DNA Barcoding of Moon Jellyfish (Cnidaria, Scyphozoa, Ulmaridae, Aurelia): Two Cryptic Species from the Azores (NE Atlantic, Macaronesia), and Evaluation of the Non-Indigenous Species (NIS)
Abstract
1. Introduction
2. Materials and Methods
3. Results
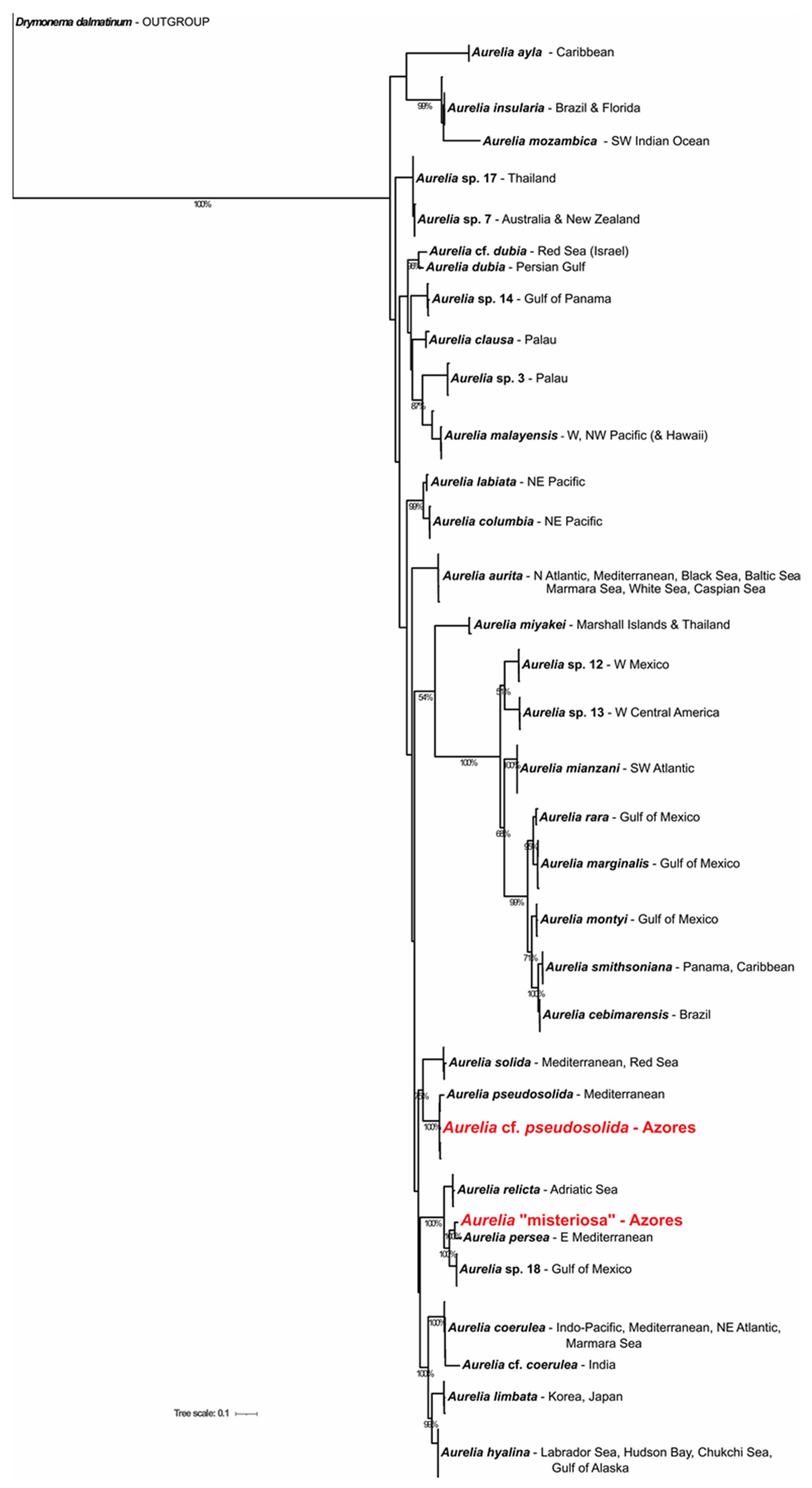
