Assessment of the Diversity, Distinctiveness and Conservation of Australia’s Central Queensland Coastal Rainforests Using DNA Barcoding
Abstract
1. Introduction
- How does the Central Queensland Coast (CQC) rainforest estate compare in terms of composition, diversity and uniqueness to other rainforest communities?
- What is the distribution of rainforest phylogenetic diversity and distinctiveness within the Central Queensland Coast (CQC) rainforest estate?
- What factors are driving differences or similarities between rainforest communities within the study area?
- Is there evidence of an overlap region or coastal corridor that may facilitate species movement in a changing climate?
- How well protected are phylogenetically diverse Central Queensland Coast (CQC) rainforests?
2. Materials and Methods
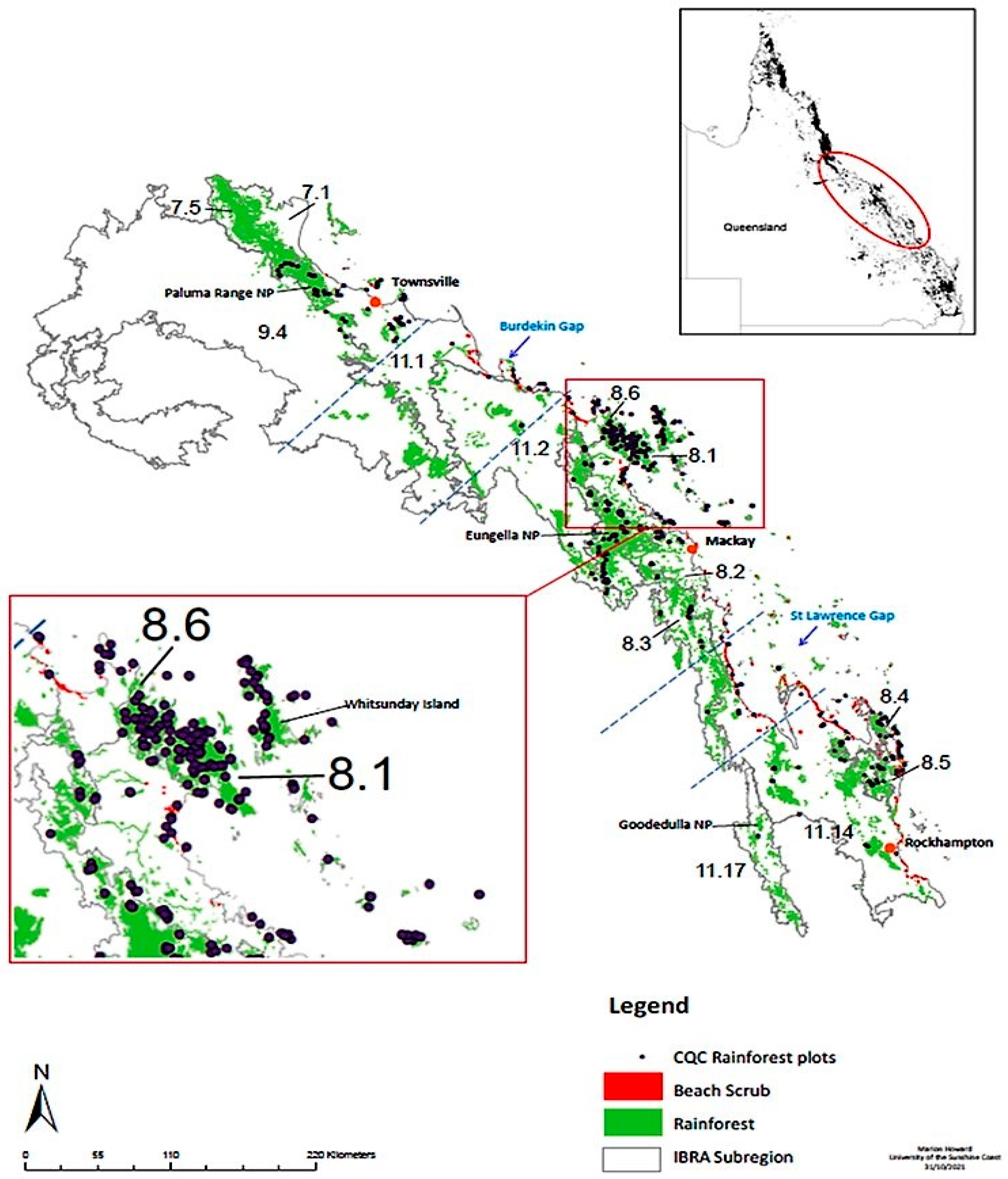
2.1. Sample Collection
2.2. Construction of the Extended DNA Barcode Library and CQC Barcoded Library
2.3. Queensland Rainforest Phylogeny
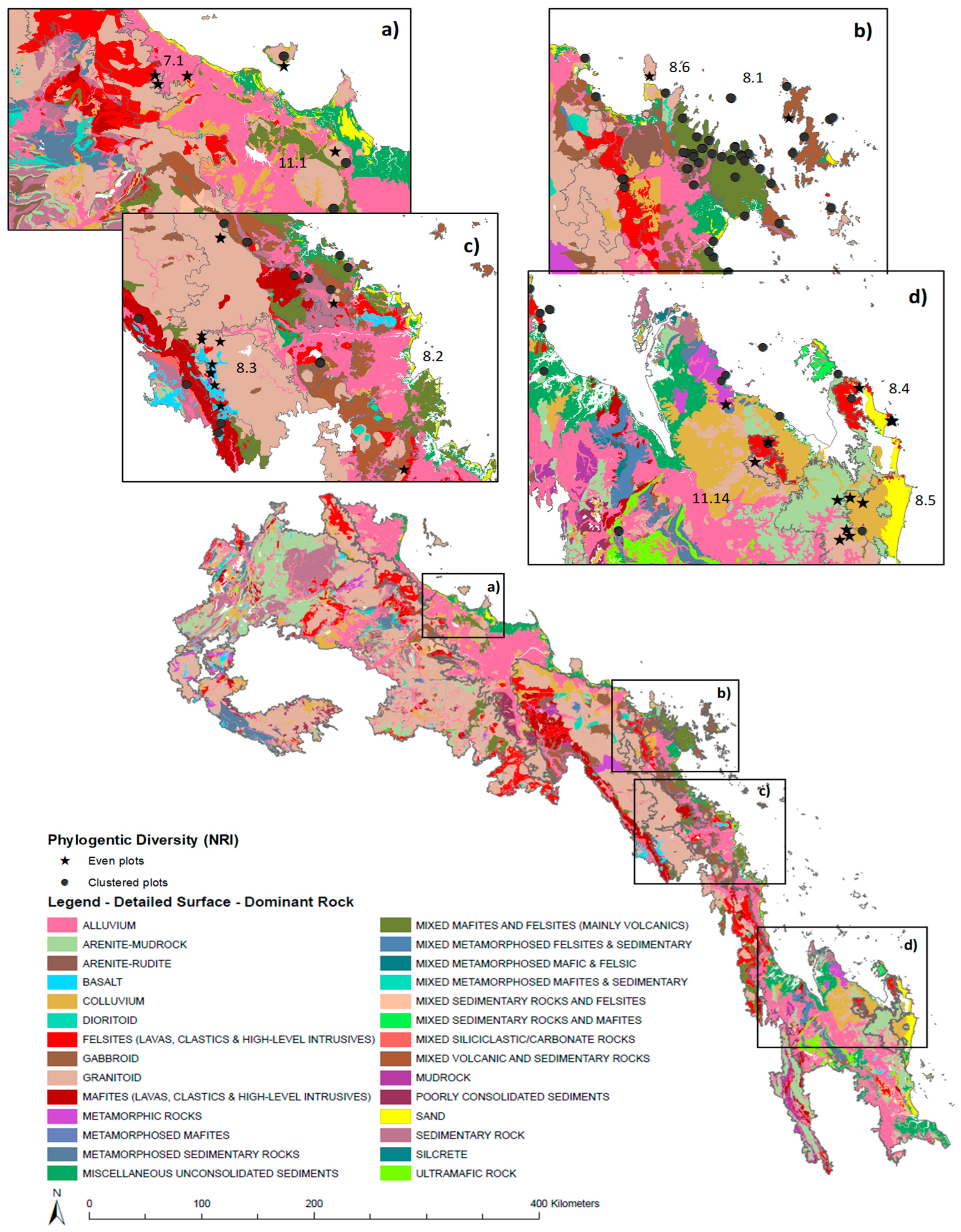
2.4. Rainforest Subregion and Regional Ecosystem (RE) Designation
2.5. Abiotic Data
2.6. Phylogenetic Analysis
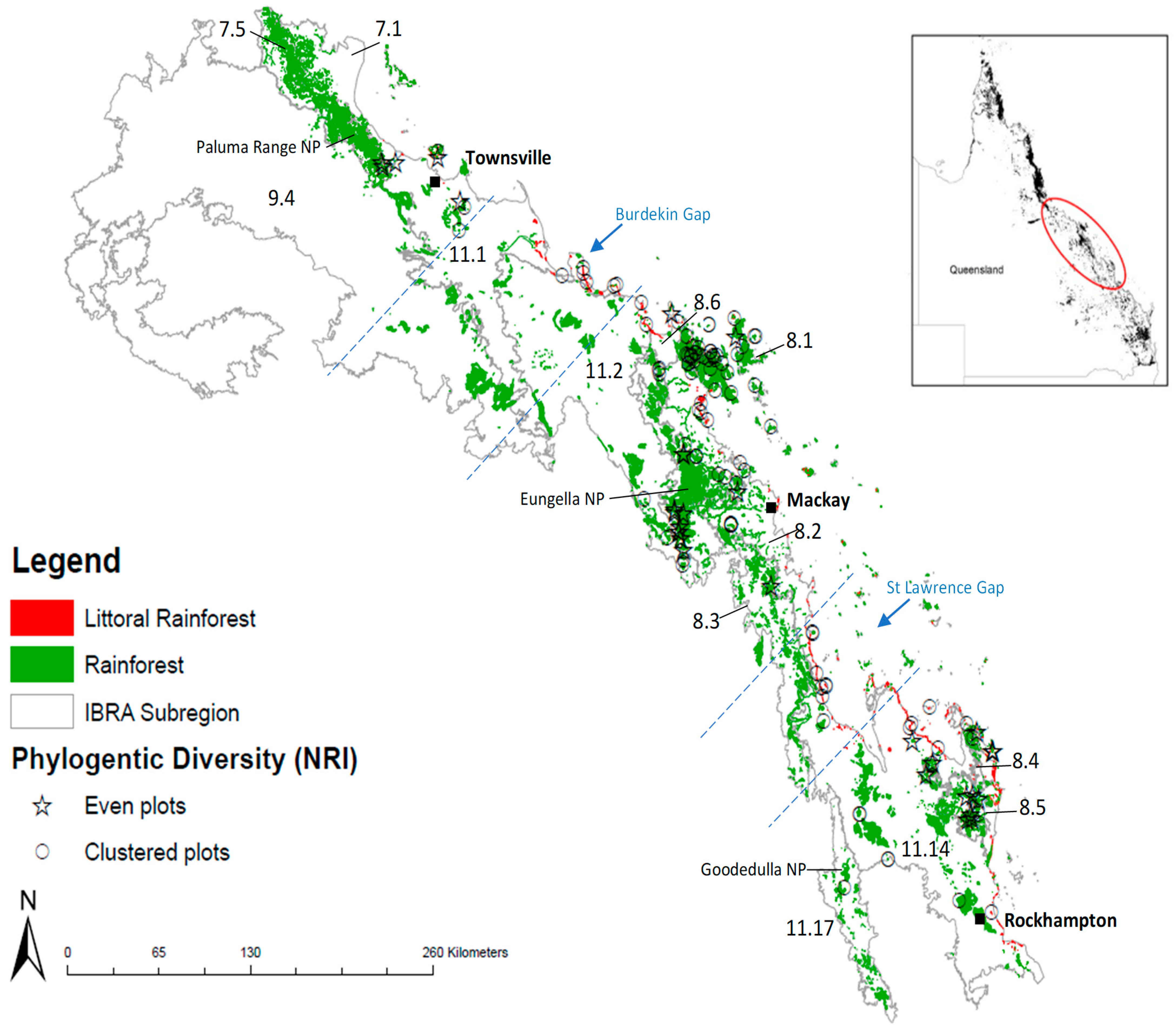
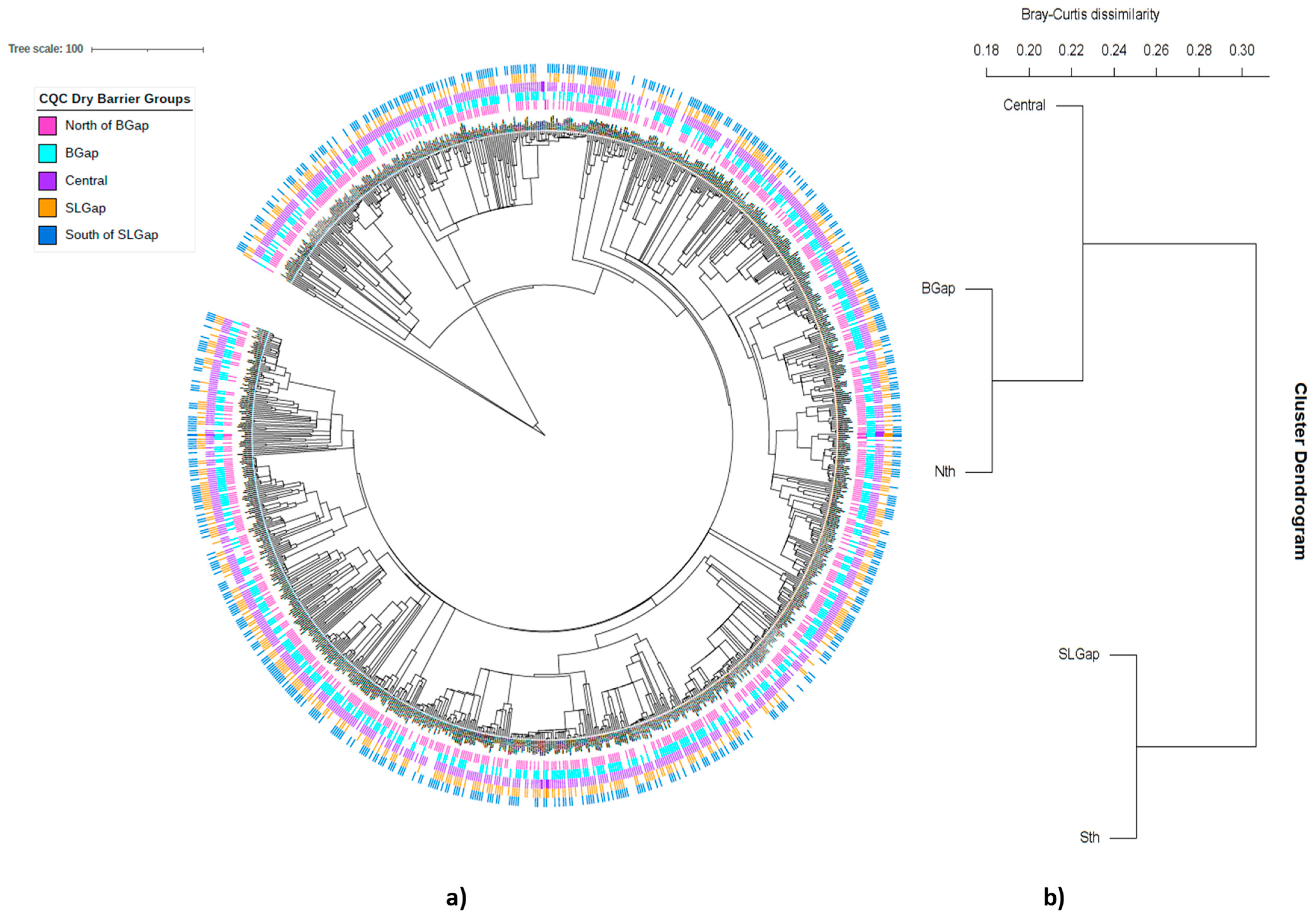
2.7. Investigation of a Corridor or Region of Overlap
2.8. Central Queensland Coast (CQC) Rainforest Protected Area Estate
3. Results
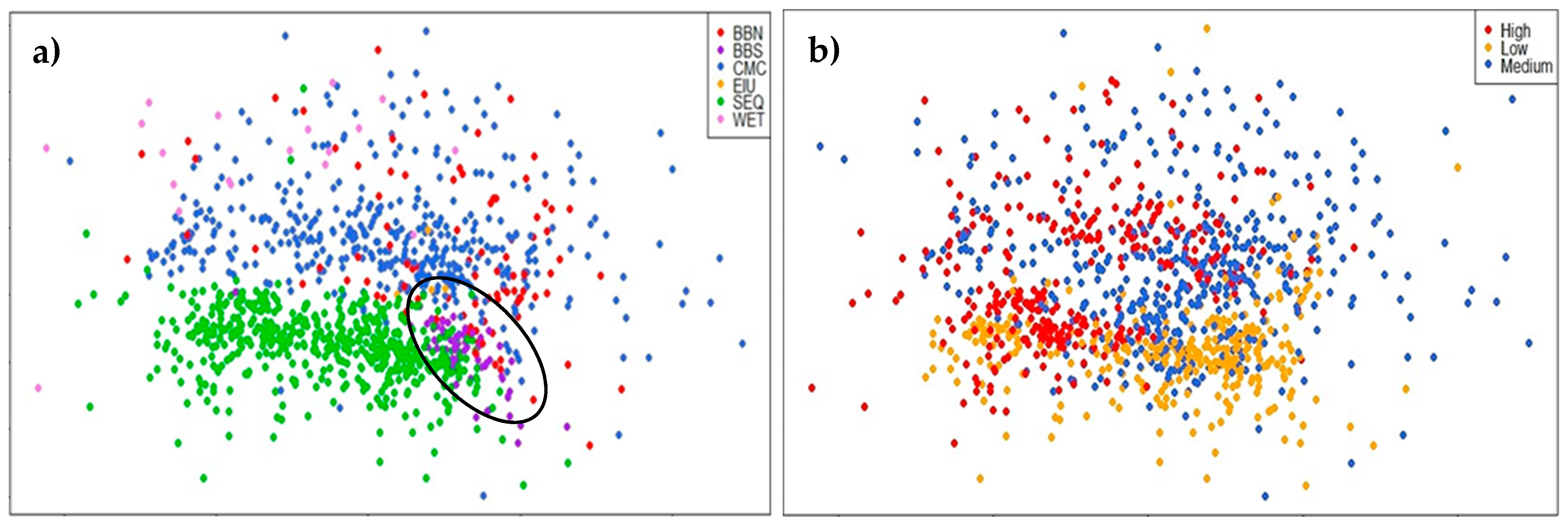
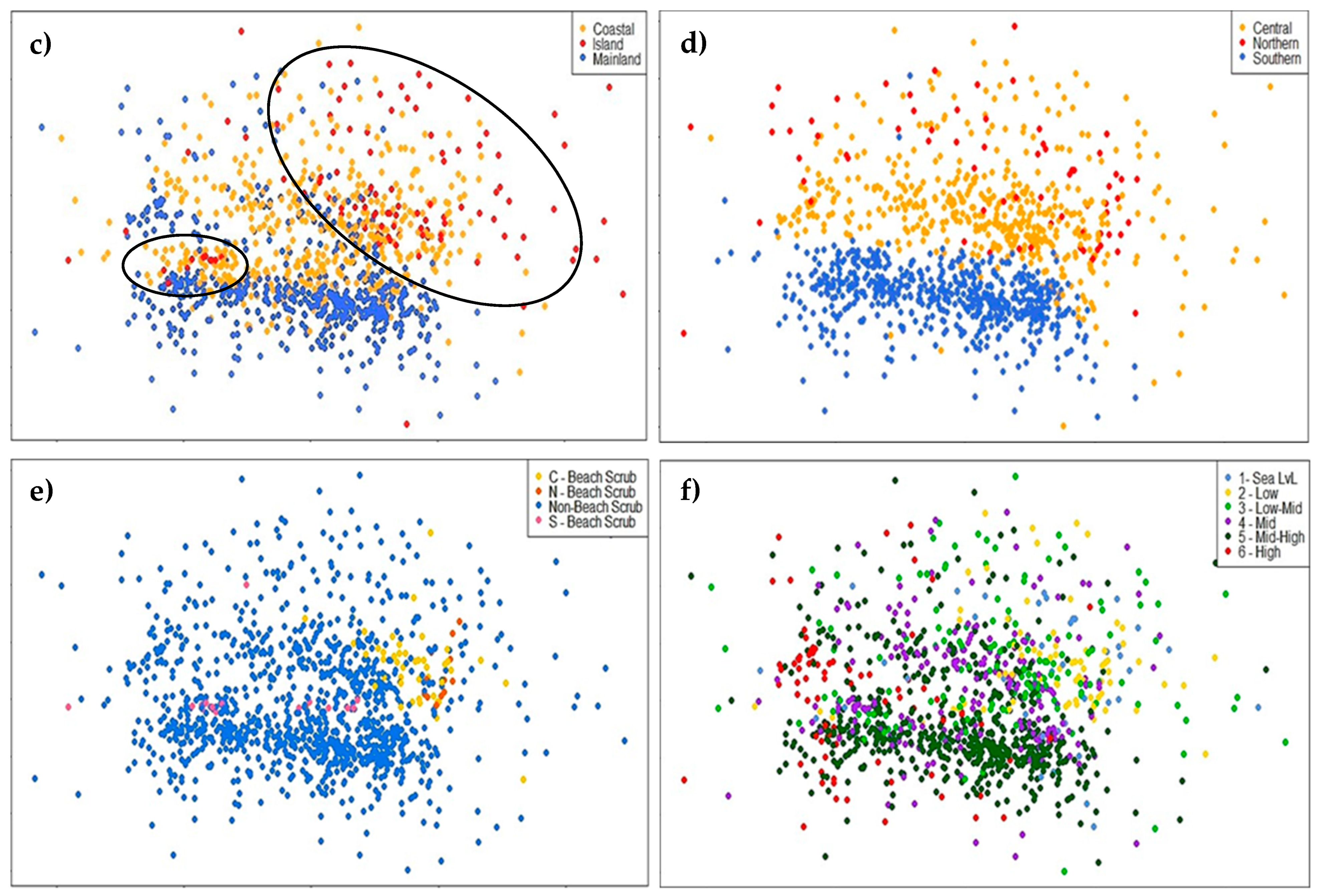
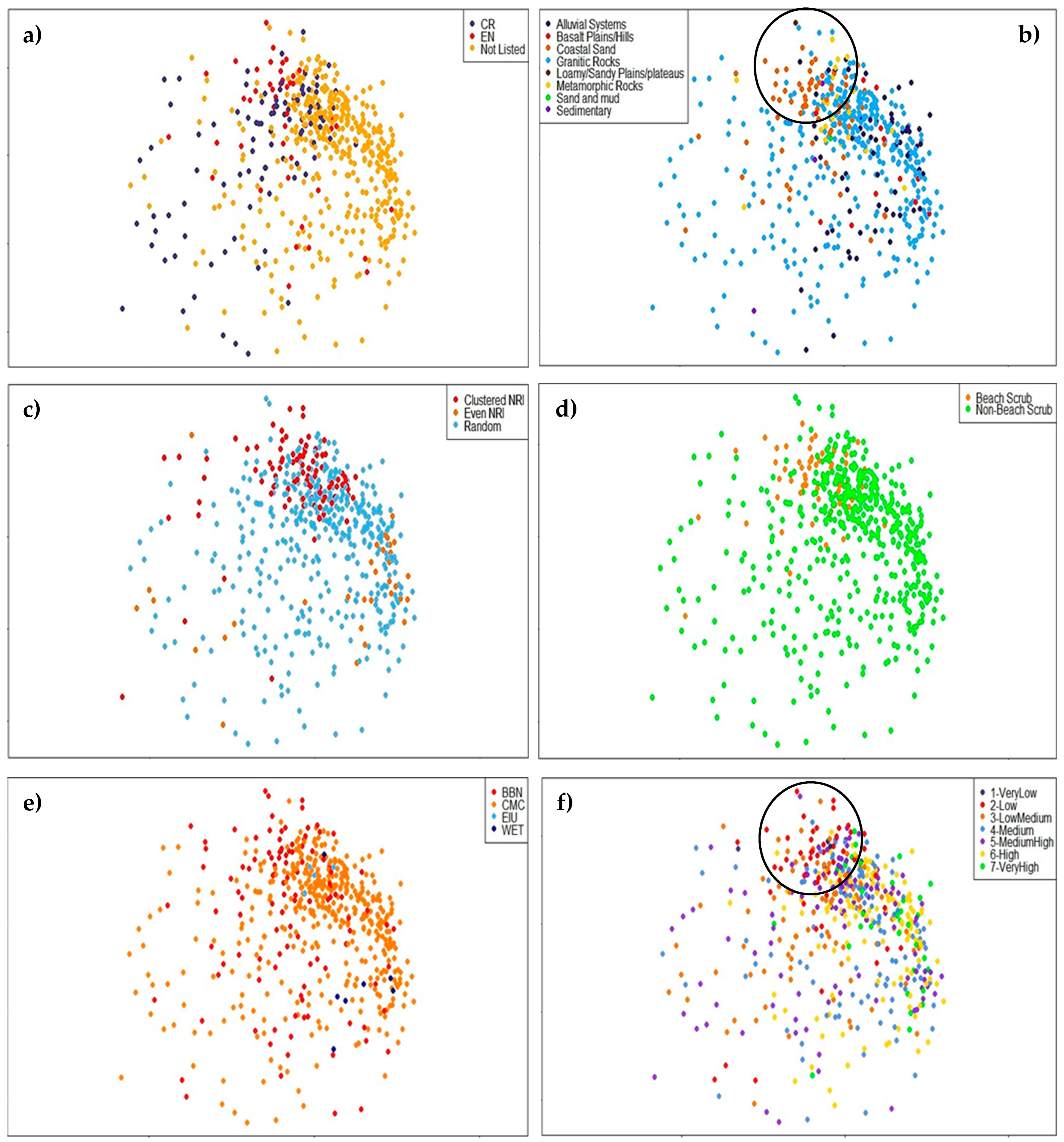
3.1. Comparison of Central and Southeast Queensland (CSEQ) Rainforest Communities
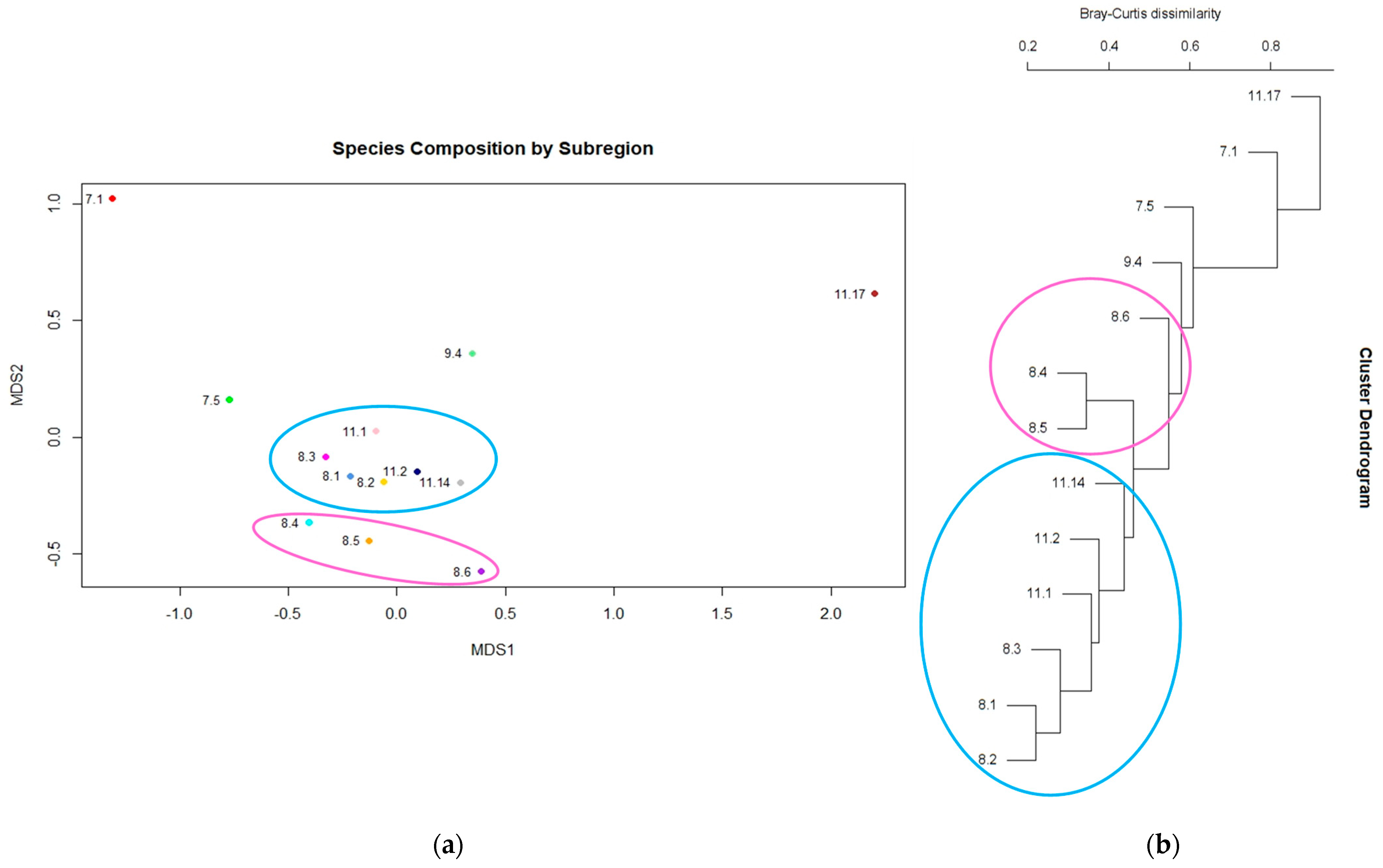
3.2. Diversity and Distinctiveness of Central Queensland Coast (CQC) Rainforest Estate
3.3. Evidence of a Corridor or Region of Overlap
3.4. Central Queensland Rainforest Protected Area Estate
4. Discussion
4.1. Comparison of Central Queensland Coast (CQC) Rainforests with Other Communities
4.2. Diversity within Central Queensland Coast (CQC) Rainforest Estate
4.3. Distinctiveness within Central Queensland Coast (CQC) Rainforest Types
4.4. Evidence of a Region of Overlap or Coastal Corridor
4.5. Conservation of Central Queensland Coast (CQC) Rainforests
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Marker | Read Direction | PCR Primers | Sequence (5′→3′) |
|---|---|---|---|
| matK | Forward | matK_xF | TAATTTACGATCAATTCATTC |
| Reverse | matK_5r | GTTCTAGCACAAGAAAGTCG | |
| psbA-trnH | Forward | psbA3_f | GTTATGCATGAACGTAATGCTC |
| Reverse | trnHf_05 | CGCGCATGGTGGATTCACAATCC | |
| rbcL | Forward | rbcLa-F | ATGTCACCACAAACAGAGACTAAAGC |
| Reverse | rbcLa-R | GTAAAATCAAGTCCACCYCG |
| RE | PD | Preclear (Ha) | Remnant (Ha) | Preclear% RfPA | Remnant %RfPA | EPBC Act (1999) | IBRA Subregion | IBRA Bioregion |
|---|---|---|---|---|---|---|---|---|
