porcupine Seabight. J. Mar. Biol. Assoc. 1991, 71, 311–328. [Google Scholar] [CrossRef]
- Udekem d’Acoz, C.D. Inventaire et distribution des crustacés décapodes de l’Atlantique nord oriental, de la Méditerranée et des eaux continentales adjacentes au nord de 25° N. Mus. Nat. Hist. Nat. 1999, 40, 383. [Google Scholar]
- WoRMS. Geryon trispinosus (Herbst, 1803). Available online: https://www.marinespecies.org/aphia.php?p=taxdetails&id=107374 (accessed on 3 February 2023).
- Abelló, P. Crustáceos. Los Decápodos. Los Geriónidos. In La Riqueza de Nuestros Mares: Especies de Interés del Sector Pesquero Español; Ministerio de Medio Ambiente y Medio Rural y Marino: Madrid, Spain, 2008; pp. 623–628. [Google Scholar]
- Amores, A.; Rueda, L.; Monserrat, S.; Guijarro, B.; Pasqual, C.; Massutí, E. Influence of the hydrodynamic conditions on the accessibility of Aristeus antennatus and other demersal species to the deep water trawl fishery off the Balearic Islands (western Mediterranean). J. Mar. Syst. 2014, 138, 203–210. [Google Scholar] [CrossRef]
- Guerao, G.; Abelló, P.; Castejón, M.R. Morphology of the larval stages of the deep-sea crab Geryon longipes (Brachyura, Geryonidae). J. Nat. Hist. 1996, 30, 505–521. [Google Scholar] [CrossRef]
- Carbonell, A.; Tor, A.; Álvarez-Berasategui, D.; Vélez-Belchi, P.; Dos Santos, A.; Balbín, R.; Alemany, F. Environmental driving forces determining the epipelagic decapod larval community distribution in the Balearic Sea (Western Mediterranean). Crustaceana 2014, 87, 686–714. [Google Scholar] [CrossRef]
- Carbonell, A.; Aparicio-González, A.; Papiol, V.; Cartes, J.E. Composition and distribution of the larval decapod community in the deep sea of the Western Mediterranean Sea Balearic Sub-basin. Fish. Oceanograp. 2021, 30, 205–218. [Google Scholar] [CrossRef]
- Perry, H.M.; Waller, R.; Stuck, L.; Stuck, K.; Erdman, R.; Blake, N.; Lockhart, F.; Lindberg, W. Occurrence of Chaceon larvae in plankton samples from slope waters of the northeastern Gulf of Mexico. Gulf Res. Rep. 1991, 8, 313–315. [Google Scholar] [CrossRef]
