Investigating an Unknown Biodiversity: Evidence of Distinct Lineages of the Endemic Chola Guitarfish Pseudobatos percellens Walbaum, 1792 in the Western Atlantic Ocean
Abstract
1. Introduction
2. Material and Methods
2.1. Sample Collection and DNA Barcoding
2.2. ddRAD Library Preparation
2.3. Population Genomics Analysis
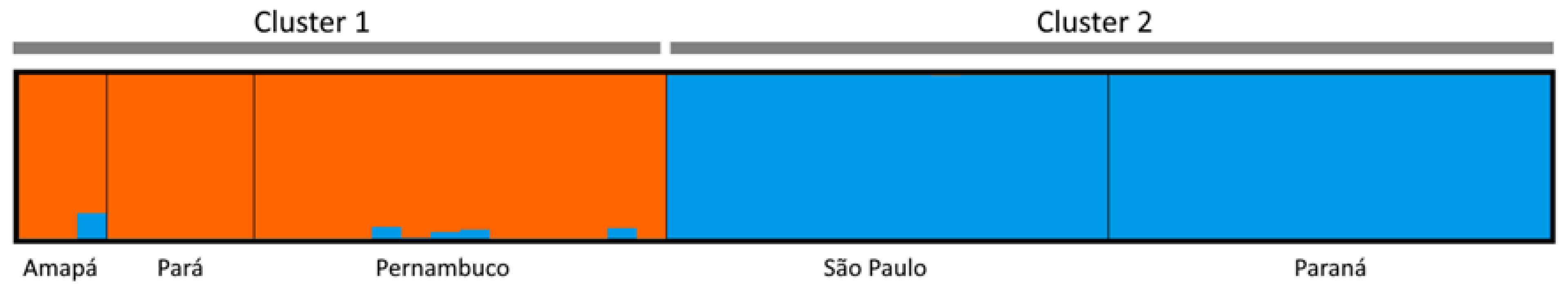
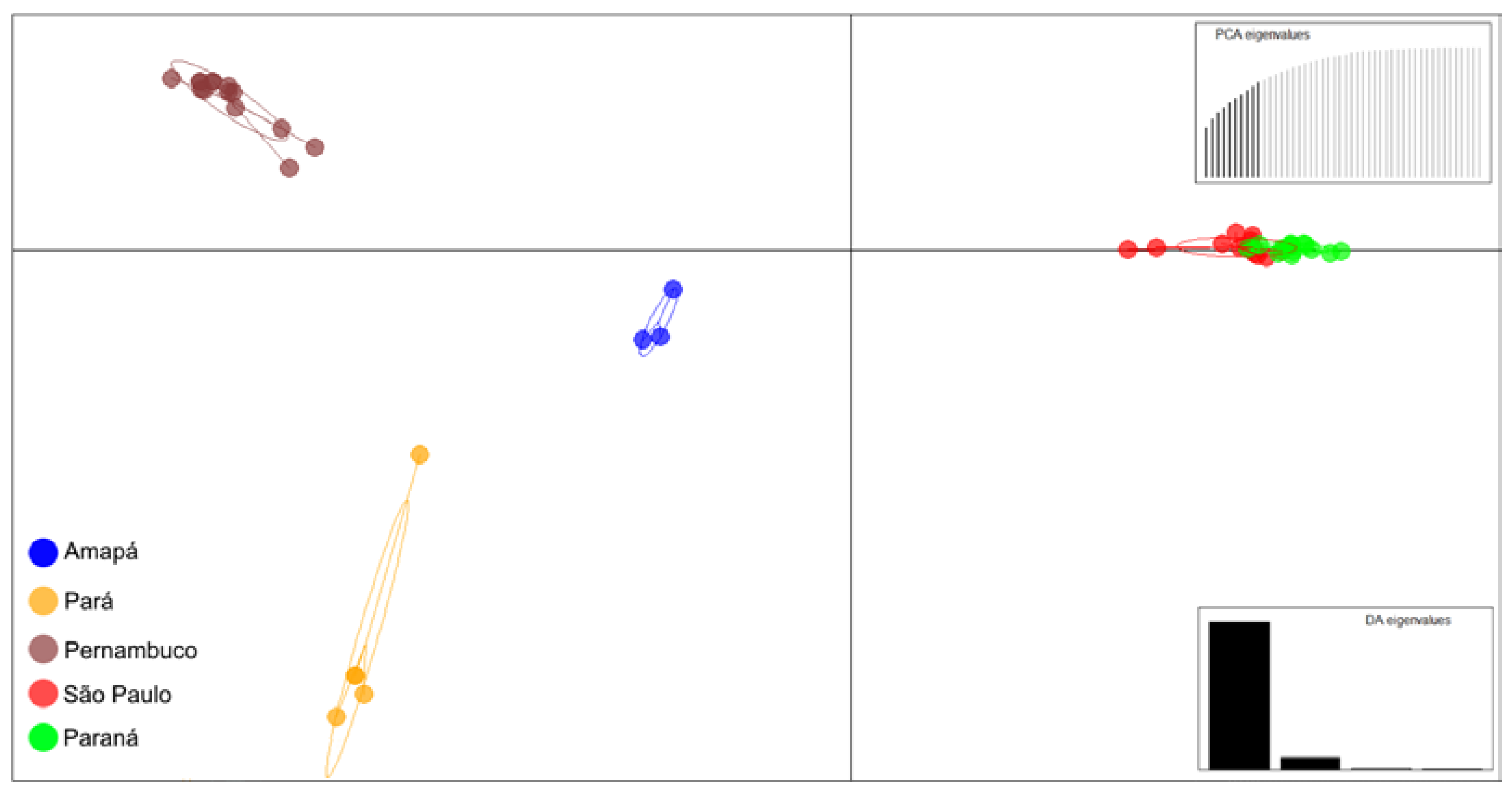
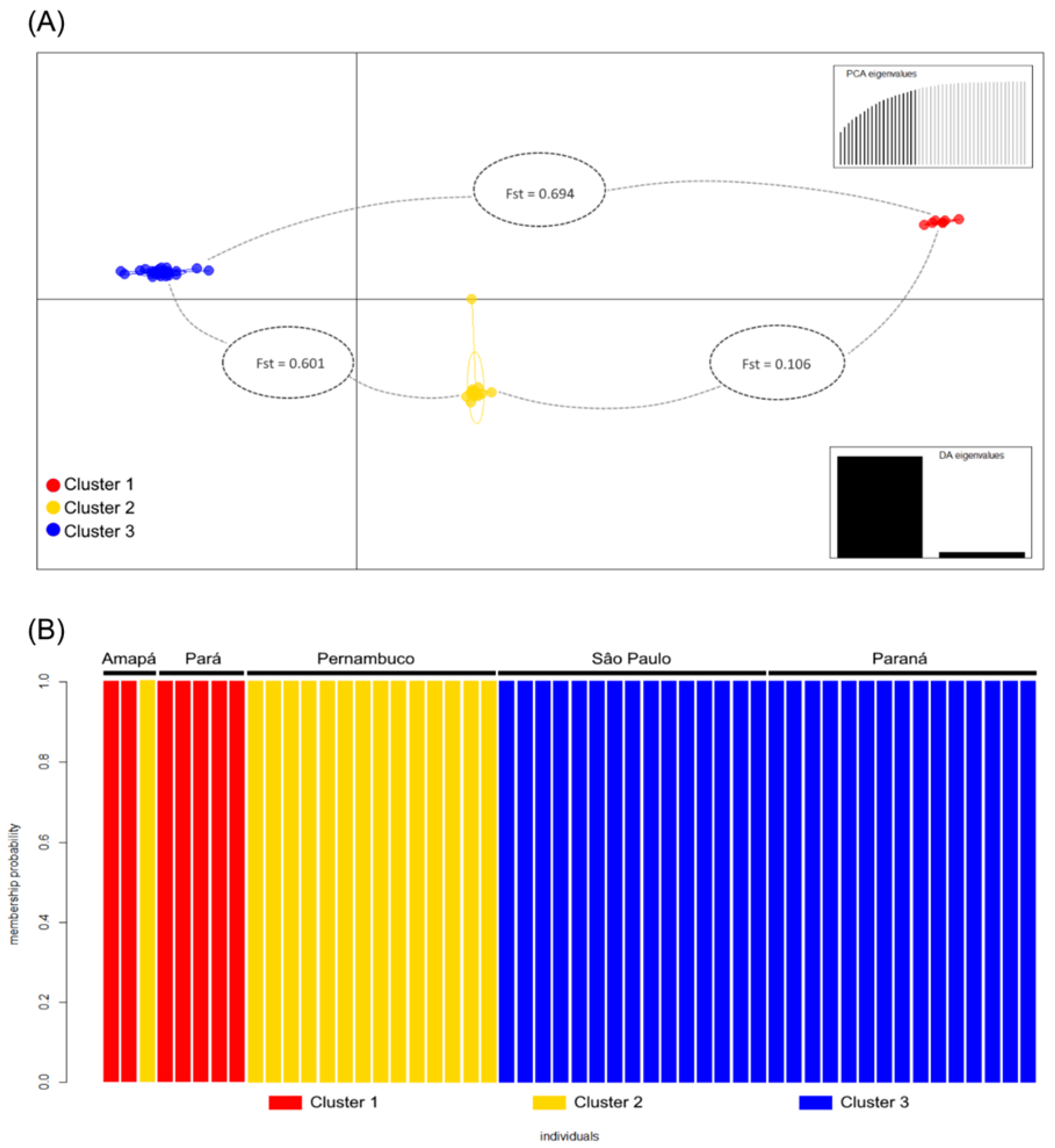
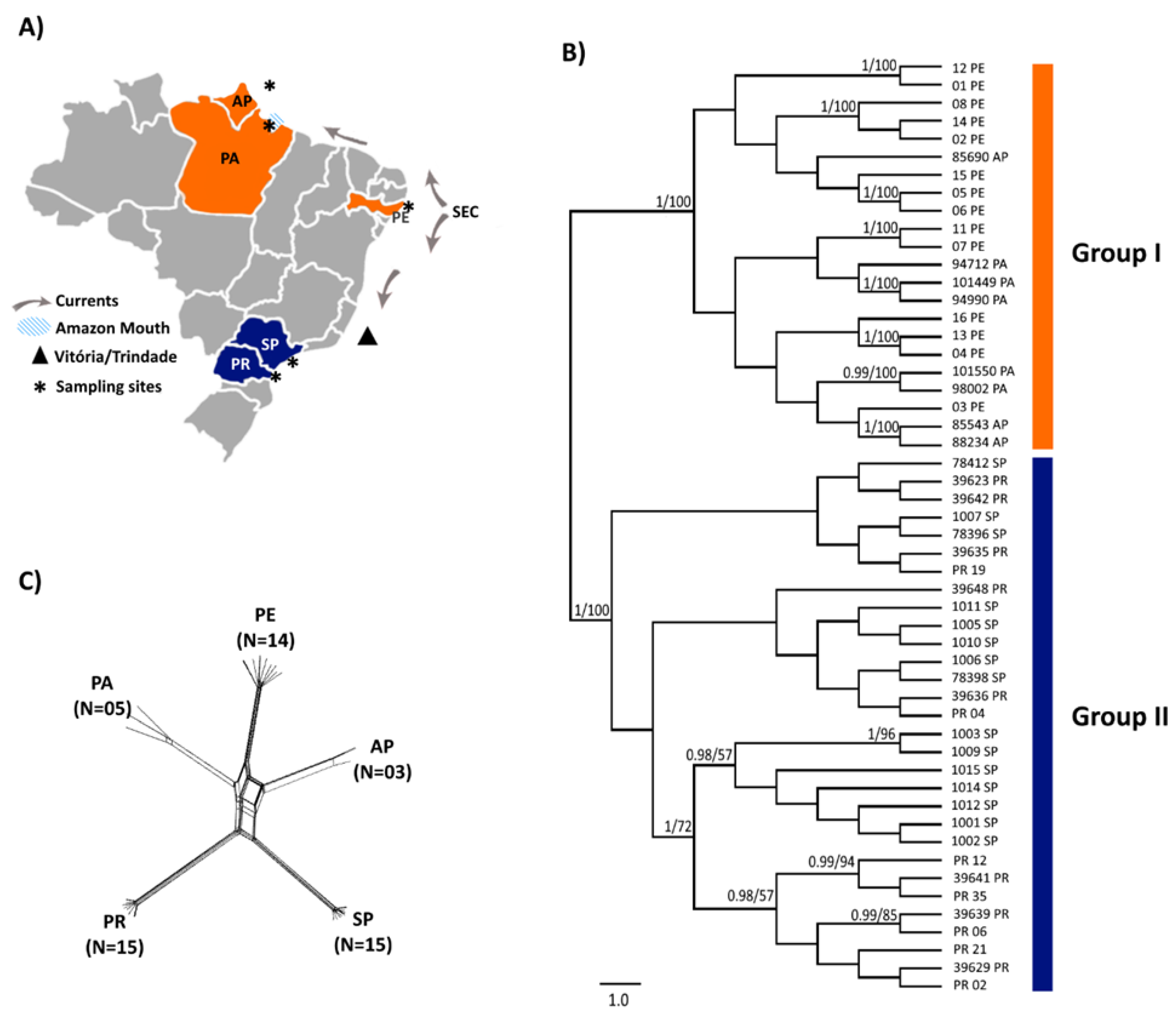
3. Results
4. Discussion
Factors Influencing Dispersal: Biogeography Barriers along the Brazilian Coast
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, H.F.; Kyne, P.M.; Jabado, R.W.; Leeney, R.H.; Davidson, L.N.; Derrick, D.H.; Finucci, B.; Freckleton, R.P.; Fordham, S.V.; Dulvy, N.K. Overfishing and habitat loss drive range contraction of iconic marine fishes to near extinction. Sci. Adv. 2021, 7, eabb6026. [Google Scholar] [CrossRef] [PubMed]
- Kappel, C.V. Losing pieces of the puzzle: Threats to marine, estuarine, and diadromous species. Front. Ecol. Environ. 2005, 3, 275–282. [Google Scholar] [CrossRef]
- Rosser, A.M.; Mainka, S.A. Overexploitation and species extinctions. Conserv. Biol. 2002, 16, 584–586. [Google Scholar] [CrossRef]
- Sala, O.E.; Stuart Chapin, F.I.; Armesto, J.J.; Berlow, E.; Bloomfield, J.; Dirzo, R.; Huber-Sanwald, E.; Huenneke, L.F.; Jackson, R.B.; Kinzig, A.; et al. Global biodiversity scenarios for the year 2100. Science 2000, 287, 1770–1774. [Google Scholar] [CrossRef]
- Tilman, D.; Clark, M.; Williams, D.R.; Kimmel, K.; Polasky, S.; Packer, C. Future threats to biodiversity and pathways to their prevention. Nature 2017, 546, 73–81. [Google Scholar] [CrossRef]
- Dulvy, N.K.; Pacoureau, N.; Rigby, C.L.; Pollom, R.A.; Jabado, R.W.; Ebert, D.A.; Simpfendorfer, C.A. Overfishing drives over one-third of all sharks and rays toward a global extinction crisis. Curr. Biol. 2021, 31, 4773–4787. [Google Scholar] [CrossRef]
- Díaz, S.; Zafra-Calvo, N.; Purvis, A.; Verburg, P.H.; Obura, D.; Leadley, P.; Zanne, A.E. Set ambitious goals for biodiversity and sustainability. Science 2020, 370, 411–413. [Google Scholar] [CrossRef]
- Hooper, D.U.; Adair, E.C.; Cardinale, B.J.; Byrnes, J.E.; Hungate, B.A.; Matulich, K.L.; O’Connor, M.I. A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature 2012, 486, 105–108. [Google Scholar] [CrossRef]
- Pasquaud, S.; Vasconcelos, R.P.; França, S.; Henriques, S.; Costa, M.J.; Cabral, H. Worldwide patterns of fish biodiversity in estuaries: Effect of global vs. local factors. Estuar. Coast. Shelf Sci. 2015, 154, 122–128. [Google Scholar] [CrossRef]
- Habibullah, M.S.; Din, B.H.; Tan, S.H.; Zahid, H. Impact of climate change on biodiversity loss: Global evidence. Environ. Sci. Pollut. Res. 2022, 29, 1073–1086. [Google Scholar] [CrossRef]
- Reader, M.O.; Eppinga, M.B.; de Boer, H.J.; Damm, A.; Petchey, O.L.; Santos, M.J. Biodiversity mediates relationships between anthropogenic drivers and ecosystem services across global mountain, island and delta systems. Glob. Environ. Change 2023, 78, 102612. [Google Scholar] [CrossRef]
- Liu, J.; Slik, F.; Zheng, S.; Lindenmayer, D.B. Undescribed species have higher extinction risk than known species. Conserv. Lett. 2022, 15, e12876. [Google Scholar] [CrossRef]