| 11.11.21 | 5109 | 1894 | 1522 | 13.2 | 16.2 | 11.14 | BBN | |
| 11.11.5 | 4747 | 37,339 | 14,165 | 7.8 | 20.0 | 11.1; 11.2; 11.14; 11.17; 8.4; 8.4 | BBN; BBS; CMC | |
| 11.11.5a | 2184 | 2448 | 2341 | 71.1 | 74.3 | 11.14 | BBN | |
| 11.12.16 | 2030 | 98 | 81 | 0.0 | 0.0 | CR | 11.2 | BBN |
| 11.12.4 | 8157 | 37,854 | 30,666 | 30.2 | 36.7 | 11.1; 11.2; 11.14; 11.17; 9.4 | BBN; BBS; EIU | |
| 11.12.4a | 7374 | 413 | 385 | 18.4 | 19.7 | 11.1; 11.14 | BBN | |
| 11.12.9 | 3643 | *H 111,558 | *H 95,779 | 7.4 | 8.6 | 11.1; 11.2; 11.14; 7.1; 7.5; 8.6; 9.4 | BBN; WET; CMC; EIU | |
| 11.2.3 | 6988 | 2915 | 2456 | 11.6 | 13.4 | EN | 11.1; 11.2; 11.14; 7.1 | BBN; WET |
| 11.3.11 | 5553 | 1809 | 309 | 2.5 | 14.8 | EN | 11.1; 11.14; 11.17; 8.4 | BBN; BBS; CMC |
| 11.3.11x1 | 2655 | 7 | 7 | 0.0 | 0.0 | 11.1 | BBN | |
| 11.3.40 | 2082 | 506 | 196 | 1.9 | 5.0 | 11.1 | BBN | |
| 11.5.15 | 3377 | 5119 | 4977 | 0.0 | 0.0 | EN | 11.14; 9.4 | BBN; EIU |
| 11.8.3 | 2264 | 510 | 394 | 1.6 | 2.1 | EN | 11.2 | BBN |
| 7.12.10a | 3625 | 1719 | 1717 | 69.9 | 70.0 | 11.1; 7.1; 7.5 | BBN; WET | |
| 7.12.11a | 4259 | 12,882 | 12,876 | 92.1 | 92.2 | EN | 11.1; 7.1; 7.5; 9.4 | BBN; WET |
| 7.12.11b | 6113 | 224 | 224 | 94.4 | 94.6 | EN | 11.1 | BBN |
| 7.12.16a | 4727 | 32,103 | 31,665 | 93.7 | 94.3 | 7.5 | WET | |
| 7.12.21b | 2226 | 5137 | 5112 | 84.4 | 84.7 | 11.1; 7.5 | BBN; WET | |
| 7.12.22b | 1911 | 3448 | 3389 | 94.5 | 95.8 | 7.5 | WET | |
| 7.12.26b | 3158 | 295 | 295 | 99.6 | 99.6 | 7.5 | WET | |
| 7.3.16b | 3108 | 11,839 | 4410 | 19.2 | 34.0 | 7.1; 7.5 | WET | |
| 7.3.26a | 2221 | 2154 | 1936 | 23.3 | 25.9 | 11.1; 7.5; 7.1 | BBN; WET | |
| 7.3.50b | 2848 | 63 | 63 | 83.2 | 83.2 | EN | 11.1; 7.5; 7.1 | BBN; WET |
| 8.10.1 | 2213 | 156 | 148 | 42.9 | 45.2 | 8.4 | CMC | |
| 8.11.2 | 7207 | 2839 | 2556 | 17.9 | 19.0 | 11.14; 8.2; 8.4; 8.5 | BBN; CMC | |
| 8.12.11 | 7358 | 116 | 116 | 98.4 | 98.4 | CR | 8.4 | CMC |
| 8.12.11a | 6730 | 14,740 | 14,665 | 90.7 | 91.5 | CR | 11.2; 8.1; 8.2; 8.4; 8.6 | BBN; CMC |
| 8.12.11c | 6110 | 1520 | 1509 | 16.8 | 17.0 | CR | 11.14; 8.4; 8.5 | BBN; CMC |
| 8.12.14b | 1310 | 8612 | 8570 | 93.6 | 94.1 | 8.1 | CMC | |
| 8.12.16 | 3562 | 4126 | 4123 | 27.1 | 27.1 | CR | 11.2; 8.3 | BBN; CMC |
| 8.12.17a | 3631 | 3655 | 3530 | 91.3 | 94.0 | 8.3 | CMC | |
| 8.12.17b | 3828 | 564 | 558 | 70.0 | 69.8 | 8.3; 8.4 | CMC | |
| 8.12.17c | 2205 | 707 | 707 | 100.0 | 100.0 | 8.1 | CMC | |
| 8.12.18 | *H 10,848 | 26,732 | 25,988 | 80.4 | 82.7 | 8.1; 8.2; 8.6 | CMC | |
| 8.12.19 | 9534 | 13,728 | 13,038 | 77.4 | 81.1 | 8.1; 8.2; 8.3 | CMC | |
| 8.12.1a | 7061 | 21,850 | 18,429 | 72.6 | 85.0 | 8.3 | CMC | |
| 8.12.1b | 6503 | 1392 | 1392 | 99.9 | 99.9 | 8.1 | CMC | |
