Abstract
On 5 September 2022, a dead baleen whale was found stranded at Laem Phak Bia, Phetchaburi, the Gulf of Thailand, Thailand but could not be identified because it was in an advanced stage of decomposition. It was first suspected to be Omura’s whale (Balaenoptera Omurai), as that is a common species in the Gulf of Thailand. However, the cranium morphology was different from B. omurai and more similar to the common minke whale (Balaenoptera acutorostrata) from the North Pacific Ocean, which has never been reported in Thai territorial waters. The mitochondrial DNA control region (D-loop) was then used to identify the species through the Basic Local Alignment Search Tool (BLAST) available at the National Center for Biotechnology Information (NCBI) GenBank, which resulted in a high percent identity, 96.49 to 98.84, with B. acutorostrata. A Bayesian phylogenetic tree was further used to confirm the species, which grouped with B. acutorostrata from the North Pacific Ocean. This study provides evidence of the first stranding event of B. acutorostrata in the Gulf of Thailand. It is new information that extends previous knowledge on the distribution of the common minke whale and raises the need for more active surveys of cetaceans in the South China Sea going forward.
1. Introduction
Cetaceans represent a group of marine mammals that include whales, dolphins, and porpoises [1]. They are divided into two parvorders, Mysticeti (baleen whale) and Odontoceti (toothed whale), with a total of 90 living species in 13 families [1]. Most of these marine mammals are distributed in oceans throughout the world, with a specific range for each species [1,2]. In Thai seas, including the Gulf of Thailand and Thai Andaman Sea, twenty cetacean species representing six families were originally reported to occur, according to Chantrapornsyl, et al., 1996 [3]. However, since 2015, the number of cetacean species found in Thailand seas has increased to 27 species, as shown in Table 1 [4,5,6].

Table 1.
The list of cetaceans found in Thai seas.
For baleen whales, 14 species have been documented in the world’s oceans [1]. Generally, most are larger than toothed whales and usually have a long-range seasonal migration between high (feeding ground)- and low (calving ground)-latitude areas [7]. To date, only five species of baleen whales from family Balaenopteridae have been reported in Thai seas: Bryde’s whale (Balaenoptera edeni), blue whale (B. musculus), Omura’s whale (B. omurai), fin whale (B. physarus), and humpback whale (Magaptera novaeangliae) [5,6]. Two species, B. edeni and B. omurai, are listed within the conserved marine mammals of Thailand [8], and their occurrence along the coastal waters of the Gulf of Thailand is well known [6]. By contrast, there are fewer sightings of the other three species.
The common minke whale, B. acutorostrata, is the smallest member of family Balaenopteridae [1,9] and is divided into three subspecies [10,11,12], two of which occur in northern oceans, namely the North Atlantic minke whale (B. a. acutorostrata) and the North Pacific minke whale (B. a. scammoni). An unnamed dwarf form (B. a. subsp.) lives primarily in the southern hemisphere together with a closely related species, the Antarctic minke whale (B. bonaerensis) [1,13]. Although B. acutorostrata is widely distributed in all oceans worldwide [1,14], they are considered rare for some areas, such as the eastern tropical Pacific and Mediterranean Sea [1,15,16]. Occasionally, this species is confused with other rorquals, such as B. edeni and B. omurai, because the body size and shape are similar at a distance, particularly in areas with high densities of B. edeni and B. omurai [1,17]. However, B. acutorostrata has never been reported in Thai territorial waters.
In the North Pacific Ocean, B. acutorostrata is the most common baleen whale, and they are observed in the waters of Korea, Japan, and the lowest latitude of the Taiwan strait [18,19]. This species has been exploited and is a target of commercial and scientific whaling in these areas [20]. Populations of B. acutorostrata inhabiting the North Pacific Ocean are genetically divided into either ‘O’ stock living in the western North Pacific and Okhotsk Sea or ‘J’ stock in the Sea of Japan, Yellow Sea, and East China Sea [19]. During spring to summer, both stocks migrate to higher latitudes to access feeding grounds in the Okhotsk Sea [21]. The South China Sea was originally thought to be the location for the overwintering of this species [19]; however, there are no reports of any populations or stranding events in this area apart from the Taiwan strait [18]. The only record of an individual skeleton was in northern Borneo, Peninsular Malaysia [22,23], with one tentative sighting in Vietnam, which might have been confused with B. edeni [24].
Up until the present time, the occurrence of B. acutorostrata in Thai territorial waters, including the Gulf of Thailand and Thai Andaman Sea, has never been documented. In this study, we report the stranding event of B. acutorostrata in the Gulf of Thailand for the first time using morphological traits of the cranial bone, genetic data from the mitochondrial DNA control region (D-loop), and a phylogenetic tree to identify the species. This is new information extends previous knowledge about the common minke whale in this area.