- Chowdhury, S. Threatened species could be more vulnerable to climate change in tropical countries. Sci. Total Environ. 2023, 858, 159989. [Google Scholar] [CrossRef] [PubMed]
- Bertorelle, G.; Bruford, M.W.; Hauffe, H.C.; Rizzoli, A.; Vernesi, C. Population Genetics for Animal Conservation; United States of America by Cambridge University Press: New York, NY, USA, 2009. [Google Scholar]
- Allendorf, F.W. Genetics and the conservation of natural populations: Allozymes to genomes. Mol. Ecol. 2017, 26, 420–430. [Google Scholar] [CrossRef]
- Hanson, J.O.; Rhodes, J.R.; Butchart, S.H.; Buchanan, G.M.; Rondinini, C.; Ficetola, G.F.; Fuller, R.A. Global conservation of species’ niches. Nature 2020, 580, 232–234. [Google Scholar] [CrossRef]
- De Kort, H.; Prunier, J.G.; Ducatez, S.; Honnay, O.; Baguette, M.; Stevens, V.M.; Blanchet, S. Life history, climate and biogeography interactively affect worldwide genetic diversity of plant and animal populations. Nat. Commun. 2021, 12, 516. [Google Scholar] [CrossRef] [PubMed]
- Stronen, A.V.; Norman, A.J.; Vander Wal, E.; Paquet, P.C. The relevance of genetic structure in ecotype designation and conservation management. Evol. Appl. 2022, 15, 185–202. [Google Scholar] [CrossRef] [PubMed]
- DeSalle, R.; Amato, G. The Expansion of Conservation Genetics. In Conservation Genetics in the Age of Genomics; Amato, G., DeSalle, R., Ryder, O., Rosenbaum, H., Eds.; Publisher Columbia University Press: New York, NY, USA, 2009; Volume 1, pp. 1–24. [Google Scholar] [CrossRef]
- Frankham, R. Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 2010, 143, 1919–1927. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Dudash, M.R.; Eldridge, M.D.; Fenster, C.B.; Lacy, R.C.; Ryder, O.A. Implications of different species concepts for conserving biodiversity. Biol. Conserv. 2012, 153, 25–31. [Google Scholar] [CrossRef]
- Bernatchez, L. On the maintenance of genetic variation and adaptation to environmental change: Considerations from population genomics in fishes. J. Fish Biol. 2016, 89, 2519–2556. [Google Scholar] [CrossRef]
- Coates, D.J.; Byrne, M.; Moritz, C. Genetic diversity and conservation units: Dealing with the species-population continuum in the age of genomics. Front. Ecol. Evol. 2018, 6, 165. [Google Scholar] [CrossRef]
- Willi, Y.; Kristensen, T.N.; Sgrò, C.M.; Weeks, A.R.; Ørsted, M.; Hoffmann, A.A. Conservation genetics as a management tool: The five best-supported paradigms to assist the management of threatened species. Proc. Natl. Acad. Sci. USA 2022, 119, e2105076119. [Google Scholar] [CrossRef]
- Cortés, E. Life history patterns and correlations in sharks. Rev. Fish. Sci. 2000, 8, 299–344. [Google Scholar] [CrossRef]
- Stevens, J.D.; Bonfil, R.; Dulvy, N.K.; Walker, P.A. The effects of fishing on sharks, rays, and chimaeras (chondrichthyans), and the implications for marine ecosystems. ICES J. Mar. Sci. 2000, 57, 476–494. [Google Scholar] [CrossRef]
- Whitenack, L.B.; Kim, S.L.; Sibert, E.C. Bridging the gap between chondrichthyan paleobiology and biology. In Biology of Sharks and Their Relatives, 3rd ed.; Carrier, J.C., Simpfendorfer, C.A., Heithaus, M.R., Yopak, K.E., Eds.; CRC Press: Boca Raton, FL, USA, 2022; pp. 1–29. [Google Scholar]
- Ferretti, F.; Worm, B.; Britten, G.L.; Heithaus, M.R.; Lotze, H.K. Patterns and ecosystem consequences of shark declines in the ocean. Ecol. Lett. 2010, 13, 1055–1071. [Google Scholar] [CrossRef] [PubMed]