- Landeria, J.M.; Tamura, H. Morphology of the first zoea of Chaceon affinis (A. Milne-Edwards and Bouvier, 1894) and occurrence of Chaceon spp. larvae (Decapoda: Brachyura: Gerynonidae) in the Canary Islands waters, Northeastern Atlantic. Zootaxa 2018, 4413, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, J.; Gil de Sola, L.; Papaconstantinou, C.; Relini, G.; Souplet, A. The general specifications of the MEDITS surveys. Sci. Mar. 2002, 66 (Suppl. 2), 9–17. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 1994, 3, 294–299. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP v6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Cadiou, Y. Karto: Programme de Représentation Géographique, Version 5.2; IFREMER: Nantes, France, 1994. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community ecology package. R Package Version 2016, 2, 321–326. [Google Scholar]
- Schizas, N.V. Misconceptions regarding nuclear mitochondrial pseudogenes (Numts) may obscure detection of mitochondrial evolutionary novelties. Aquat. Biol. 2012, 17, 91–96. [Google Scholar] [CrossRef]
- Williams, S.T.; Knowlton, N. Mitochondrial pseudogenes are pervasive and often insidious in the snapping shrimp genus Alpheus. Mol. Biol. Evol. 2001, 18, 1484–1493. [Google Scholar] [CrossRef]
- Buhay, J.E. ‘COI-like’ sequences are becoming problematic in molecular systematic and DNA barcoding studies. J. Crust. Biol. 2009, 29, 96–110. [Google Scholar] [CrossRef]
- Schubart, C.D. Mitochondrial DNA and decapod phylogenies: The importance of pseudogenes and primer optimization. In Decapod Crustacean Phylogenetics; Martin, J.W., Crandall, K.A., Felder, D.L., Eds.; CRC Press: Boca Raton, FL, USA, 2009; pp. 47–65. [Google Scholar]
- Roldán, M.I.; Heras, S.; Patellani, R.; Maltagliati, F. Analysis of genetic structure of the red shrimp Aristeus antennatus from the Western Mediterranean employing two mitochondrial regions. Genetica 2009, 136, 1–4. [Google Scholar] [CrossRef]