| 8.12.2 | 8876 | 34,039 | 31,234 | 61.4 | 66.4 | 8.2; 8.3 | CMC | |
| 8.12.26 | 3193 | 4525 | 3112 | 24.3 | 38.0 | 8.1; 8.2; 8.4; 8.6 | CMC | |
| 8.12.28 | 6164 | 1320 | 1298 | 58.5 | 59.4 | 8.1 | CMC | |
| 8.12.29 | 5731 | 217 | 216 | 86.8 | 87.0 | 8.4 | CMC | |
| 8.12.30 | 2361 | 487 | 487 | 100.0 | 100.0 | 8.1 | CMC | |
| 8.12.3a | *H 11,518 | *H 61,830 | *H 57,428 | 44.8 | 48.0 | 11.14; 11.2; 8.2; 8.3; 8.4; 8.6 | BBN; CMC | |
| 8.12.3b | 1946 | 1894 | 1892 | 88.0 | 88.0 | 8.3 | CMC | |
| 8.12.3c | 8166 | 2127 | 1935 | 11.1 | 12.1 | 11.14; 8.4; 8.5 | BBN; CMC | |
| 8.2.2 | *H 10,341 | 2305 | 1981 | 8.3 | 9.7 | CR | 11.14; 8.1; 8.2; 8.3; 8.4; 8.5; 8.6 | BBN; CMC |
| 8.2.5 | 4501 | 215 | 215 | 8.9 | 8.9 | 8.5 | CMC | |
| 8.2.6b | 2715 | 1255 | 930 | 2.3 | 3.1 | 11.14; 8.5; 8.4 | BBN; CMC | |
| 8.3.10 | 10,176 | 2042 | 1664 | 50.6 | 61.9 | 8.1; 8.2; 8.3; 8.4; 8.6 | CMC | |
| 8.3.1a | 8392 | 11,835 | 5891 | 1.2 | 2.6 | 11.14; 8.1; 8.2; 8.3; 8.6 | BBN; CMC | |
| 8.3.1b | 8478 | 2313 | 1781 | 24.6 | 16.6 | 11.14; 8.4; 8.5 | BBN; CMC | |
| 8.3.9 | 7594 | 1277 | 1277 | 100.0 | 100.0 | 8.1 | CMC | |
| 8.8.1a | 4507 | 2340 | 1222 | 29.9 | 56.5 | 8.3 | CMC | |
| 8.8.1b | 6632 | 946 | 946 | 86.0 | 86.0 | 8.3 | CMC | |
| 9.12.34 | 5103 | 12,161 | 12,151 | 8.0 | 8.0 | 11.1; 11.2; 7.5; 9.4 | BBN; WET; EIU | |
| Mean | 5179 | 9385 | 7926 | 49.0 | 52.0 | |||
| (Stdv) | (2679) | (18,537) | (16,031) | (37.5) | (37.0) | |||
| Total | 18,422 | 516,195 | 435,956 | 37.7 | 48.9 | 13 | 5 |
References
- Webb, L.J.; Tracey, J.G. Australian Rainforest: Pattern and change. In Ecological Biogeography of Australia, 1st ed.; Keast, A., Ed.; Dr. W. Junk: The Hague, The Netherlands, 1981; pp. 605–694. [Google Scholar]
- Fajardo, L.; González, V.; Nassar, J.M.; Lacabana, P.; Portillo Q., C.A.; Carrasquel, F.; Rodríguez, J.P. Tropical Dry Forests of Venezuela: Characterization and Current Conservation Status. Biotropica 2005, 37, 531–546. [Google Scholar] [CrossRef]
- Koenen, E.J.; Clarkson, J.J.; Pennington, T.D.; Chatrou, L.W. Recently evolved diversity and convergent radiations of rainforest mahoganies (Meliaceae) shed new light on the origins of rainforest hyperdiversity. New Phytol. 2015, 207, 327–339. [Google Scholar] [CrossRef]
- Beard, J.S. The Classification of Tropical American Vegetation-Types. Ecology 1995, 36, 89–100. [Google Scholar] [CrossRef]
- Webb, L.J.; Tracey, J.G. The rainforests of northern Australia. In Australian Vegetation; Groves, R.H., Ed.; University Press: Cambridge, UK, 1994. [Google Scholar]
- Gillespie, T.W.; O’Neill, K.; Keppel, G.; Pau, S.; Meyer, J.-Y.; Price, J.P.; Jaffré, T. Prioritizing conservation of tropical dry forests in the Pacific. Oryx 2014, 48, 337–344. [Google Scholar] [CrossRef]