- Navia, A.F.; Cortés, E.; Mejía-Falla, P.A. Topological analysis of the ecological importance of elasmobranch fishes: A food web study on the Gulf of Tortugas, Colombia. Ecol. Modell. 2010, 221, 2918–2926. [Google Scholar] [CrossRef]
- Bornatowski, H.; Navia, A.F.; Braga, R.R.; Abilhoa, V.; Corrêa, M.F.M. Ecological importance of sharks and rays in a structural foodweb analysis in southern Brazil. ICES J. Mar. Sci. 2014, 71, 1586–1592. [Google Scholar] [CrossRef]
- Rupp, A.; Bornatowski, H. Food web model to assess the fishing impacts and ecological role of elasmobranchs in a coastal ecosystem of Southern Brazil. Environ. Biol. Fishes 2021, 104, 905–921. [Google Scholar] [CrossRef]
- Froese, R.; Pauly, D. Editors. FishBase. World Wide Web Electronic Publication. 2022. Available online: www.fishbase.org,version (accessed on 1 August 2022).
- Castillo-Páez, A.; Sosa-Nishizaki, O.; Sandoval-Castillo, J.; Galván-Magaña, F.; Blanco-Parra, M.D.P.; Rocha-Olivares, A. Strong population structure and shallow mitochondrial phylogeny in the banded guitarfish, Zapteryx exasperata (Jordan y Gilbert, 1880), from the northern Mexican Pacific. J. Hered. 2014, 105, 91–100. [Google Scholar] [CrossRef]
- Dulvy, N.K.; Fowler, S.L.; Musick, J.A.; Cavanagh, R.D.; Kyne, P.M.; Harrison, L.R.; White, W.T. Extinction risk and conservation of the world’s sharks and rays. Elife 2014, 3, e00590. [Google Scholar] [CrossRef]
- Shiffman, D.S.; Bittick, S.J.; Cashion, M.S.; Colla, S.R.; Coristine, L.E.; Derrick, D.H.; Dulvy, N.K. Inaccurate and biased global media coverage underlies public misunderstanding of shark conservation threats and solutions. Iscience 2020, 23, 101205. [Google Scholar] [CrossRef]
- Cardeñosa, D.; Hyde, J.; Caballero, S. Genetic diversity and population structure of the pelagic thresher shark (Alopias pelagicus) in the Pacific Ocean: Evidence for two evolutionarily significant units. PLoS ONE 2014, 9, e110193. [Google Scholar] [CrossRef]
- Carmo, C.B.; Ferrette, B.L.; Camargo, S.M.; Roxo, F.F.; Coelho, R.; Garla, R.C.; Mendonça, F.F. A new map of the tiger shark (Galeocerdo cuvier) genetic population structure in the western Atlantic Ocean: Hypothesis of an equatorial convergence centre. Aquat. Conserv. 2019, 29, 760–772. [Google Scholar] [CrossRef]
- Bargnesi, F.; Lucrezi, S.; Ferretti, F. Opportunities from citizen science for shark conservation, with a focus on the Mediterranean Sea. Eur. Zool. J. 2020, 87, 20–34. [Google Scholar] [CrossRef]
- Bernard, A.M.; Finnegan, K.A.; Pavinski Bitar, P.; Stanhope, M.J.; Shivji, M.S. Genomic assessment of global population structure in a highly migratory and habitat versatile apex predator, the tiger shark (Galeocerdo cuvier). J. Hered. 2021, 112, 497–507. [Google Scholar] [CrossRef]
- Hammerschlag, N.; McDonnell, L.H.; Rider, M.J.; Street, G.M.; Hazen, E.L.; Natanson, L.J.; Kirtman, B. Ocean warming alters the distributional range, migratory timing, and spatial protections of an apex predator, the tiger shark (Galeocerdo cuvier). Glob. Change Biol. 2022, 28, 1990–2005. [Google Scholar] [CrossRef] [PubMed]
- Veríssimo, A.; Sampaio, Í.; McDowell, J.R.; Alexandrino, P.; Mucientes, G.; Queiroz, N.; Noble, L.R. World without borders—Genetic population structure of a highly migratory marine predator, the blue shark (Prionace glauca). Ecol. Evol. 2017, 7, 4768–4781. [Google Scholar] [CrossRef]