- Cartes, J.E.; Maynou, F.; Moranta, J.; Massutí, E.; Lloris, D.; Morales-Nin, B. Patterns of bathymetric distribution among deep-sea fauna at local spatial scale: Comparison of mainland vs. insular areas. Prog. Oceanogr. 2004, 60, 29–45. [Google Scholar] [CrossRef]
- Politou, C.Y.; Maiorano, P.; D’Onghia, G.; Mytilineou, C. Deep-water decapod crustacean fauna of the Eastern Ionian Sea. Belg. J. Zool. 2005, 135, 235–241. [Google Scholar]
- Etter, R.J.; Rex, M.A.; Chase, M.R.; Quattro, J.M. Population differentiation decreases with depth in deep-sea bivalves. Evolution 2005, 59, 1479–1491. [Google Scholar] [CrossRef]
- Gaither, M.R.; Violi, B.; Gray, H.W.I.; Neat, F.; Drazen, J.C.; Grubbs, R.D.; Roa-Varón, A.; Sutton, T.; Hoelzel, A.R. Depth as a driver of evolution in the deep sea: Insights from grenadiers (Gadiformes: Macrouridae) of the genus Coryphaenoides. Mol. Phylogenet. Evol. 2016, 48, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Kalkan, E.; Karhan, S.Ü.; Bilgin, R. Population genetic structure of the marbled crab, Pachygrapsus marmoratus from Turkish coasts of the Black Sea and the Eastern Mediterranean. Rapp. Comm. Int. Mer Médit. 2013, 40, 713. [Google Scholar]
- Deli, T.; Said, K.; Chatti, N. Genetic differentiation among populations of the green crab Carcinus aestuarii (Nardo, 1847) (Brachyura, Carcinidae) from the eastern and western Mediterranean coast of Tunisia. Acta Zool. Bulg. 2015, 67, 327–335. [Google Scholar]
- Tavares, A.I.; Cabezas, M.P.; Xavier, R.; Branco, M.; Lima, F.P.; Seabra, R.; Ribeiro, P.A.; Lopes, E.P.; Santos, A.M. Phylogeography and phylogeny of the genus Acanthonyx (Decapoda, Epialtidae) in the north-east Atlantic and Mediterranean. Zool. Scr. 2017, 46, 571–583. [Google Scholar] [CrossRef]
- Fernández, M.V.; Heras, S.; Maltagliati, F.; Rodán, M.I. Deep genetic divergence in giant red shrimp Aristaeomorpha foliacea (Risso, 1827) across a wide distributional range. J. Sea Res. 2013, 76, 146–153. [Google Scholar] [CrossRef]