2. Materials and Methods
On 5 September 2022, an unidentified dead whale was found stranded at Laem Phak Bia, Phetchaburi, the Gulf of Thailand, Thailand (13.060762, 100.105103) (Figure 1). This unknown whale was transported to an open municipal site to be examined, where the carcass condition was scored according to established criteria [25,26]. The decomposed carcass was identified as a male, with a total length of around 5.27 m (Figure 2). A necropsy was conducted by personnel from the Department of Marine and Coastal Resources, Thailand, after which the carcass was cleaned and kept for further study. Photos of the cranial bone were taken from the dorsal and ventral views for species comparison using a standardized protocol [27].
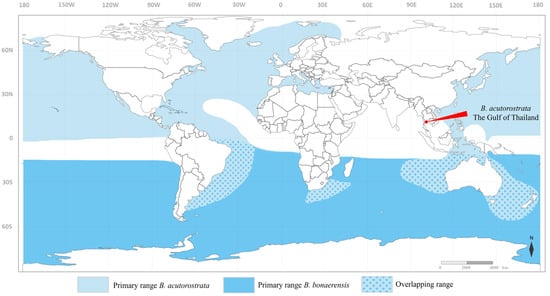
Figure 1.
Map showing the stranding site of Balaenoptera acutorostrata in the Gulf of Thailand and the normal distribution range of B. acutorostrata and B. boenarensis in the world’s oceans.
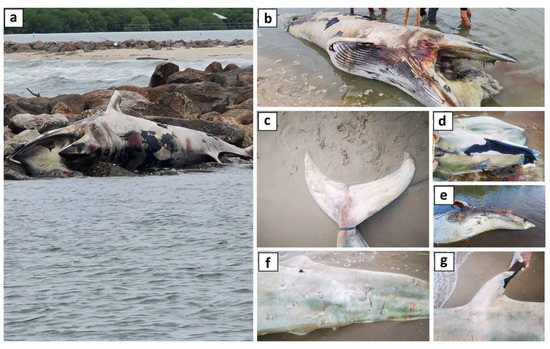
Figure 2.
External morphology of a stranded, unknown male whale, 5.27 m, found at Laem Phak Bia, Phetchaburi, the Gulf of Thailand, Thailand (13.060762, 100.105103) in September 2022. (a) Whale at the stranding site; (b) overall appearance; (c) fluke; (d) dorsal view of the head; (e) ventral view of the body; (f) lateral view of the caudal part of the body; and (g) dorsal fin.
The tissues of non-decomposed organs were collected, with muscle tissue preserved in 95% ethanol for DNA extraction according to the manufacturer’s instructions (DNeasy Blood & Tissue Kit, QIAGEN, Hilden, Germany). The extracted DNA, diluted to 50 ng/μL, was measured qualitatively and quantitatively using 2% agarose gel electrophoresis and absorbance at A260. The D-loop was chosen as a marker for identifying the unknown whale species, as it provides a better phylogenetic resolution for many taxa of cetaceans and has been widely acknowledged [10,28,29,30]. The D-loop of this sample was amplified from the extracted DNA using PCR primers: forward, 5′-CAT ATT ACA ACG GTC TTG TAA ACC-3′; and reverse, 5′-GTC ATA AGT CCA TCG AGA TGT C-3′ [31] as the universal primers. This pair of primers has the ability to amplify the tRNA-Pro gene to the middle of D-loop, as shown for other cetacean species [32,33,34]. PCR reactions were conducted in 25 μL reaction volumes using Platinum Taq DNA polymerase (Invitrogen) consisting of 1× reaction buffer, 2 mM MgCl2, 0.4 mg/mL bovine serum albumin, 0.25 mM dNTPs, 0.4 μM of both forward and reverse primers, and 2 μL of the DNA sample (10 ng/μL). The PCR conditions were performed as follows: 95 °C for 5 min, 40 cycles of 95 °C for 30 s, 50 °C for 45 s, 72 °C for 1 min, and 72 °C for 10 min. The PCR product obtained from the amplification was sequenced by ATGC CO., Ltd., Pathum Thani, Thailand. Complementary sequences were assembled. The sequence identities were checked for identifying species using the Basic Local Alignment Search Tool (BLAST) available at the National Center for Biotechnology Information (NCBI) GenBank. The sequence of D-loop from this study was then deposited in GenBank (accession number OQ446815).