3.1. DNA Analyses
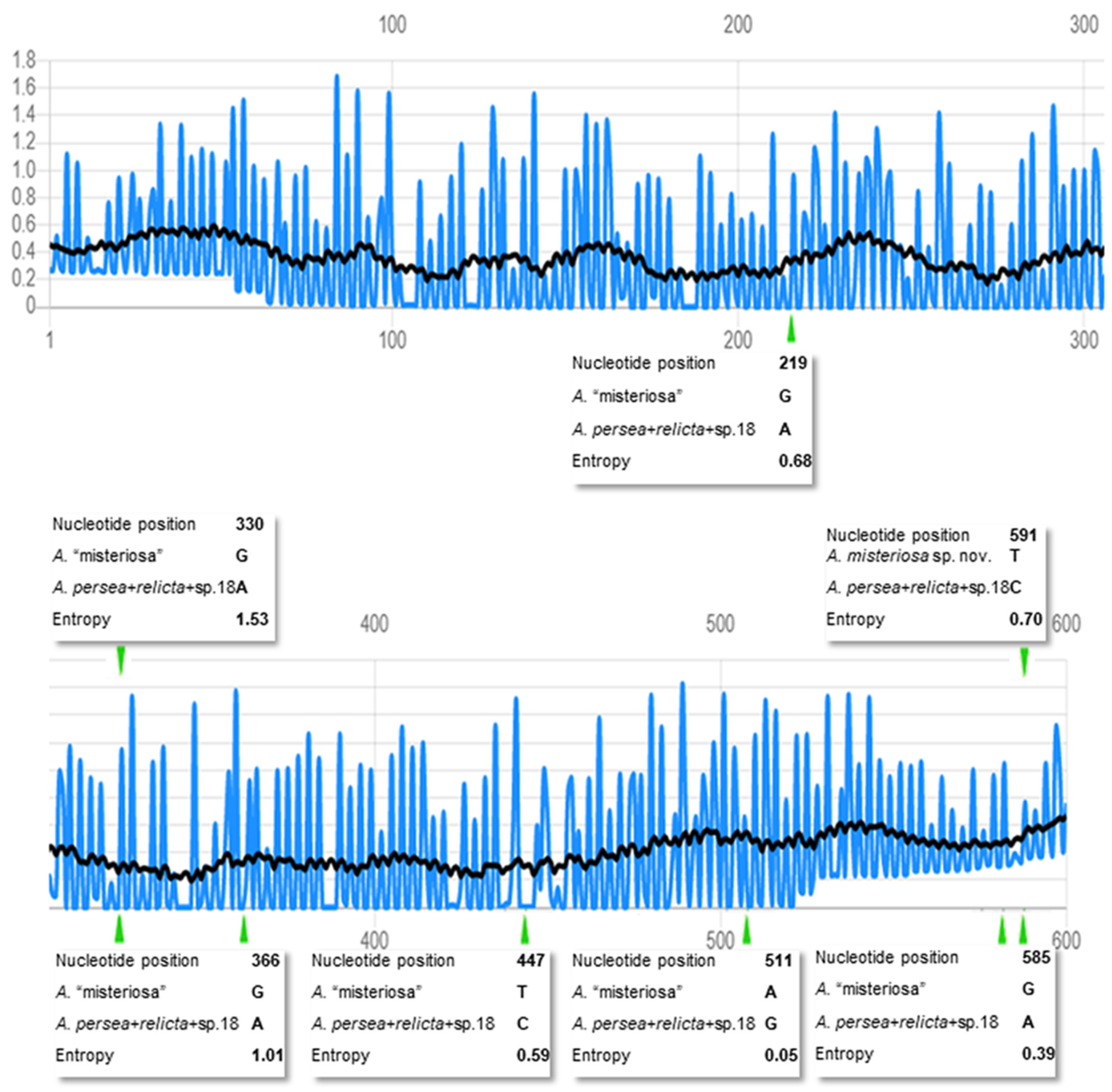
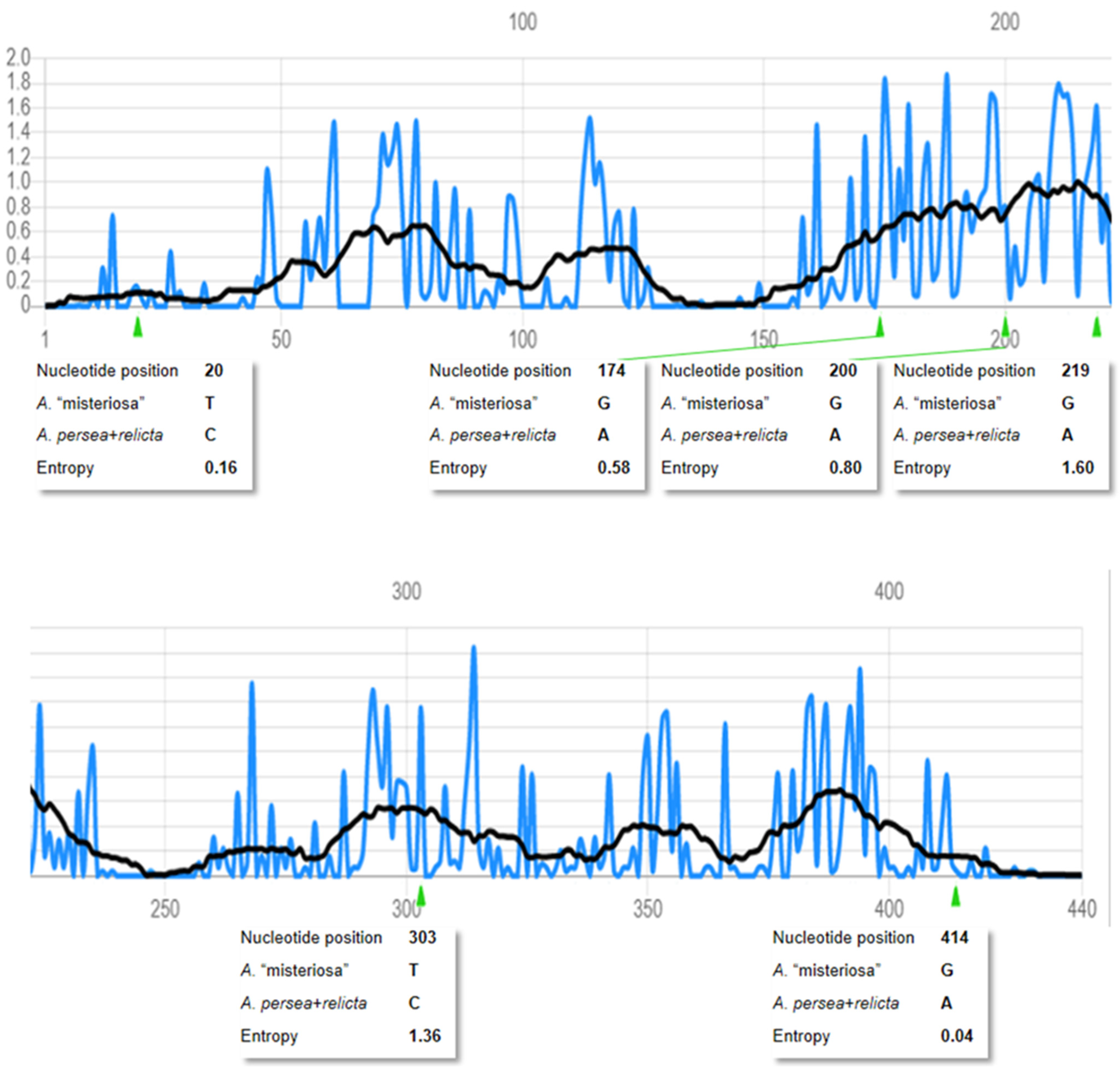