- Janzen, D.H. Management of habitat fragments in a tropical dry forest: Growth. Ann. Missouri. Bot. Garden. 1988, 75, 105–116. [Google Scholar] [CrossRef]
- Mayle, F.E. Assessment of the Neotropical dry forest refugia hypothesis in the light of palaeoecological data and vegetation model simulations. J. Quat. Sci. 2004, 19, 713–720. [Google Scholar] [CrossRef]
- Consiglio, T.; Schatz, G.E.; McPherson, G.; Lowry, P.P., 2nd; Rabenantoandro, J.; Rogers, Z.S.; Rabevohitra, R.; Rabehevitra, D. Deforestation and plant diversity of Madagascar’s littoral forests. Conserv. Biol. 2006, 20, 1799–1803. [Google Scholar] [CrossRef]
- Gillespie, L.J.; Grijalva, A.; Farris, C.N. Diversity, Composition and structure of tropical dry forest in Central america. Plant Ecol. 2000, 147, 37–47. [Google Scholar] [CrossRef]
- Tng, D. Australia’s Seasonally Dry Tropical Forests need attention. Austral. Ecol. 2019, 44, 552–554. [Google Scholar] [CrossRef]
- Blanchard, G.; Birnbaum, P.; Munoz, F. Extinction–immigration dynamics lag behind environmental filtering in shaping the composition of tropical dry forests within a changing landscape. Ecography 2020, 43, 869–881. [Google Scholar] [CrossRef]
- Laidlaw, M.J.; McDonald, W.J.F.; Hunter, R.J.; Putland, D.A.; Kitching, R.L. The potential impacts of climate change on Australian subtropical rainforest. Aust. J. Bot. 2011, 59, 440–449. [Google Scholar] [CrossRef]
- Shimizu-Kimura, Y.; Accad, A.; Shapcott, A. The relationship between climate change and the endangered rainforest shrub Triunia robusta (Proteaceae) endemic to southeast Queensland, Australia. Sci. Rep. 2017, 7, 46399. [Google Scholar] [CrossRef]
- Wilson, B.A.; Nelder, V.J.; Accad, A. The extent and status of remnant vegetation in Queensland and its implications for statewide vegetation management and legislation. Rangel. J. 2002, 24, 6–35. [Google Scholar] [CrossRef]
- Hilbert, D.W.; Graham, A.; Hopkins, M.S. Glacial and interglacial refugia within a long-term rainforest refugium: The Wet Tropics Bioregion of NE Queensland, Australia. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2007, 251, 104–118. [Google Scholar] [CrossRef]
- Kahler, C.P. NQDT Report Beach Scrub BioCondition Assessments November 2016; Ecological Interpretation: Townsville, Australia, 2016. [Google Scholar]
- Attiwill, P.M. The disturbance of forest ecosystems: The ecological basis for conservative management. For. Ecol. Manag. 1994, 63, 247–300. [Google Scholar] [CrossRef]
- Noss, R.F. Beyond Kyoto: Forest Management in a time of rapid climate change. Conserv. Biol. 2001, 15, 578–590. [Google Scholar] [CrossRef]
- Natural Resource Management Ministerial Council. Australia’s Biodiversity Conservation Strategy 2010–2030; Natural Resource Management Ministerial Council: Sydney, Australia, 2010.