A phylogenetic tree of the D-loop sequence was constructed using Bayesian analysis implemented in the program MrBayes version 3.2.7 [35]. Other D-loop sequences of B. acutorostrata, B. bonaerensis, and B. omurai were retrieved from the NCBI and Pastene, et al., 2007 [12]. The pygmy right whale (Caperea marginata) and bowhead whale (Balaena mysticetus) were used as the outgroups. The total length of the alignment sequences was 330 base pairs. To select the best tree evolutionary models, program jModelTest version 2.1.10 [36] was used, which is defined as HKY + G. The phylogenetic tree was constructed on the run length of Markov Chain Monte Carlo (MCMC) at 2,000,000 iterations, using the average standard deviation of split frequencies below 0.01 as the convergence diagnostic. The first 100,000 iterations were discarded as burn-in. The robustness of each branch was assessed by the posterior probabilities (PP). The phylogenetic tree was then illustrated using iTOL version 6.1.1 [35]. Note that cytochrome oxidase subunit 1 (COI) was not used in this study because of the insufficient B. acutorostrata sequences from different locations deposited in GenBank.
3. Results
The species of the unknown whale could not be identified using only external morphology because of the advanced stage of decomposition (Figure 2a–g). However, it was suspected to be B. omurai or B. acutorostrata because it was a small baleen whale with a single prominent dorsal ridge on the head and other distinguished cranium morphologies. Scars present on the caudal part of the body matched bite marks formed by the cookiecutter shark (Isistius brasiliensis).
3.1. Cranial Bone Morphology
The cranial bone of this sample was used to compare with two species, B. omurai and B. acutorostrata, using figures published by Yamada, et al., 2006 [27] and this study. From the dorsal view of the cranial bone (Figure 3a–c), the mid-premaxilla bone in B. omurai was concave and slightly convex at the caudal end. The premaxilla bones of B. acutorostrata and the unknown whale were straight from the rostral to the middle part, with a higher degree of convex at the caudal area, compared with B. omurai. In B. omurai, the cranial border of the temporal fossa was in a transverse plane of the long axis of the cranial bone, while this border was in an oblique plane for B. acutorostrata and the unknown whale. The lateral border of the maxilla bone in B. omurai was convex, but in B. acutorostrata and the unknown whale, it was straight. The parietal bone was clearly visible in the dorsal view in B. omurai but not in B. acutorostrata and the unknown whale.
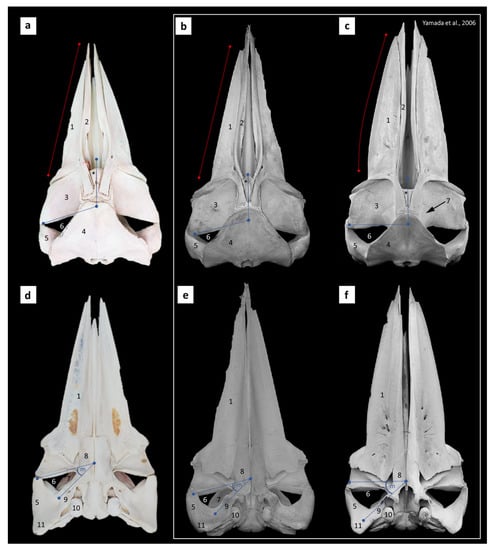
Figure 3.
Dorsal (a–c) and ventral (d–f) views of the cranial bone of the unknown whale in this study (a,d), compared to B. acutorostrata (b,e) and B. omurai (c,f) from a previous study by Yamada, et al., 2006 [27]. Labels: 1 = maxilla bone, 2 = premaxilla bone, 3 = supraorbital process of frontal bone, 4 = supraoccipital bone, 5 = zygomatic process, 6 = temporal fossa, 7 = parietal bone, 8 = palatine bone, 9 = squamosal bone, 10 = tympanic bulla, 11 = postglenoid process, m = median angle of cranial border of the temporal bone, and asterisk = nasal bone.
From the ventral view of the cranial bone (Figure 3d–f), the palatine bone in B. omurai was a triangle shape, whereas in B. acutorostrata and the unknown whale, it was rectangular-shaped. The squamosal bone in B. omurai was a V shape; in B. acutorostrata and the unknown whale, it was a rectangular shape. In this view, the lateral border of the maxilla bone in B. omurai was also convex, but in B. acutorostrata and the unknown whale, it was straight. The cranial border of the temporal fossa in B. omurai was also in a transverse plane to the long axis of the cranium bone, while in B. acutorostrata and the unknown whale, the median angle of this fossa pointed to the rostral. The postglenoid process of B. acutorostrata was rounder than the postglenoid process of B. omurai; however, for the unknown whale, this process was sharper and narrower in angle, which is more similar to B. omurai.