- Bailleul, D.; Mackenzie, A.; Sacchi, O.; Poisson, F.; Bierne, N.; Arnaud-Haond, S. Large-scale genetic panmixia in the blue shark (Prionace glauca): A single worldwide population, or a genetic lag-time effect of the “grey zone” of differentiation? Evol. Appl. 2018, 11, 614–630. [Google Scholar] [CrossRef] [PubMed]
- Boldrocchi, G.; Storai, T. Data-mining social media platforms highlights conservation action for the Mediterranean Critically Endangered blue shark Prionace glauca. Aquat. Conserv. 2021, 31, 3087–3099. [Google Scholar] [CrossRef]
- Godínez-Padilla, C.J.; Castillo-Géniz, J.L.; Hernandez de la Torre, B.; González-Ania, L.V.; Román-Verdesoto, M.H. Marine-climate interactions with the blue shark (Prionace glauca) catches in the western coast of Baja California Peninsula, Mexico. Fish. Oceanogr. 2022, 31, 291–318. [Google Scholar] [CrossRef]
- Last, P.R.; Henderson, A.C.; Naylor, G.J. Acroteriobatus omanensis (Batoidea: Rhinobatidae), a new guitarfish from the Gulf of Oman. Zootaxa 2016, 4144, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Last, P.R.; Seret, B.; Naylor, G.J. A new species of guitarfish, Rhinobatos borneensis sp. nov. with a redefinition of the family-level classification in the order Rhinopristiformes (Chondrichthyes: Batoidea). Zootaxa 2016, 4117, 451–475. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, J.L. Manual dos Peixes Marinhos do Sudeste do Brasil. I. Introdução. Cações, Raias e Quimeras. Museu de Zoologia; Universidade de São Paulo: São Paulo, Brasil, 1977; p. 104. [Google Scholar]
- Cervigón, F.; Alcalá, A. Los peces marinos de Venezuela, Vol. I. Segunda edición; Fundación Museo del Mar: Caracas, Venezuela, 1999; p. 425. [Google Scholar]
- McEachran, J.D.; de Carvalho, M.R. Batoid fishes. In The Living Marine Resources of the Western Central Atlantic; Kent E. Carpenter: Rome, Italy, 2002. [Google Scholar]
- Rocha, F.; Gadig, O.B.F. Reproductive biology of the guitarfish Rhinobatos percellens (Chondrichthyes, Rhinobatidae) from the São Paulo Coast, Brazil, western South Atlantic Ocean. J. Fish Biol. 2013, 82, 306–317. [Google Scholar] [CrossRef]
- Pinheiro, H.T.; Bernardi, G.; Simon, T.; Joyeux, J.C.; Macieira, R.M.; Gasparini, J.L.; Rocha, L.A. Island biogeography of marine organisms. Nature 2017, 549, 82–85. [Google Scholar] [CrossRef]
- Hirschfeld, M.; Dudgeon, C.; Sheaves, M.; Barnett, A. Barriers in a sea of elasmobranchs: From fishing for populations to testing hypotheses in population genetics. Glob. Ecol. Biogeogr. 2021, 30, 2147–2163. [Google Scholar] [CrossRef]
- Sullivan-Sealy, K.; Bustamante, G. Setting Geographic Priorities for Marine Conservation in Latin America and the Caribbean; The Nature Conservancy: Arlington, VA, USA, 1999; p. 125. [Google Scholar]
- Miloslavich, P.; Klein, E.; Díaz, J.M.; Hernandez, C.E.; Bigatti, G.; Campos, L.; Martín, A. Marine biodiversity in the Atlantic and Pacific coasts of South America: Knowledge and gaps. PLoS ONE 2011, 6, e14631. [Google Scholar] [CrossRef]
- Giachini-Tosetto, E.; Bertrand, A.; Neumann-Leitão, S.; Nogueira Júnior, M. The Amazon River plume, a barrier to animal dispersal in the Western Tropical Atlantic. Sci. Rep. 2022, 12, 537. [Google Scholar] [CrossRef]