- Byrne, M.; Steane, D.A.; Joseph, L.; Yeates, D.K.; Jordan, G.J.; Crayn, D.; Aplin, K.; Cantrill, D.J.; Cook, L.G.; Crisp, M.D.; et al. Decline of a biome: Evolution, contraction, fragmentation, extinction and invasion of the Australian mesic zone biota. J. Biogeogr. 2011, 38, 1635–1656. [Google Scholar] [CrossRef]
- Powell, M.; Accad, A.; Shapcott, A. Where they are, why they are there, and where they are going: Using niche models to assess impacts of disturbance on the distribution of three endemic rare subtropical rainforest trees of Macadamia (Proteaceae) species. Aust. J. Bot. 2014, 62, 322–334. [Google Scholar] [CrossRef]
- Metcalfe, D.J.; Lawson, T.J. An International Union for Conservation of Nature risk assessment of coastal lowland rainforests of the Wet Tropics Bioregion, Queensland, Australia. Austral. Ecol. 2015, 40, 373–385. [Google Scholar] [CrossRef]
- Weber, E.T.; Catterall, C.P.; Locke, J.; Ota, L.S.; Prideaux, B.; Shirreffs, L.; Talbot, L.; Gordon, I.J. Managing a World Heritage Site in the Face of Climate Change: A Case Study of the Wet Tropics in Northern Queensland. Earth 2021, 2, 248–271. [Google Scholar] [CrossRef]
- Colhoun, E.A.; Barrows, T.T. The Glaciation of Australia. In Quaternary Glaciations—Extent and Chronology—A Closer Look; Developments in Quaternary Sciences; Elsevier: Amsterdam, The Netherlands, 2011; pp. 1037–1045. [Google Scholar]
- Kooyman, R.M.; Rossetto, M.; Allen, C.; Cornwell, W. Australian Tropical and Subtropical Rain Forest Community Assembly: Phylogeny, Functional Biogeography, and Environmental Gradients. Biotropica 2012, 44, 668–679. [Google Scholar] [CrossRef]
- De Deckker, P.; Moros, M.; Perner, K.; Blanz, T.; Wacker, L.; Schneider, R.; Barrows, T.T.; O’Loingsigh, T.; Jansen, E. Climatic evolution in the Australian region over the last 94 ka—Spanning human occupancy -, and unveiling the Last Glacial Maximum. Quat. Sci. Rev. 2020, 249, 106593. [Google Scholar] [CrossRef]
- Adam, P. Australian Rainforests; Clarendon Press: Oxford, UK, 1992. [Google Scholar]
- Bowman, D.M.J.S.; Prior, L.D. TURNER REVIEW No. 10. Why do evergreen trees dominate the Australian seasonal tropics? Aust. J. Bot. 2005, 53, 379–399. [Google Scholar] [CrossRef]
- Crayn, D.M.; Costion, C.; Harrington, M.G.; Richardson, J. The Sahul-Sunda floristic exchange: Dated molecular phylogenies document Cenozoic intercontinental dispersal dynamics. J. Biogeogr. 2015, 42, 11–24. [Google Scholar] [CrossRef]
- Keast, A. Ecological Biogeography of Australia, 1st ed.; Keast, A., Ed.; W. Junk: The Hague, The Netherlands, 1981. [Google Scholar]
- Barlow, B.A. Phytogeography of the Australian region. In Australian Vegetation, 2nd ed.; Groves, R.H., Ed.; Univesity Press: Cambridge, UK, 1994; pp. 3–35. [Google Scholar]
- Shapcott, A.; Forster, P.I.; Guymer, G.P.; McDonald, W.J.F.; Faith, D.P.; Erickson, D.; Kress, W.J. Mapping Biodiversity and Setting Conservation Priorities for SE Queensland’s Rainforests Using DNA Barcoding. PLoS ONE 2015, 10, e0122164. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.G.; McDonald, W.J.; Forster, P.I.; Kress, W.J.; Erickson, D.; Faith, D.P.; Shapcott, A. Patterns of Phylogenetic Diversity of Subtropical Rainforest of the Great Sandy Region, Australia Indicate Long Term Climatic Refugia. PLoS ONE 2016, 11, e0153565. [Google Scholar] [CrossRef]
- IBRA. Revision of the Interim Biogeographic Regionalisation of Australia (IBRA) and the Development of Version 5.1.—Summary Report; Department of Climate Change, Energy, the Environment and Water: Canberra, Australia, 2000.
- Neldner, V.J.; Butler, D.W.; Guymer, G.P. Queensland’s Regional Ecosystems: Building a Maintaining a Biodiversity Inventory, Planning Framework and Information System for Queensland, Version 2.0; The State of Queensland (Department of Environment and Science): Manly, Australia, 2019; p. 55.
- Bryan, S.E.; Ewart, A.; Stephens, C.J.; Parianos, J.; Downes, P.J. The Whitsunday Volcanic Province, Central Queensland, Australia: Lithological and stratigraphic investigations of a silicic-dominated large igneous province. J. Volcanol. Geotherm. Res. 2000, 99, 55–78. [Google Scholar] [CrossRef]
- Willmott, W.F. Rock and Landscapes of the National Parks Central Queensland; Geological Society of Australia Incorporated: Brisbane, Australia, 2006. [Google Scholar]
- Cresswell, I.D.; Janke, T.; Johnston, E.L. Australia State of the Environment 2021: Overview, Independent Report to the Australian; Government Minister for the Environment: Wellington, Australia, 2021; p. 274. [CrossRef]
- Shapcott, A.; Liu, Y.; Howard, M.; Forster, P.I.; Kress, W.J.; Erickson, D.L.; Faith, D.P.; Shimizu, Y.; McDonald, W.J.F. Comparing Floristic Diversity and Conservation Priorities across South East Queensland Regional Rain Forest Ecosystems Using Phylodiversity Indexes. Int. J. Plant Sci. 2017, 178, 211–229. [Google Scholar] [CrossRef]
- MacArthur, R.H.; Wilson, E.O. An Equilibrium Theory of Insular Zoogeography. Evolution 1963, 17, 373–387. [Google Scholar] [CrossRef]
- Worboys, G.L.; Pulsford, I. Connectivity Conservation in Australian Landscapes; Department of Sustainability, Environment, Water, Populations and Communities: Canberra, Australia, 2011.
- Pulsford, I.; Lindenmayer, D.; Wyborn, C.; Lausche, B.; Vasilijević, M.; Worboys, G.L.; Lefroy, T. Connectivity Conservation Management. In Protected Area Governance and Management; Worboys, G.L., Lockwood, M., Kothari, A., Feary, S., Pulsford, I., Eds.; ANU Press: Canberra, Australia, 2015; pp. 851–888. [Google Scholar]
- DEE. National Recovery Plan for the Littoral Rainforest and Coastal Vine Thickets of Eastern Australia Ecological Community; Department of the Environment and Energy: Canberra, Australia, 2019.