3.2. The Sequence of D-Loop and Phylogenetic Tree
The D-loop sequence of this unknown whale (GenBank accession number OQ446815) was similar to the nucleotide sequence of B. acutorostrata, with high percent identity values between 96.49 and 98.84 (Table 2). The unknown sample was grouped with the clade of B. acutorostrata from the North Pacific Ocean as a monophyletic clade with a high value of posterior probability at 1.00 and was clearly separate from B. omurai (Figure 4). Thus, this information confirmed this whale was not B. omurai, but B. acutorostrata.

Table 2.
Percent identity, based on the Basic Local Alignment Search Tool (BLAST) available at the National Center for Biotechnology Information (NCBI) GenBank, of D-loop.
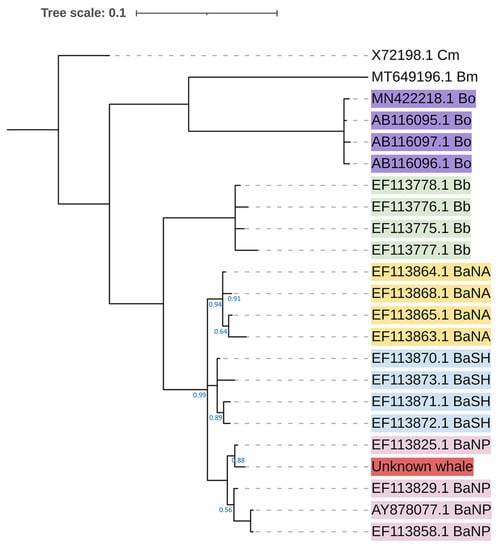
Figure 4.
Bayesian phylogenetic tree of the unknown whale (accession number OQ446815), based on the HKY + G evolutionary model. Posterior probability (PP) value is shown below each branch. All branches had a PP value greater than 0.5. The branch without numbering indicates a PP value greater than 0.99. The accession number of each sequence is labeled at the tips. Cm = Caperea marginata, Bo = Balaenoptera omurai, Bb = Balaenoptera bonaerensis, BaNA = Balaenoptera acutorostrata from North Atlantic Ocean, BaSH = Balaenoptera acutorostrata from the southern hemisphere, BaNP = Balaenoptera acutorostrata from North Pacific Ocean, unknown whale = sample from this study.
4. Discussion
Originally, this unknown whale was believed to be B. omurai because of its small body size and its prominent median head ridge, which are also characteristics of that species [1]. Additionally, it is a common baleen whale found in the Gulf of Thailand, while the stranding or sighting of B. acutorostrata had never been reported in this area [6]. Here, data from this study, including the cranium morphology and D-loop sequence, were sufficient to support the conclusion that this unknown whale stranded on the Gulf of Thailand was a common minke whale, B. acutorostrata. Its D-loop data matched closely with other sequences obtained from the B. acutorostrata of North Pacific origin (reference to accession number EF113825).
The cranial bone of the unknown whale had more similarities to B. acutorostrata than B. omurai in many aspects, which were reported by Yamada, et al., 2006 [27]. Although there was a slight difference found in the postglenoid process between this sample and B. acutorostrata (code NMNS0999), this could be because of the variation in skull morphologies within the species and subspecies. From a previous study, the sample code NMNS0999 was collected from Ilan, Taiwan [27], which is considered the standard form of the North Pacific minke whale (B. a. scammoni), given that there are subspecies found at the higher latitudes [1,14]. However, the postglenoid process of that sample was more similar to the dwarf common minke whale from the southern hemisphere, i.e., rounder at the caudal end, as shown in Appendix 1 in the study of Kato, et al., 2021 [37]. The study of Yamato, et al., 2012 [38] showed the ventral view of the cranial bone of B. acutorostrata collected from the northeast region of the United States, which was a North Atlantic minke whale (B. a. acutorostrata). The postglenoid process of this form also had a similar shape to our sample. Unfortunately, the ventral view of the other cranial bone of B. a. scammoni was unavailable for comparison. Further, the shape of the nasal bone can also be used for identifying the dwarf form from the standard form minke whale [37]. The nasal bone of the dwarf common minke whale is narrow, elongated, and almost reaches the caudal end of the maxilla bone, while the nasal bone of the standard form minke whale is not elongated but larger in the rostral area [37]. Similarly, our cranium sample had a shortened nasal bone that did not reach the caudal end of the maxilla bone, in line with the characteristics of a standard-form minke whale from the North Pacific region [37]. Thus, for our unknown baleen whale, there was a high possibility of it being a standard-form minke whale from the North Pacific region, using cranial bone morphology.