- Menni, R.C.; Jaureguizar, A.J.; Stehmann, M.F.; Lucifora, L.O. Marine biodiversity at the community level: Zoogeography of sharks, skates, rays and chimaeras in the southwestern Atlantic. Biodivers. Conserv. 2010, 19, 775–796. [Google Scholar] [CrossRef]
- Caires, S.A.; Wuddivira, M.N.; Bekele, I. Assessing the Temporal Stability of Spatial Patterns of Soil Apparent Electrical Conductivity using Geophysical Methods; International Agrophysics: Lublin, Poland, 2014; Volume 28, pp. 423–433. [Google Scholar] [CrossRef]
- Séret, B.; Last, P.R.; Naylor, G.J.P. Guitarfishes. Family Rhinobatidae. Rays of the World; Cornell University Press: Ithaca, NY, USA, 2016; pp. 77–109. [Google Scholar]
- Morley, S.A.; Abele, D.; Barnes, D.K.; Cárdenas, C.A.; Cotté, C.; Gutt, J.; Henley, S.F.; Höfer, J.; Hughes, K.A.; Martin, S.M.; et al. Global drivers on Southern Ocean ecosystems: Changing physical environments and anthropogenic pressures in an earth system. Front. Mar. Sci. 2020, 7, 547188. [Google Scholar] [CrossRef]
- Rocha, L.A. Patterns of distribution and processes of speciation in Brazilian reef fishes. J. Biogeogr. 2003, 30, 1161–1171. [Google Scholar] [CrossRef]
- Floeter, S.R.; Rocha, L.A.; Robertson, D.R.; Joyeux, J.C.; Smith-Vaniz, W.F.; Wirtz, P.; Bernardi, G. Atlantic reef fish biogeography and evolution. J. Biogeogr. 2008, 35, 22–47. [Google Scholar] [CrossRef]
- Rodríguez-Rey, G.T.; Carvalho Filho, A.; De Araújo, M.E.; Solé-Cava, A.M. Evolutionary history of Bathygobius (Perciformes: Gobiidae) in the Atlantic biogeographic provinces: A new endemic species and old mitochondrial lineages. Zool. J. Linn. Soc. 2018, 182, 360–384. [Google Scholar] [CrossRef]
- Araujo, G.S.; Rocha, L.A.; Lastrucci, N.S.; Luiz, O.J.; Di Dario, F.; Floeter, S.R. The Amazon-Orinoco Barrier as a driver of reef-fish speciation in the Western Atlantic through time. J. Biogeogr. 2022, 49, 1407–1419. [Google Scholar] [CrossRef]
- Yokota, L.; Lessa, R.P. A nursery area for sharks and rays in Northeastern Brazil. Environ. Biol. Fishes 1998, 75, 349–360. [Google Scholar] [CrossRef]
- Caltabellotta, F.P.; Siders, Z.A.; Murie, D.J.; Motta, F.S.; Cailliet, G.M.; Gadig, O.B. Age and growth of thre endemic threatened guitarfishes Pseudobatos horkelii, P. percellens and Zapteryx brevirostris in the western South Atlantic Ocean. J. Fish Biol. 2019, 95, 1236–1248. [Google Scholar] [CrossRef] [PubMed]
- Cruz, V.P.; Adachi, A.M.C.L.; Oliveira, P.H.; Ribeiro, G.S.; Paim, F.G.; Souza, B.C.; Foresti, F. Genetic diversity in two threatened species of guitarfish (Elasmobranchii: Rhinobatidae) from the Brazilian and Argentinian coasts: An alert for conservation. Neotrop. Ichthyol. 2021, 19, 1–15. [Google Scholar] [CrossRef]
- Barrett, S.C.; Kohn, J.R. Genetic and evolutionary consequences of small population size in plants: Implications for Conservation. In Genetics and Conservation of Rare Plants; Oxford University Press: New York, NY, USA, 1991; pp. 3–30. [Google Scholar]
- Gonçalves-Silva, F.; Vianna, M. Use of a species-rich and degraded tropical estuary by Elasmobranchs. Braz. J. Oceanogr. 2018, 66, 339–346. [Google Scholar] [CrossRef]
- Pollom, R.; Charvet, P.; Avalos, C.; Blanco-Parra, M.P.; Derrick, D.; Espinoza, E.; Faria, V.; Herman, K.; Mejía-Falla, P.A.; Motta, F.; et al. Pseudobatos percellens. The IUCN Red List of Threatened Species 2020-1.2020. E.T161373A887217. Available online: https://www.iucnredlist.org (accessed on 21 March 2021).