- Li, D.; Olden, J.D.; Lockwood, J.L.; Record, S.; McKinney, M.L.; Baiser, B. Changes in taxonomic and phylogenetic diversity in the Anthropocene. Proc. Biol. Sci. 2020, 287, 20200777. [Google Scholar] [CrossRef]
- Park, D.S.; Willis, C.G.; Xi, Z.; Kartesz, J.T.; Davis, C.C.; Worthington, S. Machine learning predicts large scale declines in native plant phylogenetic diversity. New Phytol. 2020, 227, 1544–1556. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P. Threatened species and the potential loss of phylogenetic diversity: Conservation scenarios based on estimated extinction probabilities and phylogenetic risk analysis. Conserv. Biol. 2008, 22, 1461–1470. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Costion, C.; Ford, A.; Cross, H.; Crayn, D.; Harrington, M.; Lowe, A. Plant DNA barcodes can accurately estimate species richness in poorly known floras. PLoS ONE 2011, 6, e26841. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Erickson, D.L.; Swenson, N.G.; Thompson, J.; Uriarte, M.; Zimmerman, J.K. Advances in the use of DNA barcodes to build a community phylogeny for tropical trees in a Puerto Rican forest dynamics plot. PLoS ONE 2010, 5, e15409. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Erickson, D.L.; Jones, F.A.; Swenson, N.G.; Perez, R.; Sanjur, O.; Bermingham, E. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc. Natl. Acad. Sci. USA 2009, 106, 18621–18626. [Google Scholar] [CrossRef] [PubMed]
- de Vere, N.; Rich, T.C.; Ford, C.R.; Trinder, S.A.; Long, C.; Moore, C.W.; Satterthwaite, D.; Davies, H.; Allainguillaume, J.; Ronca, S.; et al. DNA barcoding the native flowering plants and conifers of Wales. PLoS ONE 2012, 7, e37945. [Google Scholar] [CrossRef]
- Erickson, D.L.; Jones, F.A.; Swenson, N.G.; Pei, N.; Bourg, N.A.; Chen, W.; Davies, S.J.; Ge, X.J.; Hao, Z.; Howe, R.W.; et al. Comparative evolutionary diversity and phylogenetic structure across multiple forest dynamics plots: A mega-phylogeny approach. Front. Genet. 2014, 5, 358. [Google Scholar] [CrossRef]
- Kooyman, R.; Rossetto, M.; Cornwell, W.; Westoby, M. Phylogenetic tests of community assembly across regional to continental scales in tropical and subtropical rain forests. Glob. Ecol. Biogeogr. 2011, 20, 707–716. [Google Scholar] [CrossRef]
- Weber, L.C.; VanDerWal, J.; Schmidt, S.; McDonald, W.J.F.; Shoo, L.P.; Ladiges, P. Patterns of rain forest plant endemism in subtropical Australia relate to stable mesic refugia and species dispersal limitations. J. Biogeogr. 2014, 41, 222–238. [Google Scholar] [CrossRef]
- Webb, C.O. Exploring the Phylogenetic Structure of Ecological Communities: An Example for Rain Forest Trees. Am. Nat. 2000, 156, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Vamosi, S.M.; Heard, S.B.; Vamosi, J.C.; Webb, C.O. Emerging patterns in the comparative analysis of phylogenetic community structure. Mol. Ecol. 2009, 18, 572–592. [Google Scholar] [CrossRef] [PubMed]
- Hampe, A.; Jump, A.S. Climate Relicts: Past, Present, Future. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 313–333. [Google Scholar] [CrossRef]
- Barthe, S.; Binelli, G.; Herault, B.; Scotti-Saintagne, C.; Sabatier, D.; Scotti, I. Tropical rainforests that persisted: Inferences from the Quaternary demographic history of eight tree species in the Guiana shield. Mol. Ecol. 2017, 26, 1161–1174. [Google Scholar] [CrossRef]
- Gallagher, R.V.; Hughes, L.; Leishman, M.R. Species loss and gain in communities under future climate change: Consequences for functional diversity. Ecography 2013, 36, 531–540. [Google Scholar] [CrossRef]
- McDonald, K.; Alford, R. A Review of Declining Frogs in Northern Queensland; Environment Australia: Canberra, Australia, 1999; pp. 14–22. [Google Scholar]
- Chapple, D.G.; Hoskin, C.J.; Chapple, S.N.J.; Thompson, M.B. Phylogeographic divergence in the widespread delicate skink (Lampropholis delicata) corresponds to dry habitat barriers in eastern Australia. BMC Evol. Biol. 2011, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Melzer, A.; Cristescu, R.; Ellis, W.; FitzGibbon, S.; Manno, G. The habitat and diet of koalas (Phascolarctos cinereus) in Queensland. Aust. Mammal. 2014, 36, 189–199. [Google Scholar] [CrossRef]
- Kier, G.; Mutke, J.; Dinerstein, E.; Ricketts, T.H.; Küper, W.; Kreft, H.; Barthlott, W. Global patterns of plant diversity and floristic knowledge. J. Biogeogr. 2005, 32, 1107–1116. [Google Scholar] [CrossRef]
- Lavorel, S.; Colloff, M.J.; McIntyre, S.; Doherty, M.D.; Murphy, H.T.; Metcalfe, D.J.; Dunlop, M.; Williams, R.J.; Wise, R.M.; Williams, K.J. Ecological mechanisms underpinning climate adaptation services. Glob. Chang. Biol. 2015, 21, 12–31. [Google Scholar] [CrossRef] [PubMed]
- Bean, T. Collection and Preserving Plant Specimens: A Manual; Department of Science, Information Technology, Innovation and the Arts: Brisbane, Australia, 2013.
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Yu, J.; Holder, M.T.; Sukumaran, J.; Mirarab, S.; Oaks, J. SATé Version 2.2.7. 2013. Available online: http://phylo.bio.ku.edu/software/sate/sate.html (accessed on 23 April 2020).
- Katoh, K.; Toh, H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 2010, 26, 1899–1900. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.O.; Donoghue, M.J. Phylomatic: Tree assembly for applied phylogenetics. Mol. Ecol. Notes 2005, 5, 181–183. [Google Scholar] [CrossRef]
- APG. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 2009, 161, 105–121. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.61. 2019. Available online: http://www.mesquiteproject.org (accessed on 1 May 2020).
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for the Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Britton, T.; Anderson, C.L.; Jacquet, D.; Lundqvist, S.; Bremer, K. Estimating divergence times in large phylogenetic trees. Syst. Biol. 2007, 56, 741–752. [Google Scholar] [CrossRef]
- Kress, W.J.; Garcia-Robledo, C.; Uriarte, M.; Erickson, D.L. DNA barcodes for ecology, evolution, and conservation. Trends Ecol. Evol. 2015, 30, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Accad, A.; Nelder, V.J.; Kelley, J.; Li, J.; Richter, D. Remnant Regional Ecosystem Vegetation in Queensland, Analyses 1997–2017. 2019. Available online: https://www.researchgate.net/publication/330725687_Remnant_regional_ecosystem_vegetation_in_Queensland_Analyses_1997-2017 (accessed on 15 August 2019).
- ESRI. ArcGIS; Version 10.7.1; ESRI Inc.: Redlands, CA, USA, 2019. [Google Scholar]
- Queensland Herbarium. Regional Ecosystem Description Database (REDD). Version 11.1 (April 2019); 2019. Available online: https://www.qld.gov.au/environment/plants-animals/plants/ecosystems/descriptions (accessed on 6 July 2020).
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [PubMed]
- Hamady, M.; Lozupone, C.; Knight, R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010, 4, 17–27. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package, R Package Version 2.5-7. 2020. Available online: https://www.researchgate.net/publication/346579465_vegan_community_ecology_package_version_25-7_November_2020 (accessed on 20 February 2023).
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Pohlert, T. The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR). R Package 2014. Available online: http://CRAN.R-project.org/package=PMCMR (accessed on 15 May 2021).
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Kooyman, R.M.; Wilf, P.; Barreda, V.D.; Carpenter, R.J.; Jordan, G.J.; Sniderman, J.M.; Allen, A.; Brodribb, T.J.; Crayn, D.; Feild, T.S.; et al. Paleo-Antarctic rainforest into the modern Old World tropics: The rich past and threatened future of the “southern wet forest survivors”. Am. J. Bot. 2014, 101, 2121–2135. [Google Scholar] [CrossRef]
- Slik, J.W.; Arroyo-Rodriguez, V.; Aiba, S.; Alvarez-Loayza, P.; Alves, L.F.; Ashton, P.; Balvanera, P.; Bastian, M.L.; Bellingham, P.J.; van den Berg, E.; et al. An estimate of the number of tropical tree species. Proc. Natl. Acad. Sci. USA 2015, 112, 7472–7477. [Google Scholar] [CrossRef]
- Ashton, P.; Zhu, H. The tropical-subtropical evergreen forest transition in East Asia: An exploration. Plant Divers 2020, 42, 255–280. [Google Scholar] [CrossRef] [PubMed]
- Yap, J.-Y.S.; Rossetto, M.; Costion, C.; Crayn, D.; Kooyman, R.M.; Richardson, J.; Henry, R. Filters of floristic exchange: How traits and climate shape the rain forest invasion of Sahul from Sunda. J. Biogeogr. 2018. [Google Scholar] [CrossRef]
- Kooyman, R.M.; Rossetto, M.; Sauquet, H.; Laffan, S.W. Landscape patterns in rainforest phylogenetic signal: Isolated islands of refugia or structured continental distributions? PLoS ONE 2013, 8, e80685. [Google Scholar] [CrossRef] [PubMed]
- Gastauer, M.; Thiele, J.; Porembski, S.; Neri, A.V. How do altitude and soil properties influence the taxonomic and phylogenetic structure and diversity of Brazilian páramo vegetation? J. Mt. Sci. 2020, 17, 1045–1057. [Google Scholar] [CrossRef]
- Specht, A. Biodiversity and Conservation. In Australian Vegetation; Groves, R.H., Ed.; University Press: Cambridge, UK, 1994. [Google Scholar]
- Echeverría-Londoño, S.; Enquist, B.J.; Neves, D.M.; Violle, C.; Boyle, B.; Kraft, N.J.B.; Maitner, B.S.; McGill, B.; Peet, R.K.; Sandel, B.; et al. Plant Functional Diversity and the Biogeography of Biomes in North and South America. Front. Ecol. Evol. 2018, 6, 219. [Google Scholar] [CrossRef]
- Xiao, Y.-m.; Yang, L.-c.; Nie, X.-q.; Li, C.-b.; Xiong, F.; Zhou, G.-y. Phylogenetic patterns of shrub communities along the longitudinal and latitudinal gradients on the northeastern Qinghai–Tibetan Plateau. J. Mt. Sci. 2020, 17, 1106–1114. [Google Scholar] [CrossRef]
- Lv, T.; Wang, N.; Xie, L.; Chen, S.; Zhao, R.; Feng, Y.; Li, Y.; Ding, H.; Fang, Y. Environmental Heterogeneity Affecting Community Assembly Patterns and Phylogenetic Diversity of Three Forest Communities at Mt. Huangshan, China. Forests 2022, 13, 133. [Google Scholar] [CrossRef]
- Melick, D.R.; Ashton, D.H. The Effects of Natural Disturbances on Warm Temperate Rainforests in South-eastern Australia. Aust. J. Bot. 1991, 39, 1–30. [Google Scholar] [CrossRef]
- Cali, M. BioCondition Methodology and Fine Scale Mapping of the Remnant Beach Scrubs of Central Queensland; Department of Environment and Science, Queensland Government: Brisbane, Australia, 2008; p. 14.
- Batianoff, G.N.; Nelder, V.J.; Singh, S. Vascular Plant Census and Floristic Analysis of Serpentine Landscapes in Central Queensland. Proc. R. Soc. Qld. 2000, 109, 11–30. [Google Scholar]
- Webb, C.O.; Ackerly, D.D.; McPeek, M.A.; Donoghue, M.J. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 2002, 33, 475–505. [Google Scholar] [CrossRef]
- Jaffré, T. Plant formations on ultramafic rocks in New Caledonia. Bot. Lett. 2022, 1–11. [Google Scholar] [CrossRef]
- Kim, H.; Lee, C.-B. On the relative importance of landscape variables to plant diversity and phylogenetic community structure on uninhabited islands, South Korea. Landsc. Ecol. 2020, 36, 209–221. [Google Scholar] [CrossRef]
- Willmott, W.F. Rocks and Landscapes of the National Parks of North Queensland; Geological Society of Australia Incorporated: Brisbane, Australia, 2009. [Google Scholar]
- Willmott, W.F. Rocks and Landscapes of the National Parks of Southern Queensland, 2nd ed.; Geological Society of Australia Incorporated: Brisbane, Australia, 2014. [Google Scholar]
- Gillison, A.N. The ‘Dry’ Rainforests of Terra Australis; Australian Public Service Commission: Canberra, Australia, 1987; pp. 305–321.