BLAST is an efficient method of determining similarities and dissimilarities of sequences that are available in an online database and can be used to confirm species when morphological appearances are not useful [39,40]. The percent identity of the D-loop sample from our unknown whale agreed with data from the cranium morphology, as it had a high value of similarity to the D-loop sequence of B. acutorostrata but not B. omurai. Moreover, the result of the phylogenetic tree also supported the contention that the unknown sample had an origin in the North Pacific Ocean, not the southern hemisphere, as it was clustered within the monophyletic clade of B. acutorostrata from the Northern Pacific region. Although both species, B. acutorostrata and B. omurai, are from the same family and share co-ancestors around 10 million years ago in the middle age of the Miocene period, some degrees of differences in the mitochondrial sequences between both species have been shown in the form of a phylogenetic tree [41]. In fact, B. omurai is in the monophyletic group with B. edeni and B. brydei, while B. acutorostrata forms the monophyletic clade with B. bonaerensis and diverged earliest within the family Balaenopteridae [41]. Thus, the high percent identity of our sequence to B. acutorostrata and the result of the phylogenetic tree in this study can be used to confirm the presence of B. acutorostrata in the Gulf of Thailand. In addition, B. acutorostrata also has genetic variations within species. In a previous study, the genetic diversities of three subspecies of B. acutorostrata were revealed using D-loop analysis [42]. A total of 70 haplotypes with a high total nucleotide diversity value of 0.0231 were found for this species without sharing the maternal lineage among subspecies that inhabited different areas, including the North Pacific, North Atlantic, western South Pacific, and western South Atlantic.
Previously, the primary range of B. acutorostrata was thought to cover the entire area of the South China Sea [1]. In fact, the southern-most distribution records of this species have it restricted to the Taiwan strait [18,19], with only a few unconfirmed occurrences in the Vietnam Sea and Borneo [22,23,24]. Thus, in our study, this stranding event in the Gulf of Thailand provides evidence of an extralimital range of distribution not documented previously for this species. The extension of the living range south into the Gulf of Thailand may be due to a reduction in the distribution range of their main prey (krill) or to competition with other baleen species for this food source, perhaps due to changing water temperatures as a result of climate change [43,44,45]. Thus, it is possible they are seeking alternative feeding habitats outside the normal range. Moreover, extensive hunting in the northern Pacific Ocean could be driving this southward migration [20]. It remains unclear, however, if this species is in the process of moving into the Gulf of Thailand or if the animal or its carcass had somehow drifted over an extensive range from other places within the South China Sea. Collaboration with neighboring countries is needed for more active surveillance of this and other cetacean species to determine how or if habitat ranges are changing, and if so, why.
5. Conclusions
This study confirmed the first stranding event of B. acutorostrata in the Gulf of Thailand. The comparison of cranium morphology and the phylogenetic tree generated by D-loop sequences were sufficient for determining the species of the unknown whale. This information will extend previous knowledge on the distribution of B. acutorostrata to Thai waters and raises the need for more active cetacean surveys in the South China Sea going forward.
Author Contributions
Data curation, R.C.; formal analysis, R.C.; funding acquisition, K.N.; investigation, R.C., P.Y., S.T., K.K. and C.T.; methodology, R.C. and P.P.; project administration, K.N. and P.P.; resources, R.C., P.Y., S.T., K.K. and C.T.; software, P.P.; supervision, J.L.B., K.N. and P.P.; validation, J.L.B., K.N. and P.P.; visualization, J.L.B., K.N. and P.P.; writing—original draft, R.C., K.N. and P.P.; writing—review and editing, R.C., J.L.B., K.N. and P.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Research Center in Veterinary Bioscience and Public Health, Chiang Mai University, Chiang Mai 50200, Thailand (number: 009/2023).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The accession numbers used in this study are shown in Figure 4.
Acknowledgments
We dedicate the value of this research to the Excellence Center in Veterinary Bioscience, Department of Veterinary Biosciences and Public Health, Faculty of Veterinary Medicine, Chiang Mai University, Chiang Mai, Thailand. To this center, we express our deepest gratitude for their helpful insights that helped us fully analyze the results of this study.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no roles in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Jefferson, T.; Webber, M.; Pitman, R. Marine Mammals of the World: A Comprehensive Guide to Their Identification; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Dold, C. Cetacea (Whales, Dolphins, Porpoises). Fowler’s Zoo Wild Anim. Med. 2014, 8, 422. [Google Scholar]
- Chantrapornsyl, S.; Adulyanukosol, K.; Kittiwathanawong, K. Records of cetaceans in Thailand. Phuket Mar. Biol. Cent. Res. Bull. 1996, 61, 39–63. [Google Scholar]
- Department of Marine and Coastal Resources. “Whale and Dolphin Species”. Central Database System and Data Standard for Marine and Coastal Resources. 2013. Available online: http://km.dmcr.go.th/th/c_7/d_2448 (accessed on 13 December 2022).