- Instituto Chico Mendes de Conservação da Biodiversidade, ICMBio. 2022. Available online: https://icmbio.gov.br (accessed on 18 October 2022).
- Martins, M.F.; Costa, P.G.; Gadig, O.B.; Bianchini, A. Metal contamination in threatened elasmobranchs from an impacted urban coast. Sci. Total Environ. 2021, 757, 143803. [Google Scholar] [CrossRef]
- Costa, L.; Chaves, P.D.T. D C. Elasmobrânquios capturados pela pesca artesanal na costa sul do Paraná e norte de Santa Catarina, Brasil. Biota Neotrop. 2006, 6, 1–10. [Google Scholar] [CrossRef]
- Rodrigues, A.F.S.; de Sousa Rangel, B.; Wosnick, N.; Bornatowski, H.; Santos, J.L.; Moreira, R.G.; de Amorim, A.F. Report of injuries in batoids caught in small-scale fisheries: Implications for management plans. Oecol. Aust. 2018, 23, 78–89. [Google Scholar] [CrossRef]
- Guimarães, N.; da Costa, F.M.; de Oliveira, F.P.; de Alcantara, A.A.; de Matos, J.G.M.V. Ocorrência de Hypanus americanus (Hildebrand and Schroeder, 1928) e Pseudobatos percellens (Walbaum, 1792) como descarte das pescas artesanais da Praia Grande da Ilha de Itacuruçá, Rio de Janeiro, Brasil. Acta Sci. Tech. 2021, 9, 81–85. [Google Scholar] [CrossRef]
- Mariguela, T.C.; De-Franco, B.; Almeida, T.V.V.; Mendonça, F.F.; Gadig, O.B.F.; Foresti, F.; Oliveira, C. Identification of guitarfish species Rhinobatos percellens, R. horkelii, and Zapteryx brevirostris (Chondrichthyes) using mitochondrial genes and RFLP technique. Conserv. Genet. Resour. 2009, 1, 393–396. [Google Scholar] [CrossRef]
- De-Franco, B.; Mendonça, F.F.; Hashimoto, D.T.; Porto-Foresti, F.; Oliveira, C.; Foresti, F. Forensic identification of the guitarfish species Rhinobatos horkelli, R. percellens and Zapteryx brevirostris using multiplex-PCR. Mol. Ecol. Resour. 2010, 10, 197–199. [Google Scholar] [CrossRef] [PubMed]
- Alexandre de-Franco, B.; Fernandes Mendonça, F.; Oliveira, C.; Foresti, F. Illegal trade of the guitarfish Rhinobatos horkelii on the coasts of central and southern Brazil: Genetic identification to aid conservation. Aquat. Conserv. 2012, 22, 272–276. [Google Scholar] [CrossRef]
- Ferrette, B.L.; Domingues, R.R.; Rotundo, M.M.; Miranda, M.P.; Bunholi, I.V.; De Biasi, J.B.; Oliveira, C.; Foresti, F.; Mendonça, F.F. DNA Barcode reveals the bycatch of endangered batoids species in the Southwest Atlantic: Implications for sustainable fisheries management and conservation efforts. Genes 2019, 10, 304. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, C.; de Lima Adachi, A.M.C.; da Cruz, V.P.; Foresti, F.; Loose, R.H.; Bornatowski, H. The label “Cação” is a shark or a ray and can be a threatened species! Elasmobranch trade in Southern Brazil unveiled by DNA barcoding. Mar. Pol. 2020, 116, 103920. [Google Scholar] [CrossRef]
- Alvarenga, M.; Solé-Cava, A.M.; Henning, F. What’s in a name? Phylogenetic species identification reveals extensive trade of endangered guitarfishes and sharks. Biol. Conserv. 2021, 257, 109119. [Google Scholar] [CrossRef]
- Hebert, P.D.; Ratnasingham, S.; De Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Drummond, A. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.; Conn, J.E.; Alonso, D.P.; Vinetz, J.M.; Emerson, K.J.; Ribolla, P.E.M. Microgeographical structure in the major Neotropical malaria vector Anopheles darlingi using microsatellites and SNP markers. Parasit. Vectors 2017, 10, 1–8. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 January 2021).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Driller, M.; Vilaca, S.T.; Arantes, L.S.; Carrasco-Valenzuela, T.; Heeger, F.; Chevallier, D.; Mazzoni, C.J. Optimization of ddRAD-like data leads to high quality sets of reduced representation single copy orthologs (R2SCOs) in a sea turtle multi-species analysis. BioRxiv 2020, 1–22. [Google Scholar] [CrossRef]
- Rochette, N.C.; Rivera-Colón, A.G.; Catchen, J.M. Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol. Ecol. 2019, 28, 4737–4754. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, s13742-015. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Rousset, F. GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Puechmaille, S.J. The program structure does not reliably recover the correct population structure when sampling is uneven: Subsampling and new estimators alleviate the problem. Mol. Ecol. Resour. 2016, 16, 608–627. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.X. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Menni, R.; Stehmann, M. Distribution, environment and biology of batoid fishes off Argentina, Uruguay and Brazil. A review. Rev. Mus. Argent. Cienc. Nat. Nueva Ser. 2000, 2, 69–109. [Google Scholar] [CrossRef]
- Miranda, L.D.; Vooren, C.M. Captura e esforço da pesca de elasmobrânquios demersais no sul do Brasil nos anos de 1975 a 1997. Frente Marít. 2003, 19, 217–231. [Google Scholar]
- Ehemann, N.R.; del V González-González, L. First record of a single-clasper specimen of Pseudobatos percellens (Elasmobranchii: Rhinopristiformes: Rhinobatidae) from the Caribbean Sea, Venezuela. Acta Ichthyol. Piscat. 2018, 48, 235–240. [Google Scholar] [CrossRef]