- Manning, R.B.; Holthuis, L.B. Two new genera and nine species of geryonid crabs (Crustacea, Decapoda, Geryonidae). Proc. Biol. Soc. Wash. 1989, 102, 50–77. [Google Scholar]
- Hernández, M.; Martín, M.V.; Herrador-Gómez, P.M.; Jiménez, S.; Hernández-González, C.; Barreiro, S.; Sarralde, S.; van Zyl, B.J.; Gamatham, J.C.; Almeida, T.; et al. Mitochondrial COI and 16S rDNA sequences support morphological identification and biogeography of deep-sea red crabs of the genus Chaceon (Crustacea, Decapoda, Geryonidae) in the Eastern Central and South Atlantic Ocean. PLoS ONE 2019, 14, e0211717. [Google Scholar] [CrossRef] [PubMed]
- Marco-Herrero, E.; Abelló, P.; Drake, P.; García-Raso, J.E.; González-Gordillo, J.I.; Guerao, G.; Palero, F.; Cuesta, J.A. Annotated checklist of brachyuran crabs (Crustacea: Decapoda) of the Iberian Peninsula (SW Europe). Sci. Mar. 2015, 79, 243–256. [Google Scholar] [CrossRef]
- Dittel, A.I.; Epifanio, C.E. Seasonal abundance and vertical-distribution of crab larvae in Delaware Bay. Estuaries 1982, 5, 197–202. [Google Scholar] [CrossRef]
- Zeng, C.; Naylor, E. Occurrence in coastal waters and endogenous tidal swimming rhythms of late megalopae of the shore crab Carcinus maenas: Implications for onshore recruitment. Mar. Ecol. Prog. Ser. 1996, 136, 69–79. [Google Scholar] [CrossRef]
- Cartes, J.E. Diets of deep-sea brachyuran crabs in the Western Mediterranean Sea. Mar. Biol. 1993, 117, 449–457. [Google Scholar] [CrossRef]
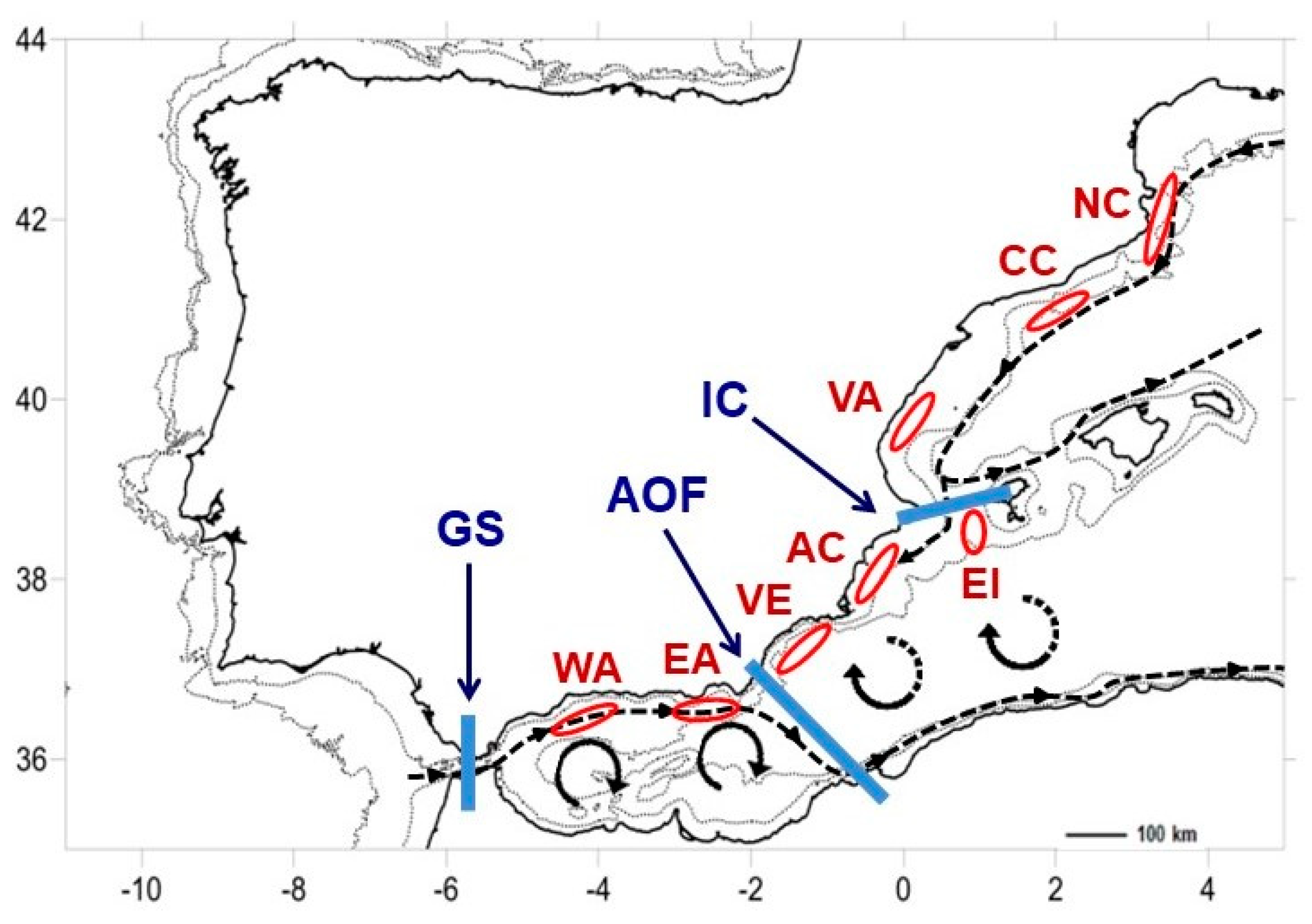



| Population | Latitude | Longitude | Depth | n | Reference |
|---|---|---|---|---|---|
| West Alboran (WA) | 36.312 N | 4.340 W | 770 | 15 | This study |
| East Alboran (EA) | 36.581 N | 2.498 W | 528 | 14 | This study |
| Vera (VE) | 36.856 N | 1.759 W | 714 | 16 | This study |
| Ibiza Is. (EI) | 38.838 N | 0.842 E | 681 | 15 | This study |