- Fensham, R.J. Floristics and Environmental Relations of inland Dry Rainforest in North Queensland. J. Biogeogr. 1995, 22, 1047–1063. [Google Scholar] [CrossRef]
- Edwards, R.D.; Crisp, M.D.; Cook, D.H.; Cook, L.G. Congruent biogeographical disjunctions at a continent-wide scale: Quantifying and clarifying the role of biogeographic barriers in the Australian tropics. PLoS ONE 2017, 12, e0174812. [Google Scholar] [CrossRef]
- Costion, C.M.; Kress, W.J.; Crayn, D.M. DNA Barcodes Confirm the Taxonomic and Conservation Status of a Species of Tree on the Brink of Extinction in the Pacific. PLoS ONE 2016, 11, e0155118. [Google Scholar] [CrossRef]
- Read, J.; Hope, G.; Hill, R. The Dynamics of Some Nothofagus-Dominated Rain Forests in Papua New Guinea. J. Biogeogr. 1990, 17, 185. [Google Scholar] [CrossRef]
- Apgaua, D.M.G.; Tng, D.Y.P.; Laurance, S.G.W. Tropical wet and dry forest tree species exhibit contrasting hydraulic architecture. Flora 2022, 291, 152072. [Google Scholar] [CrossRef]
- DES. Queensland State of the Environment 2020, Summary; Department of Environment and Science: Manly, Australia, 2021; p. 96.
- Sattler, P.S.; Williams, R. The Conservation Status of Queensland’s Bioregional Ecosystems; Environmental Protection Agency, Queensland Government: Brisbane, Australia, 1999.

| Region | PD | SR | GR | FR | Distinctive Families | Family Distribution |
|---|---|---|---|---|---|---|
| CQC | *H 18,118 | 870 | 467 | 127 | Balanopaceae | CQC, Northern QLD |
| Cochlospermaceae | Northern Australia, NG | |||||
| Connaraceae | CQC; FNQ | |||||
| Maesaceae | CQC, FNQ, NG, Malesia, Melanesia | |||||
| Opiliaceae | Northern Australia, NG, Malesia | |||||
| Sphenostemonaceae | CQC, FNQ | |||||
| SEQ | *L 15,228 | 759 | 388 | 117 | Akaniaceae | SEQ, NSW |
| Aphanopetalaceae | SEQ, NSW | |||||
| Berberidopsidaceae | SEQ, NSW | |||||
| Luzuriagaceae | SEQ, NSW | |||||
| Nothofagaceae | SEQ, NSW, VIC, TAS | |||||
| Petermanniaceae | SEQ, NSW |
| Subregion ID | IBRA Subregion | IBRA Bioregion | SR | PD | MPD | NRI | MNTD | NTI | No. Plots | No. REs |
|---|---|---|---|---|---|---|---|---|---|---|
| 8.1 | Whitsunday | CMC | 535 | 14,018 | 206.5 | 0.85 | 31.4 | *E −2.23 | 167 | 16 |
| 8.3 | Clarke–Connors Ranges | CMC | 555 | 13,544 | 208.6 | −0.06 | 27.7 | 0.19 | 110 | 16 |
| 8.2 | Proserpine–Sarina Lowlands | CMC | 422 | 11,965 | 205.2 | 1.19 | 34.4 | −1.34 | 67 | 10 |
| 11.14 | Marlborough Plains | BBN | 395 | 11,571 | 208.6 | −0.01 | 33.1 | 0.06 | 49 | 12 |
| 11.1 | Townsville Plains | BBN | 367 | 10,970 | 208.2 | 0.05 | 36.7 | −1.19 | 41 | 10 |
| 8.5 | Manifold | CMC | 259 | 9437 | 218.5 | *E −2.48 | 42.8 | −0.76 | 35 | 7 |
| 8.4 | Byfield | CMC | 272 | 9353 | 217.3 | *E −2.22 | 39.4 | 0.12 | 23 | 8 |
| 11.2 | Bogie River Hills | CMC | 262 | 8624 | 203.0 | 1.37 | 39.3 | 0.43 | 15 | 7 |
| 7.5 | Paluma–Seaview | WET | 252 | 8477 | 213.9 | −1.29 | 39.1 | 0.81 | 16 | 8 |
| 8.6 | Debella | CMC | 110 | 5393 | 215.4 | −1.03 | 66.2 | −0.92 | 8 | 2 |
| 9.4 | Broken River | EIU | 102 | 5018 | 203.8 | 0.69 | 60.8 | 0.31 | 4 | 1 |
| 7.1 | Herbert | WET | 94 | 4561 | 223.3 | *E −2.07 | 60.3 | 0.65 | 3 | 3 |
| 11.17 | Boomer Range | BBS | 90 | 4648 | 184.7 | *C 3.38 | 69.3 | −0.54 | 3 | 2 |
| 11.12 | Nebo–Connors Ranges | BBN | 64 | 3496 | 181.5 | *C 3.16 | 73.4 | 0.32 | 1 | 1 |
| 12.3 | Burringbar–Conondale Ranges | SEQ | 460 | 11,756 | 206.6 | 0.69 | 29.0 | 1.08 | 135 | 17 |
| 12.1 | Scenic Rim | SEQ | 478 | 11,572 | 205.8 | 0.99 | 26.5 | *C 2.44 | 55 | 12 |
| 12.4 | Sunshine Coast–Gold Coast Lowlands | SEQ | 441 | 11,442 | 213.0 | *E −1.71 | 29.0 | 1.43 | 46 | 16 |
| 12.1 | Burnett–Curtis Hills and Ranges | SEQ | 436 | 10,906 | 208.3 | 0.05 | 26.9 | *C 2.81 | 75 | 21 |
| 12.7 | Gympie Block | SEQ | 402 | 10,318 | 207.7 | 0.25 | 28.4 | *C 2.41 | 58 | 14 |
| 11.18 | Mount Morgan Ranges | BBS | 288 | 8859 | 199.2 | *C 2.54 | 34.6 | 1.65 | 39 | 12 |
| 12.5 | Brisbane–Barambah Volcanics | SEQ | 299 | 8472 | 200.2 | *C 2.32 | 30.8 | *C 3.05 | 33 | 12 |
| 11.22 | Banana–Auburn Ranges | BBS | 184 | 6506 | 189.5 | *C 3.88 | 44.9 | 0.77 | 12 | 7 |
| 12.8 | Burnett–Curtis Coastal Lowlands | SEQ | 255 | 7798 | 203.7 | 1.14 | 35.5 | *C 2.07 | 13 | 7 |
| 12.6 | South Burnett | SEQ | 239 | 7526 | 197.6 | *C 2.70 | 34.4 | *C 2.93 | 44 | 9 |
| 12.9 | Great Sandy | SEQ | 209 | 7266 | 223.5 | *E −3.25 | 37.8 | *C 2.20 | 47 | 10 |
| 12.2 | Moreton Basin | SEQ | 122 | 5234 | 190.9 | *C 2.82 | 55.1 | 0.55 | 3 | 2 |
| 11.19 | Callide Creek Downs | BBS | 113 | 4836 | 190.6 | *C 2.84 | 54.5 | 0.83 | 8 | 2 |
| 11.27 | Barakula | BBS | 106 | 4547 | 185.3 | *C 3.51 | 52.3 | 1.49 | 2 | 1 |
| 11.31 | Eastern Darling Downs | BBS | 93 | 3946 | 186.3 | *C 3.11 | 46.2 | *C 2.82 | 2 | 2 |
| Mean | 8347 | 203.7 | 42.0 | |||||||
| (Stdv) | (3033) | (11.3) | (13.5) | |||||||
| CSEQ | Total | 1227 | 21,244 |
| Subregion ID | IBRA Subregion | PD | GR | FR | SR | MPD | NRI | MNTD | NTI | Rainfall (mm) |
|---|---|---|---|---|---|---|---|---|---|---|
| 7.1 | Herbert | 4574 | 75 | 42 | 88 | 220.3 | −1.73 | 71.1 | −0.37 | 1224 (184) |
| 7.5 | Paluma–Seaview | 10,973 | 250 | 130 | 389 | 214.9 | *E −2.25 | 31.6 | *C 2.07 | 1600 (354) |
| 8.1 | Whitsunday | 15,310 | 370 | 114 | 616 | 207.3 | 0.69 | 29.8 | −1.39 | 1707 (305) |
| 8.2 | Proserpine–Sarina Lowlands | 14,089 | 351 | 113 | 527 | 205.8 | 1.27 | 33.2 | *E −2.04 | 1435 (258) |
| 8.3 | Clark–Connors Ranges | 14,891 | 368 | 117 | 617 | 209.1 | −0.32 | 27.9 | 0.33 | 1506 (277) |
| 8.4 | Byfield | 10,529 | 230 | 86 | 330 | 214.5 | *E −1.91 | 36.9 | 0.53 | 1201 (117) |
| 8.5 | Manifold | 11,340 | 256 | 98 | 363 | 214.1 | *E −1.92 | 36.6 | −0.14 | 1161 (157) |
| 8.6 | Debella | 8240 | 175 | 69 | 228 | 205.7 | 0.74 | 44.5 | 0.25 | 1260 (97) |
| 9.4 | Broken River | 7630 | 167 | 70 | 199 | 202.8 | 1.44 | 48.1 | −0.01 | 1594 (199) |
| 11.1 | Townsville Plains | 13,250 | 314 | 107 | 468 | 206.7 | 0.82 | 34.6 | −1.54 | 1223 (231) |
| 11.14 | Marlborough Plains | 10,239 | 236 | 81 | 336 | 199.9 | *C 2.84 | 38.3 | −0.26 | 1049 (197) |
| 11.17 | Boomer Range | 1870 | 28 | 19 | 32 | 173.8 | *C 2.83 | 69.8 | *C 2.35 | 839 (24) |
| 11.2 | Bogie River Hills | 12,052 | 284 | 97 | 422 | 202.8 | *C 2.23 | 34.2 | −0.22 | 972 (100) |
| Total | CQC | 19,916 | 525 | 146 | 996 |
| Subregion ID | Dominant Rock | Geological Period | ~Ma | SR Mean (STDv) | FR Mean (STDv) | PD Mean (STDv) | NRI Mean (STDv) | NTI Mean (STDv) |
|---|---|---|---|---|---|---|---|---|
| 8.3 | * Volcanic/Sedimentary (VS) | Early Permian (EP) | 273–299 | 104 (8)FC;GT;GP | 50 (5)GR;GP | 5017 (193)FC;GT;GR;GP | −0.18 (1.16) | −0.28 (0.14) |