- Hines, E.; Ponnampalam, L.; Jamal Hisne, F.; Tara, S.; Jackson-Ricketts, J.; Kuit, S.; Acebes, J. Report of the Third Southeast Asian Marine Mammal Symposium (SEAMAM III). CMS Tech. Ser. 2015, 32, 105–109. [Google Scholar]
- Kanjana, A.; Surasak, T.; Theerawat, P.; Phaothep, C. Field Guide to Marine Mammals and Sea Turtles of Thailand; Marine and Coastal Resources Research Center, Upper Gulf of Thailand, National Office of Buddhism: Nakhon Pathom, Thailand, 2014.
- Roman, J.; Estes, J.A.; Morissette, L.; Smith, C.; Costa, D.; McCarthy, J.; Nation, J.; Nicol, S.; Pershing, A.; Smetacek, V. Whales as marine ecosystem engineers. Front. Ecol. Environ. 2014, 12, 377–385. [Google Scholar] [CrossRef]
- Department of National Parks Wildlife and Plant Conservation. “Wildlife Conservation and Protection Act, B.E. 2562 (2019)”. Thai Government Gazette. 2019. Available online: http://portal.dnp.go.th/Law (accessed on 28 April 2021).
- Segre, P.S.; Gough, W.T.; Roualdes, E.A.; Cade, D.E.; Czapanskiy, M.F.; Fahlbusch, J.; Kahane-Rapport, S.R.; Oestreich, W.K.; Bejder, L.; Bierlich, K. Scaling of maneuvering performance in baleen whales: Larger whales outperform expectations. J. Exp. Biol. 2022, 225, jeb243224. [Google Scholar] [CrossRef]
- Pastene, L.A.; Acevedo, J.; Goto, M.; Zerbini, A.N.; Acuna, P.; Aguayo-Lobo, A. Population structure and possible migratory links of common minke whales, Balaenoptera acutorostrata, in the Southern Hemisphere. Conserv. Genet. 2010, 11, 1553–1558. [Google Scholar] [CrossRef]
- Glover, K.A.; Kanda, N.; Haug, T.; Pastene, L.A.; Øien, N.; Seliussen, B.B.; Sørvik, A.G.; Skaug, H.J. Hybrids between common and Antarctic minke whales are fertile and can back-cross. Bmc Genet. 2013, 14, 25. [Google Scholar] [CrossRef]
- Pastene, L.A.; Goto, M.; Kanda, N.; Zerbini, A.N.; Kerem, D.; Watanabe, K.; Bessho, Y.; Hasegawa, M.; Nielsen, R.; Larsen, F. Radiation and speciation of pelagic organisms during periods of global warming: The case of the common minke whale, Balaenoptera acutorostrata. Mol. Ecol. 2007, 16, 1481–1495. [Google Scholar] [CrossRef]
- Arnold, P.W.; Birtles, R.A.; Dunstan, A.; Lukoschek, V.; Matthews, M. Colour patterns of the dwarf minke whale Balaenoptera acutorostrata sensu lato: Description, cladistic analysis and taxonomic implications. Mem. Qld. Mus. 2005, 51, 277–307. [Google Scholar]
- Perrin, W.F.; Mallette, S.D.; Brownell, R.L., Jr. Minke whales: Balaenoptera acutorostrata and B. bonaerensis. In Encyclopedia of Marine Mammals; Elsevier: Amsterdam, The Netherlands, 2018; pp. 608–613. [Google Scholar]
- Fraija-Fernández, N.; Crespo-Picazo, J.L.; Domènech, F.; Míguez-Lozano, R.; Palacios-Abella, J.F.; Rodríguez-González, A.; Villar-Torres, M.; Gozalbes, P. First stranding event of a common minke whale calf, Balaenoptera acutorostrata Lacépède, 1804, reported in Spanish Mediterranean waters. Mammal Study 2015, 40, 95–100. [Google Scholar] [CrossRef]
- Öztürk, A.A.; Tonay, A.M.; Dede, A.; Ayhan, D. Strandings of the beaked whales, Risso’s dolphins, and a minke whale on the Turkish coast of the Eastern Mediterranean Sea. J. Black Sea/Mediterr. Environ. 2011, 17, 269–274. [Google Scholar]
- Rudolph, P.; Smeenk, C. Indo-West Pacific marine mammals. In Encyclopedia of Marine Mammals; Elsevier: Amsterdam, The Netherlands, 2009; pp. 608–616. [Google Scholar]