| Alicante (AC) | 38.069 N | 0.040 W | 581 | 16 | This study |
| Valencia (VA) | 39.453 N | 0.156 E | 561 | 16 | This study |
| Central Catalonia (CC) | 41.161 N | 2.358 E | 665 | 17 | This study |
| North Catalonia (NC) | 41.391 N | 3.269 E | 622.5 | 15 | This study |
| Nahariyya, Israel (IS) | 33.050 N | 34.830 E | 1043 | 1 | Tel Aviv University |
| Castellammare del Golfo, Sicily (CG) | 35.730 N | 14.050 E | 605 | 2 | Matzen da Silva et al., 2011 |
| South coast of Portugal (SP) | 36.600 N | 8.030 W | 752 | 5 | Matzen da Silva et al., 2011 |
| SW coast of Portugal (WP) | 37.540 N | 9.190 W | 612 | 2 | Matzen da Silva et al., 2011 |
| NW of St. Kilda, Scotland (SC) | 58.170 N | 9.000 W | 600 | 3 | Matzen da Silva et al., 2011 |
| Population | n | h | S | Hd | π × 100 |
|---|---|---|---|---|---|
| WA | 15 | 4 | 2 | 0.714 ± 0.081 | 0.223 ± 0.020 |
| EA | 14 | 4 | 2 | 0.495 ± 0.151 | 0.146 ± 0.044 |
| VE | 16 | 5 | 5 | 0.708 ± 0.094 | 0.246 ± 0.065 |
| EI | 15 | 3 | 2 | 0.590 ± 0.106 | 0.117 ± 0.027 |
| AC | 16 | 2 | 1 | 0.458 ± 0.095 | 0.080 ± 0.017 |
| VA | 16 | 3 | 2 | 0.658 ± 0.075 | 0.137 ± 0.024 |
| CC | 17 | 3 | 2 | 0.699 ± 0.049 | 0.154 ± 0.020 |
| NC | 15 | 2 | 1 | 0.343 ± 0.128 | 0.060 ± 0.022 |
| Population | Species | n | h | h/n | Hd | π × 100 |
|---|---|---|---|---|---|---|
| WA | G. long. | 15 | 4 | 0.267 | 0.714 ± 0.081 | 0.223 ± 0.020 |
| L. dep. | 24 | 11 | 0.458 | 0.815 ± 0.063 | 0.431 ± 0.055 | |
| EA | G. long. | 14 | 4 | 0.286 | 0.495 ± 0.151 | 0.146 ± 0.044 |
| L. dep. | 23 | 8 | 0.348 | 0.581 ± 0.120 | 0.246 ± 0.075 | |
| AC | G. long. | 16 | 2 | 0.125 | 0.458 ± 0.095 | 0.080 ± 0.017 |
| L. dep. | 25 | 7 | 0.280 | 0.633 ± 0.104 | 0.301 ± 0.402 | |
| VA | G. long. | 16 | 3 | 0.188 | 0.658 ± 0.075 | 0.137 ± 0.024 |
| L. dep. | 41 | 8 | 0.195 | 0.316 ± 0.095 | 0.093 ± 0.039 | |
| CC | G. long. | 17 | 3 | 0.176 | 0.699 ± 0.049 | 0.154 ± 0.020 |
| L. dep. | 6 | 2 | 0.333 | 0.333 ± 0.215 | 0.063 ± 0.083 | |
| TOTAL | G. long | 78 | 5 | 0.064 | 0.734 ± 0.024 | 0.203 ± 0.011 |