- Lessa, R.P.; Barreto, R.R.; Quaggio, A.L.C.; Valença, L.R.; Santana, F.; Yokota, L.; Gianetti, M.D. Levantamento das espécies de elasmobrânquios capturados por aparelhos-de-pesca que atuam no berçário de Caiçara do Norte (RN). Arquiv. Cienc. Mar. 2008, 41, 58–64. [Google Scholar]
- Rosa, R.S.; Gadig, O.B.F. Conhecimento da diversidade dos Chondrichthyes marinhos no Brasil: A contribuição de José Lima de Figueiredo. Arq. Zool. 2014, 45, 89–104. [Google Scholar] [CrossRef]
- Ferrari, A.; Di Crescenzo, S.; Cariani, A.; Crobe, V.; Benvenuto, A.; Piattoni, F.; Tinti, F. Puzzling over spurdogs: Molecular taxonomy assessment of the Squalus species in the Strait of Sicily. Eur. Zool. J. 2021, 88, 181–190. [Google Scholar] [CrossRef]
- Toffoli, D.; Hrbek, T.; Araújo, M.L.G.D.; Almeida, M.P.D.; Charvet-Almeida, P.; Farias, I.P. A test of the utility of DNA barcoding in the radiation of the freshwater stingray genus Potamotrygon (Potamotrygonidae, Myliobatiformes). Genet. Mol. Biol. 2008, 31, 324–336. [Google Scholar] [CrossRef]
- Hendry, A.P.; Bolnick, D.I.; Berner, D.; Peichel, C.L. Along the speciation continuum in sticklebacks. J. Fish Biol. 2009, 75, 2000–2036. [Google Scholar] [CrossRef]
- Prado, H.M.; Schlindwein, M.N.; Murrieta, R.S.S.; Nascimento, D.R.D.; Souza, E.P.D.; Cunha-Lignon, M.; Contente, R.F. The Valo Grande channel in the Cananéia-Iguape estuary-lagoon complex (SP, Brazil): Environmental history, ecology, and future perspectives. Ambient. Soc. 2019, 22, 1–22. [Google Scholar] [CrossRef]
- Ferrol-Schulte, D.; Wolff, M.; Ferse, S.; Glaser, M. Sustainable Livelihoods Approach in tropical coastal and marine social–ecological systems: A review. Mar. Pol. 2013, 42, 253–258. [Google Scholar] [CrossRef]
- Castro, R.M.C.; Menezes, N.A. Estudo Diagnóstico de Diversidade de Peixes do Estado de São Paulo. In Biodiversidade do Estado de São Paulo, Brasil: Síntese do Conhecimento ao Final do Século XX; Joly, C.A., Bicudo, C.E.M., Eds.; Vertebrados-6; FAPESP: São Paulo, Brazil, 1998; pp. 1–13. [Google Scholar]
- Menezes, N.A.; Buckup, P.A.; de Figueiredo, J.L.; de Moura, R.L. (Eds.) Catálogo das Espécies de Peixes Marinhos do Brasil; Museu de Zoologia da Universidade de São Paulo: São Paulo, Brazil, 2003; Volume 1. [Google Scholar]
- Schlaff, A.M.; Heupel, M.R.; Simpfendorfer, C.A. Influence of environmental factors on shark and ray movement, behaviour and habitat use: A review. Rev. Fish Biol. Fish. 2014, 24, 1089–1103. [Google Scholar] [CrossRef]
- Machado, L.F.; de Souza Damasceno, J.; Bertoncini, Á.A.; Farro, A.P.C.; Hostim-Silva, M.; Oliveira, C. Population genetic structure and demographic history of the spadefish, Chaetodipterus faber (Ephippidae) from Southwestern Atlantic. J. Exp. Mar. Biol. Ecol. 2017, 487, 45–52. [Google Scholar] [CrossRef]
- Little, A.G.; Loughland, I.; Seebacher, F. What do warming waters mean for fish physiology and fisheries? J. Fish Biol. 2020, 97, 328–340. [Google Scholar] [CrossRef]
- Lindmark, M.; Audzijonyte, A.; Blanchard, J.L.; Gårdmark, A. Temperature impacts on fish physiology and resource abundance lead to faster growth but smaller fish sizes and yields under warming. Glob. Change Biol. 2022, 28, 6239–6253. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Briceño, J.D.; Villafaña, J.A.; De Gracia, C.; Flores-Alcívar, F.F.; Kindlimann, R.; Abella, J. Diversity and paleoenvironmental implications of an elasmobranch assemblage from the Oligocene–Miocene boundary of Ecuador. PeerJ. 2020, 8, e9051. [Google Scholar] [CrossRef] [PubMed]
- Dennis, A.B.; Hellberg, M.E. Ecological partitioning among parapatric cryptic species. Mol. Ecol. 2010, 19, 3206–3225. [Google Scholar] [CrossRef] [PubMed]
- Barreto, G.C.; Di Domenico, M.; Medeiros, R.P. Human dimensions of marine protected areas and small-scale fisheries management: A review of the interpretations. Mar. Pol. 2020, 119, 104040. [Google Scholar] [CrossRef]
- Collette, B.B.; Rützler, K. Reef fishes over sponge bottoms off the mouth of the Amazon river. In Proceedings of the Third International Coral Reef Symposium, Miami, FL, USA, 1 May 1977; Volume 1, pp. 305–310. [Google Scholar]
- Luiz, O.J.; Madin, J.S.; Robertson, D.R.; Rocha, L.A.; Wirtz, P.; Floeter, S.R. Ecological traits influencing range expansion across large oceanic dispersal barriers: Insights from tropical Atlantic reef fishes. Proc. R. Soc. B Biol. Sci. 2012, 279, 1033–1040. [Google Scholar] [CrossRef]
- Bornatowski, H.; Abilhoa, V.; Charvet-Almeida, P. Elasmobranchs of the Paraná Coast, southern Brazil, south-western Atlantic. Mar. Biodiv. Rec. 2009, 158, 1–6. [Google Scholar] [CrossRef]
- Hackradt, C.W.; Félix-Hackradt, F.C.; Pichler, H.A.; Spach, H.L.; Santos, L.D.O. Factors influencing spatial patterns of the ichthyofauna of low energy estuarine beaches in southern Brazil. J. Mar. Biol. Assoc. UK 2011, 91, 1345–1357. [Google Scholar] [CrossRef]