- Zhao, L.; Zhu, Q.; Miao, X.; Xu, M.; Wu, F.; Dai, Y.; Tao, C.; Mou, J.; Wang, X. An overview of cetacean strandings, bycatches and rescues along the western coast of the Taiwan Strait, China: 2010–2015. Acta Oceanol. Sin. 2017, 36, 31–36. [Google Scholar] [CrossRef]
- Lee, D.; An, Y.R.; Park, K.J.; Kim, H.W.; Lee, D.; Joo, H.T.; Oh, Y.G.; Kim, S.M.; Kang, C.K.; Lee, S.H. Spatial distribution of common Minke whale (Balaenoptera acutorostrata) as an indication of a biological hotspot in the East Sea. Deep. Sea Res. Part II Top. Stud. Oceanogr. 2017, 143, 91–99. [Google Scholar] [CrossRef]
- Risch, D.; Norris, T.; Curnock, M.; Friedlaender, A. Common and Antarctic minke whales: Conservation status and future research directions. Front. Mar. Sci. 2019, 6, 247. [Google Scholar] [CrossRef]
- Pastene, L.A.; Goto, M.; Taguchi, M.; Kitakado, T. Temporal and Spatial Distribution of the ‘J’ and ‘O’ Stocks of Common Minke Whale in Waters around Japan Based on Microsatellite DNA; Paper SC/F16/JR38 Presented to the JARPNII Special Permit Expert Panel Review Workshop; Paper SC: Tokyo, Japan, 2016. [Google Scholar]
- Jaaman, S.A.; Najib, R.; Syuhaime, A.; Yuhana, U. Records of Marine Mammals in Peninsular Malaysia: A Review. Tropical Marine Environment: Charting Strategies for the Millennium; Malacca Straits Research and Development Centre (MASDEC), Universiti Putra Malaysia: Serdang, Malaysia, 2002; pp. 499–515. [Google Scholar]
- Mazlan, A.; Zaidi, C.; Wan-Lotfi, W.; Othman, B. On the Current Status of Coastal Marine Biodiversity in Malaysia. Indian J. Mar. Sci. 2005, 34, 76–87. [Google Scholar]
- Smith, B.D.; Jefferson, T.A.; Leatherwood, S.; Ho, D.; Thuoc, C.V.; Quang, L.H. Investigations of marine mammals in Vietnam. Asian Mar. Biol. 1997, 14, 145–172. [Google Scholar]
- Geraci, J.R.; Lounsbury, V.J. Marine Mammals Ashore: A Field Guide for Strandings; National Aquarium in Baltimore: Baltimore, MD, USA, 2005. [Google Scholar]
- Pugliares, K.R.; Bogomolni, A.; Touhey, K.M.; Herzig, S.M.; Harry, C.T. Marine Mammal Necropsy: An Introductory Guide for Stranding Responders and Field Biologists; MBLWHOI Library: Woods Hole, MA, USA, 2007. [Google Scholar]
- Yamada, T.K.; Chou, L.-S.; Chantrapornsyl, S.; Adulyanukosol, K.; Chakravarti, S.K.; Oishi, M.; Wada, S.; Yao, C.-J.; Kakuda, T.; Tajima, Y. Middle-sized balaenopterid whale specimens (Cetacea: Balaenopteridae) preserved at several institutions in Taiwan, Thailand, and India. Mem. Natl. Sci.Mus. 2006, 44, 1–10. [Google Scholar]
- Dalebout, M.; Baker, C.; Mead, J.; Cockcroft, V.; Yamada, T. A comprehensive and validated molecular taxonomy of beaked whales, family Ziphiidae. J. Hered. 2004, 95, 459–473. [Google Scholar] [CrossRef]
- Oremus, M.; Gales, R.; Dalebout, M.L.; Funahashi, N.; Endo, T.; Kage, T.; Steel, D.; Baker, S.C. Worldwide mitochondrial DNA diversity and phylogeography of pilot whales (Globicephala spp.). Biol. J. Linn. Soc. 2009, 98, 729–744. [Google Scholar] [CrossRef]
- Kraft, S.; Pérez-Álvarez, M.; Olavarría, C.; Poulin, E. Global phylogeography and genetic diversity of the long-finned pilot whale Globicephala melas, with new data from the southeastern Pacific. Sci. Rep. 2020, 10, 1769. [Google Scholar] [CrossRef]
- Blair, D.; McMahon, A.; McDonald, B.; Tikel, D.; Waycott, M.; Marsh, H. Pleistocene sea level fluctuations and the phylogeography of the dugong in Australian waters. Mar. Mammal Sci. 2014, 30, 104–121. [Google Scholar] [CrossRef]