| L. dep. | 119 | 24 | 0.202 | 0.592 ± 0.052 | 0.300 ± 0.034 |
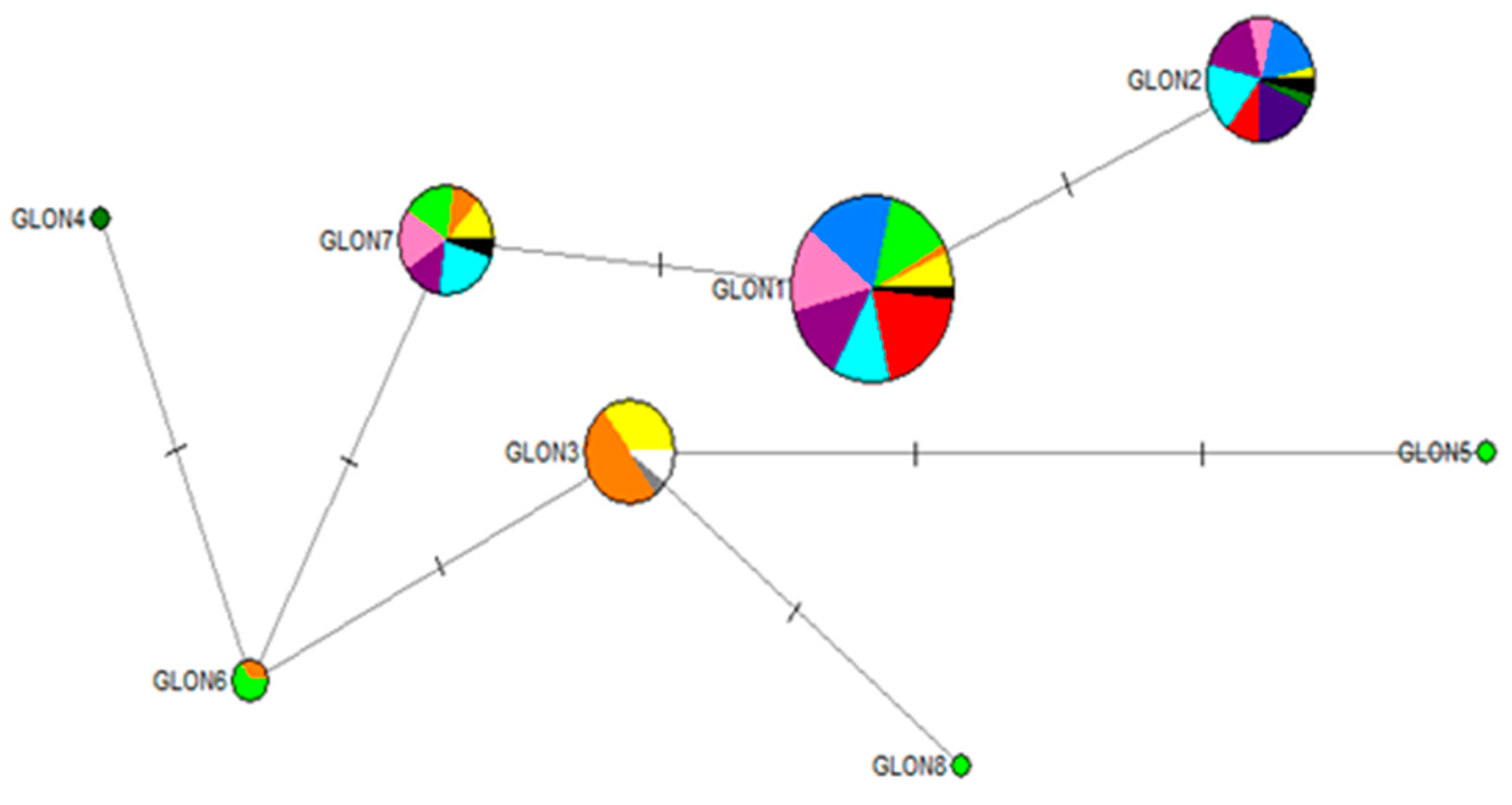
| Population | Haplotype | |||||||
|---|---|---|---|---|---|---|---|---|
| Glon_1 | Glon_2 | Glon_3 | Glon_4 | Glon_5 | Glon_6 | Glon_7 | Glon_8 | |
| WA | + | + | + | − | − | − | + | − |
| EA | + | − | + | − | − | + | + | − |
| VE | + | − | − | − | + | + | + | + |
| EI | + | + | − | − | − | − | + | − |
| AC | + | + | − | − | − | − | − | − |
| VA | + | + | − | − | − | − | + | − |
| CC | + | + | − | − | − | − | + | − |
| NC | + | + | − | − | − | − | − | − |
| IS | − | − | + | − | − | − | − | − |
| CG | − | + | − | + | − | − | − | − |
| SP | − | + | + | − | − | − | − | − |
| WP | − | − | + | − | − | − | − | − |
| SC | + | + | − | − | − | − | + | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colmenero, A.; Serra, B.; Lagares, C.; Rojo-Francàs, E.; Pérez-Gil, J.L.; Mestres, F.; Abelló, P. Genetic Variability and Connectivity in the Western Mediterranean Populations of the Bathyal Crab Geryon longipes. Diversity 2023, 15, 534. https://doi.org/10.3390/d15040534
Colmenero A, Serra B, Lagares C, Rojo-Francàs E, Pérez-Gil JL, Mestres F, Abelló P. Genetic Variability and Connectivity in the Western Mediterranean Populations of the Bathyal Crab Geryon longipes. Diversity. 2023; 15(4):534. https://doi.org/10.3390/d15040534
Chicago/Turabian StyleColmenero, Ariadna, Bruna Serra, Clàudia Lagares, Eva Rojo-Francàs, José L. Pérez-Gil, Francesc Mestres, and Pere Abelló. 2023. "Genetic Variability and Connectivity in the Western Mediterranean Populations of the Bathyal Crab Geryon longipes" Diversity 15, no. 4: 534. https://doi.org/10.3390/d15040534
APA StyleColmenero, A., Serra, B., Lagares, C., Rojo-Francàs, E., Pérez-Gil, J. L., Mestres, F., & Abelló, P. (2023). Genetic Variability and Connectivity in the Western Mediterranean Populations of the Bathyal Crab Geryon longipes. Diversity, 15(4), 534. https://doi.org/10.3390/d15040534