| 8.3 | * Mafites (MC) | Early Carboniferous (EA) | 323–359 | 63 (19) | 39 (10)GR | 3497 (774) | −0.57 (0.45) | 0.64 (0.28) |
| 8.3 | * Volcanics (VP)GR | Early Permian (EP) | 273–299 | 60 (18)GR | 37 (8)GR | 3425 (744)GR | 0.46 (0.99) | 0.12 (0.61) |
| 8.1; 8.2 | * Mafites/Felsites (MF) | Early Permian (EP) | 273–299 | 61 (19)FM;FC;GR;GL;GT | 35 (7)FM;FC;GR;GL | 3366 (757)FM;FC;GR;GL;GT | 0.83 (1.06)GC | 0.52 (0.88) |
| 8.4; 11.14 | * Granitoid (GL) | Late Permian (LP) | 252–259 | 55 (14)FC | 35 (7)FC | 3295 (659)FC | −0.94 (1.38) | −0.14 (1.10) |
| 8.1; 8.3 | * Sedimentary (SQ) | Quaternary (Q) | 0–2.6 | 56 (20)GR | 33 (8)GL | 3174 (790)GR;GC | 0.82 (1.11) | 0.30 (0.96) |
| 8.2; 8.3; 11.14 | Felsites (FE) | Late Permian (LP) | 273–299 | 51 (16) | 33 (9) | 3107 (755) | 0.06 (0.96) | −0.19 (0.92) |
| 8.3; 11.2 | Basalt (BT) | Tertiary (T) | 2.6–66 | 51 (16) | 31 (10) | 2994 (720) | −0.41 (1.25) | 0.45 (0.71) |
| 8.2; 8.3; 11.1; 11.2; 7.5 | * Granitoid (GC) | * Carboniferous–Cretaceous (CC) | 101–323 | 54 (19)FM;FC | 33 (9)FC | 2946 (753) | −1.19 (0.67)CQ;EQ;G | 1.04 (1.25)EP;Q;EC;LP;MP;P |
| 11.14 | * Ultramafic Rock (UN) | Neoproterozoic–Early Palaeozoic (NP) | 485–1000 | 48 (8) | 29 (6) | 2850 (489) | *C 2.33 (0.34) GC | 0.41 (0.37) |
| 7.1; 8.1; 8.2; 8.4; 8.5; 11.1; 11.14 | Alluvium (AQ) | Quaternary (Q) | 0–2.6 | 44 (19) | 28 (8) | 2735 (756) | −0.23 (1.26) | 0.34 (0.74) |
| 8.1; 8.2; 8.3; 8.6 | * Volcanic/Sedimentary (VR) | Early Cretaceous (EC) | 66–101 | 45 (22)GR;GP;MF | 28 (11)GR | 2693 (972)GR;GP | 0.18 (1.06)GC;SL | 0.43 (0.99) |
| 8.1; 8.2; 8.4; 8.5; 11.14 | Colluvium (CQ) | Quaternary (Q) | 0–2.6 | 43 (12) | 29 (7) | 2658 (564) | 0.36 (1.04) | 0.54 (0.55) |
| 8.1 | * Volcanic/Sedimentary (VA) | Early Carboniferous (CA) | 323–359 | 47 (9) | 26 (4) | 2646 (368) | 1.02 (0.88)GC | 1.35 (0.67) |
| 8.2 | Gabbroid (G) | Early Cretaceous (EC) | 66–101 | 43 (5) | 27 (4) | 2633 (311) | *C 2.05 (0.32) | 0.48 (0.76) |
| 11.1; 11.2; 11.14; 8.3 | Granitoid (GE) | Early Permian (EP) | 273–299 | 42 (25) | 27 (13) | 2626 (114) | −0.27 (1.59) | 0.09 (1.11) |
| 8.1; 8.2; 11.1; 11.2; 11.14 | * Sand (SL) | Late Quaternary (LQ) | 0–0.01 | 42 (18) | 27 (9) | 2556 (816) | 1.45 (0.86)AQ;BT;GC;FM;GE | 0.63 (0.84) |
| 8.2; 11.14 | Estuarine (EQ) | Late Quaternary (LQ) | 0–0.01 | 43 (4) | 26 (2) | 2528 (206) | *C 2.21 (0.79) | 0.93 (0.52) |
| 8.5; 11.14 | * Sand (SE) | Early Quaternary (LQ) | 0.01–2.6 | 30 (7)MC | 24 (6) | 2200 (408)MF | −0.58 (0.68)SL | −0.01 (0.95) |
| 8.5; 11.14 | Felsites (FM) | Early Mid Permian (MP) | 259–299 | 34 (27) | 23 (13) | 2198 (118) | −0.34 (1.29) | 0.16 (0.86) |
| 8.2 | * Volcanic/Sedimentary (VT) | Mid-Tertiary (MT) | 23–56 | 28 (21)GC;MF | 19 (12)GC;MF | 1879 (107)MF | 0.44 (0.74)MF | 0.59 (0.74) |
| 11.1; 8.3 | * Granitoid (GP) | Permian (P) | 252–299 | 24 (5)GC;GL;FE | 19 (3)GC;GL;FE | 1834 (275)GC;GL;FE | 0.22 (1.29)GC | −0.07 (0.86) |
| 8.6 | Granitoid (GT) | Mid-Triassic (MR) | 237–247 | 19 (5) | 18 (6) | 1554 (262) | −0.09 (1.27) | 0.49 (0.57) |
| 7.5 | Felsites (FC) | Carboniferous (CA) | 229–359 | 18 (4) | 15 (3) | 1512 (232) | −0.33 (0.55) | 0.34 (0.84) |
| 8.2; 8.3 | * Granitoid (GR) | Early Cretaceous (EC) | 66–101 | 13 (9)BT;FE;GC;GL | 11 (7)GL;GC;FE | 1171 (514)BT;FE;GC | −0.01 (0.78) | 0.06 (0.51) |
| F | 2.922 ** |
| Group | PD | SR | GR | FR | MPD | NRI | MNTD | NTI | No. REs |
|---|---|---|---|---|---|---|---|---|---|
| Central Coast (Central) | 18,254 | 833 | 471 | 133 | 207.9 | 0.07 | 24.9 | −1.59 | 40 |
| Burdekin Gap (BGap) | 15,302 | 607 | 376 | 118 | 205.5 | 1.36 | 31.03 | *E −2.06 | 13 |
| North of BGap | 13,276 | 519 | 326 | 114 | 208.5 | −1.53 | 27.3 | 1.17 | 23 |
| South of SLGap | 13,199 | 466 | 313 | 108 | 205.4 | *E −1.72 | 29.1 | 0.07 | 20 |
| St Lawrence Gap (SLGap) | 10,271 | 338 | 237 | 81 | 208.3 | *C 2.82 | 28.7 | −0.32 | 8 |
| Subregion ID | IBRA Subregion | Preclear Area (Ha) | Remnant Area (Ha) | Preclear Rf Res (Ha) | Remnant Rf Res (Ha) | Preclear Rf PA (Ha) | Remnant Rf PA (Ha) | % Remnant Rf PA |
|---|---|---|---|---|---|---|---|---|
| 9.4 | Broken River | *H 3,309,590 | *H 3,222,744 | 30,036 | 29,127 | 1075 | 1075 | 4 |
| 11.14 | Marlborough Plains | 1,179,472 | 632,749 | 34,153 | 17,953 | 6686 | 6551 | 36 |
| 11.2 | Bogie River Hills | 1,054,241 | 840,695 | 22,606 | 17,551 | 3448 | 3389 | 19 |
| 11.1 | Townsville Plains | 760,555 | 525,994 | 11,402 | 10,755 | 5668 | 5668 | 53 |
| 8.3 | Clarke–Connors Ranges | 631,881 | 542,056 | *H 134,461 | *H 122,310 | *H 78,396 | *H 77,827 | 64 |
| 8.2 | Proserpine–Sarina Lowlands | 463,733 | 156,347 | 19,088 | 12,258 | 2593 | 2578 | 21 |
| 7.5 | Paluma–Seaview | 233,751 | 230,215 | 56,320 | 56,015 | 52,213 | 50,054 | *H 89 |
| 11.17 | Boomer Range | 220,541 | 95,187 | 35,487 | 10,919 | 3561 | 3507 | 32 |
| 7.1 | Herbert | 219,426 | 104,995 | 12,608 | 6033 | 1464 | 1456 | 24 |
| 8.4 | Byfield | 127,034 | 118,630 | 7968 | 7486 | 1661 | 1463 | 20 |
| 8.1 | Whitsunday | 88,503 | 82,430 | 50,737 | 49,509 | 42,357 | 42,411 | 86 |
| 8.6 | Debella | 80,853 | 53,664 | 999 | 989 | 166 | 174 | 18 |
| 8.5 | Manifold | 67,850 | 63,298 | 2585 | 2407 | 367 | 281 | 12 |
| Mean | 649,033 | 513,000 | 32,188 | 26,409 | 15,358 | 15,110 | 32 | |
| (Stdv) | (848,487) | (821,246) | (33,878) | (32,160) | (24,360) | (24,026) | (23) | |
| Total | CQC | 8,437,430 | 6,669,006 | 533,856 | 442,232 | 216,668 | 214,440 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Howard, M.; Pearl, H.; McDonald, W.J.F.; Shimizu, Y.; Srivastava, S.K.; Shapcott, A. Assessment of the Diversity, Distinctiveness and Conservation of Australia’s Central Queensland Coastal Rainforests Using DNA Barcoding. Diversity 2023, 15, 378. https://doi.org/10.3390/d15030378
Howard M, Pearl H, McDonald WJF, Shimizu Y, Srivastava SK, Shapcott A. Assessment of the Diversity, Distinctiveness and Conservation of Australia’s Central Queensland Coastal Rainforests Using DNA Barcoding. Diversity. 2023; 15(3):378. https://doi.org/10.3390/d15030378
Chicago/Turabian StyleHoward, Marion, Hilary Pearl, William J. F. McDonald, Yoko Shimizu, Sanjeev Kumar Srivastava, and Alison Shapcott. 2023. "Assessment of the Diversity, Distinctiveness and Conservation of Australia’s Central Queensland Coastal Rainforests Using DNA Barcoding" Diversity 15, no. 3: 378. https://doi.org/10.3390/d15030378
APA StyleHoward, M., Pearl, H., McDonald, W. J. F., Shimizu, Y., Srivastava, S. K., & Shapcott, A. (2023). Assessment of the Diversity, Distinctiveness and Conservation of Australia’s Central Queensland Coastal Rainforests Using DNA Barcoding. Diversity, 15(3), 378. https://doi.org/10.3390/d15030378