- Kriangwanich, W.; Buddhachat, K.; Poommouang, A.; Chomdej, S.; Thitaram, C.; Kaewmong, P.; Kittiwattanawong, K.; Nganvongpanit, K. Feasibility of melting fingerprint obtained from ISSR-HRM curves for marine mammal species identification. PeerJ 2021, 9, e11689. [Google Scholar] [CrossRef]
- Piboon, P.; Kriengsakpichit, N.; Poommouang, A.; Buddhachat, K.; Brown, J.L.; Kampuansai, J.; Chomdej, S.; Kaewmong, P.; Kittiwattanawong, K.; Nganvongpanit, K. Relationship of stranded cetaceans in Thai territorial waters to global populations: Mitochondrial DNA diversity of Cuvier’s beaked whale, Indo Pacific finless porpoise, pygmy sperm whale, and dwarf sperm whale. Sci. Prog. 2022, 105, 00368504221103776. [Google Scholar] [CrossRef]
- Piboon, P.; Poommouang, A.; Buddhachat, K.; Kampuansai, J.; Chomdej, S.; Kaewmong, P.; Kittiwattanawong, K.; Nganvongpanit, K. The Potential Distribution and Maternal Lineage of Two Cetaceans Species (Grampus griseus and Pseudorca crassidens) in the Subfamily Globicephalinae from the Thai Andaman Sea and the Gulf of Thailand. Diversity 2022, 14, 257. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.; Teslenko, M.; Nylander, J. MrBayes Version 3.2 Manual: Tutorials and Model Summaries. 2011. Available online: https://bioweb.pasteur.fr/docs/modules/mrbayes/3.1.2/Manual_MrBayes_v3.2.0_draft.pdf (accessed on 31 December 2022).
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Kato, H.; Fujise, Y.; Nakamura, G.; Hakamada, T.; Pastene, L.A.; Best, P.B. Dwarf Minke Whales: Morphology, Growth and Life History Based on Samples Collected from the Higher Latitudes in the Southern Hemisphere. Cetacean Popul. Stud. 2021, 3, 93–128. [Google Scholar]
- Yamato, M.; Ketten, D.R.; Arruda, J.; Cramer, S.; Moore, K. The auditory anatomy of the minke whale (Balaenoptera acutorostrata): A potential fatty sound reception pathway in a baleen whale. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 2012, 295, 991–998. [Google Scholar] [CrossRef]
- Li, T.; Wu, H.; Wu, C.; Yang, G.; Chen, B. Molecular Identification of Stranded Cetaceans in Coastal China. Aquat. Mamm. 2019, 45, 525–532. [Google Scholar] [CrossRef]
- FalcÃ, L.; Campos, A.; Freitas, J.; Furtado-Neto, M.; Faria, V. Molecular identification of cetaceans from the West Atlantic using the E3-I5 region of COI. Genet. Mol. Res. 2017, 16, 2. [Google Scholar] [CrossRef]
- McGowen, M.R.; Spaulding, M.; Gatesy, J. Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol. Phylogenetics Evol. 2009, 53, 891–906. [Google Scholar] [CrossRef]
- Milmann, L.; Taguchi, M.; Siciliano, S.; Baumgarten, J.E.; Oliveira, L.R.; Valiati, V.H.; Goto, M.; Ott, P.H.; Pastene, L.A. New genetic evidences for distinct populations of the common minke whale (Balaenoptera acutorostrata) in the Southern Hemisphere. Polar Biol. 2021, 44, 1575–1589. [Google Scholar] [CrossRef]
- Tulloch, V.J.; Plagányi, É.E.; Brown, C.; Richardson, A.J.; Matear, R. Future recovery of baleen whales is imperiled by climate change. Glob. Change Biol. 2019, 25, 1263–1281. [Google Scholar] [CrossRef]
- MacLeod, C.D. Global climate change, range changes and potential implications for the conservation of marine cetaceans: A review and synthesis. Endanger. Species Res. 2009, 7, 125–136. [Google Scholar] [CrossRef]
- Owen, K.; Jenner, K.C.S.; Jenner, M.-N.M.; McCauley, R.D.; Andrews, R.D. Water temperature correlates with baleen whale foraging behaviour at multiple scales in the Antarctic. Mar. Freshw. Res. 2018, 70, 19–32. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).