- Carmo, W.P.; Bornatowski, H.; Oliveira, E.C.; Fávaro, L.L. Diet of the chola guitarfish, Rhinobatos percellens (Rhinobatidae), in the Paranaguá Estuarine complex. An. Acad. Bras. Ciên. 2015, 87, 721–731. [Google Scholar] [CrossRef]
- Possatto, F.E.; Broadhurst, M.K.; Spach, H.L.; Winemiller, K.O.; Millar, R.B.; Santos, K.M.; Lamour, M.R. Mapping the spatio-temporal distribution of threatened batoids to improve conservation in a subtropical estuary. J. Fish Biol. 2016, 89, 1098–1104. [Google Scholar] [CrossRef]
- Fratantoni, D.M.; Glickson, D.A. North Brazil Current ring generation and evolution observed with SeaWiFS. J. Phys. Oceanogr. 2002, 32, 1058–1074. [Google Scholar] [CrossRef]
- Roberts, A.M. Effect of Heinrich Events on Ocean Circulation and Sediment Input on the Nordeste Brazilian Continental Margin; University of Southampton: Southampton, UK, 2017. [Google Scholar]
- Pires-Vanin, A.M.S.; Rossi-Wongtschowski, C.L.D.B.; Aidar, E.; Mesquita, H.D.S.; Soares, L.S.H.; Katsuragawa, M.; Matsuura, Y. Estrutura e função do ecossistema de plataforma continental do Atlântico Sul brasileiro: Síntese dos resultados. Publicação Espec. Inst. Oceanográfico 1993, 10, 217–231. [Google Scholar]
- Braga, E.D.S.; Niencheski, L.F.H. Composição das Massas de Água e Seus Potenciais Produtivos na Área Entre o Cabo de São Tomé (RJ) e o Chuí (RS); O ambiente Oceanografia da plataforma continental e do talude na região Sudeste-Sul do Brasil: São Paulo, Brasil, 2006. [Google Scholar]
- Palma, E.D.; Matano, R.P.; Piola, A.R. A numerical study of the Southwestern Atlantic Shelf circulation: Stratified ocean response to local and offshore forcing. J. Geophys. Res. Ocean. 2008, 113, C11. [Google Scholar] [CrossRef]
- Rotundo, M.M.; Severino-Rodrigues, E.; Barrella, W.; Petrere-Junior, M.; Ramires, M. Checklist of marine demersal fishes captured by the pair trawl fisheries in Southern (RJ-SC) Brazil. Biota Neotrop. 2019, 19, e20170432. [Google Scholar] [CrossRef]
- Rutledge, K.M. A new guitarfish of the genus Pseudobatos (Batoidea: Rhinobatidae) with key to the guitarfishes of the Gulf of California. Copeia 2019, 107, 451–463. [Google Scholar] [CrossRef]
- Weigmann, S. Reply to Borsa 2017: Comment on ‘Annotated checklist of the living sharks, batoids and chimaeras (Chondrichthyes) of the world, with a focus on biogeographical diversity by Weigmann (2016)’. J. Fish Biol. 2017, 90, 1176–1181. [Google Scholar] [CrossRef] [PubMed]
- Boussarie, G.; Momigliano, P.; Robbins, W.D.; Bonnin, L.; Cornu, J.F.; Fauvelot, C.; Vigliola, L. Identifying barriers to gene flow and hierarchical conservation units from seascape genomics: A modelling framework applied to a marine predator. Ecography 2022, 2022, 06158. [Google Scholar] [CrossRef]
- Bunholi, I.V.; da Silva Ferrette, B.L.; Domingues, R.R.; Rotundo, M.M.; Cuevas, J.M.; García, M.; Mendonça, F.F. Multilocus phylogeography of the endemic and endangered angular angelshark (Squatina guggenheim) in the Southwest Atlantic Ocean. Hydrobiologia 2022, 849, 2177–2192. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
| Localities | ID | N | Ap | Np | Ho | He | FIS | (π) |
|---|---|---|---|---|---|---|---|---|
| Amapá | AP | 03 | 83 | 216 | 0.577 | 0.487 | −0.542 | 0.115 ± 0.067 |
| Pará | PA | 05 | 191 | 448 | 0.471 | 0.415 | −0.216 | 0.108 ± 0.057 |
| Pernambuco | PE | 14 | 389 | 331 | 0.315 | 0.298 | −0.088 | 0.102 ± 0.050 |
| São Paulo | SP | 15 | 140 | 115 | 0.281 | 0.239 | −0.213 | 0.031 ± 0.015 |
| Paraná | PR | 15 | 192 | 158 | 0.333 | 0.273 | −0.233 | 0.038 ± 0.019 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz, V.P.; Rotundo, M.M.; Charvet, P.; Boza, B.R.; Souza, B.C.; Cerqueira, N.N.C.D.; Oliveira, C.; Lessa, R.; Foresti, F. Investigating an Unknown Biodiversity: Evidence of Distinct Lineages of the Endemic Chola Guitarfish Pseudobatos percellens Walbaum, 1792 in the Western Atlantic Ocean. Diversity 2023, 15, 344. https://doi.org/10.3390/d15030344
Cruz VP, Rotundo MM, Charvet P, Boza BR, Souza BC, Cerqueira NNCD, Oliveira C, Lessa R, Foresti F. Investigating an Unknown Biodiversity: Evidence of Distinct Lineages of the Endemic Chola Guitarfish Pseudobatos percellens Walbaum, 1792 in the Western Atlantic Ocean. Diversity. 2023; 15(3):344. https://doi.org/10.3390/d15030344
Chicago/Turabian StyleCruz, Vanessa P., Matheus M. Rotundo, Patrícia Charvet, Beatriz R. Boza, Bruno C. Souza, Najila N. C. D. Cerqueira, Claudio Oliveira, Rosângela Lessa, and Fausto Foresti. 2023. "Investigating an Unknown Biodiversity: Evidence of Distinct Lineages of the Endemic Chola Guitarfish Pseudobatos percellens Walbaum, 1792 in the Western Atlantic Ocean" Diversity 15, no. 3: 344. https://doi.org/10.3390/d15030344
APA StyleCruz, V. P., Rotundo, M. M., Charvet, P., Boza, B. R., Souza, B. C., Cerqueira, N. N. C. D., Oliveira, C., Lessa, R., & Foresti, F. (2023). Investigating an Unknown Biodiversity: Evidence of Distinct Lineages of the Endemic Chola Guitarfish Pseudobatos percellens Walbaum, 1792 in the Western Atlantic Ocean. Diversity, 15(3), 344. https://doi.org/10.3390/d15030344