3.1.1. Phylogenetic Position of Aurelia “Misteriosa”
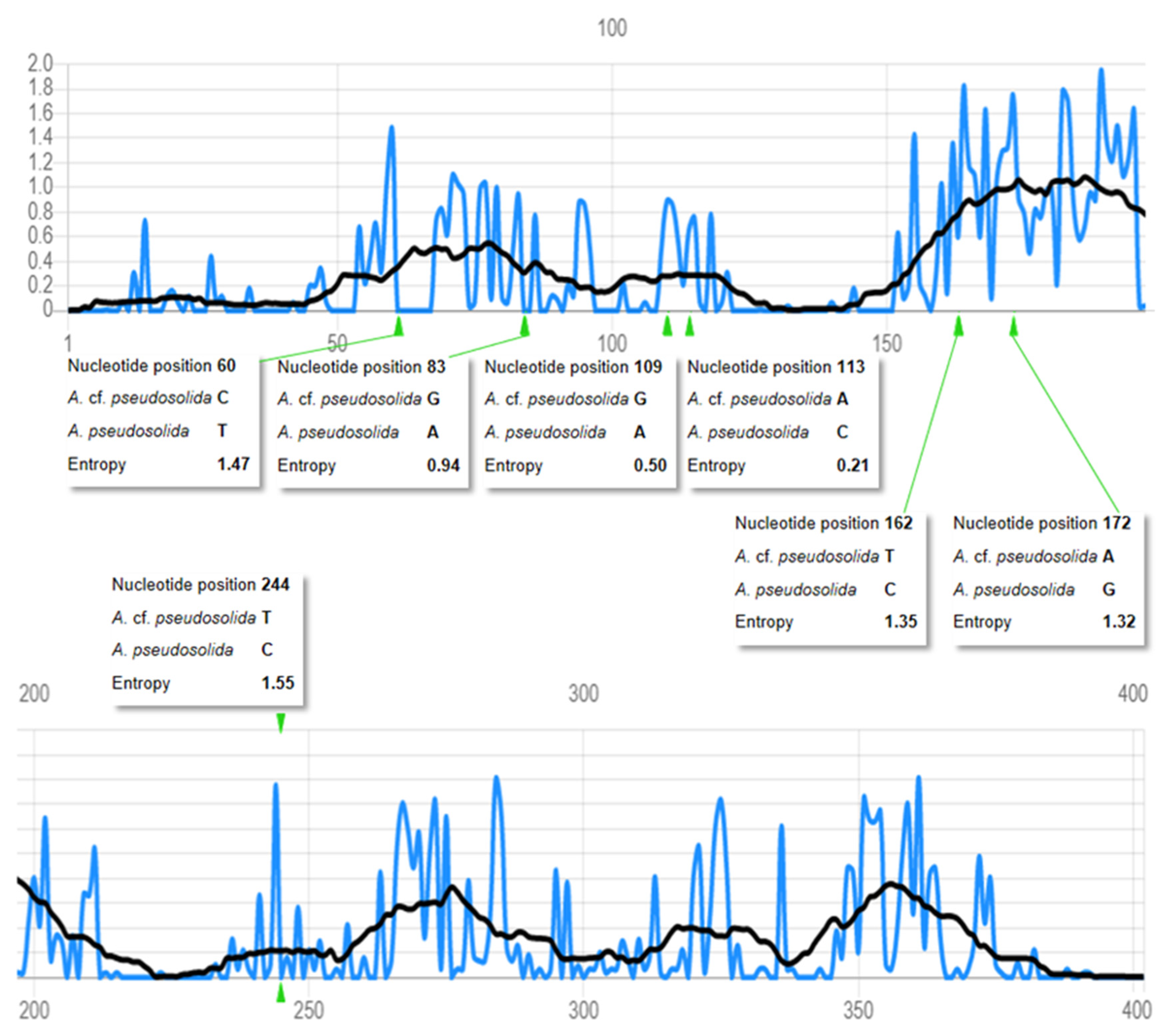
3.1.2. Phylogenetic Position of Aurelia “cf. pseudosolida”
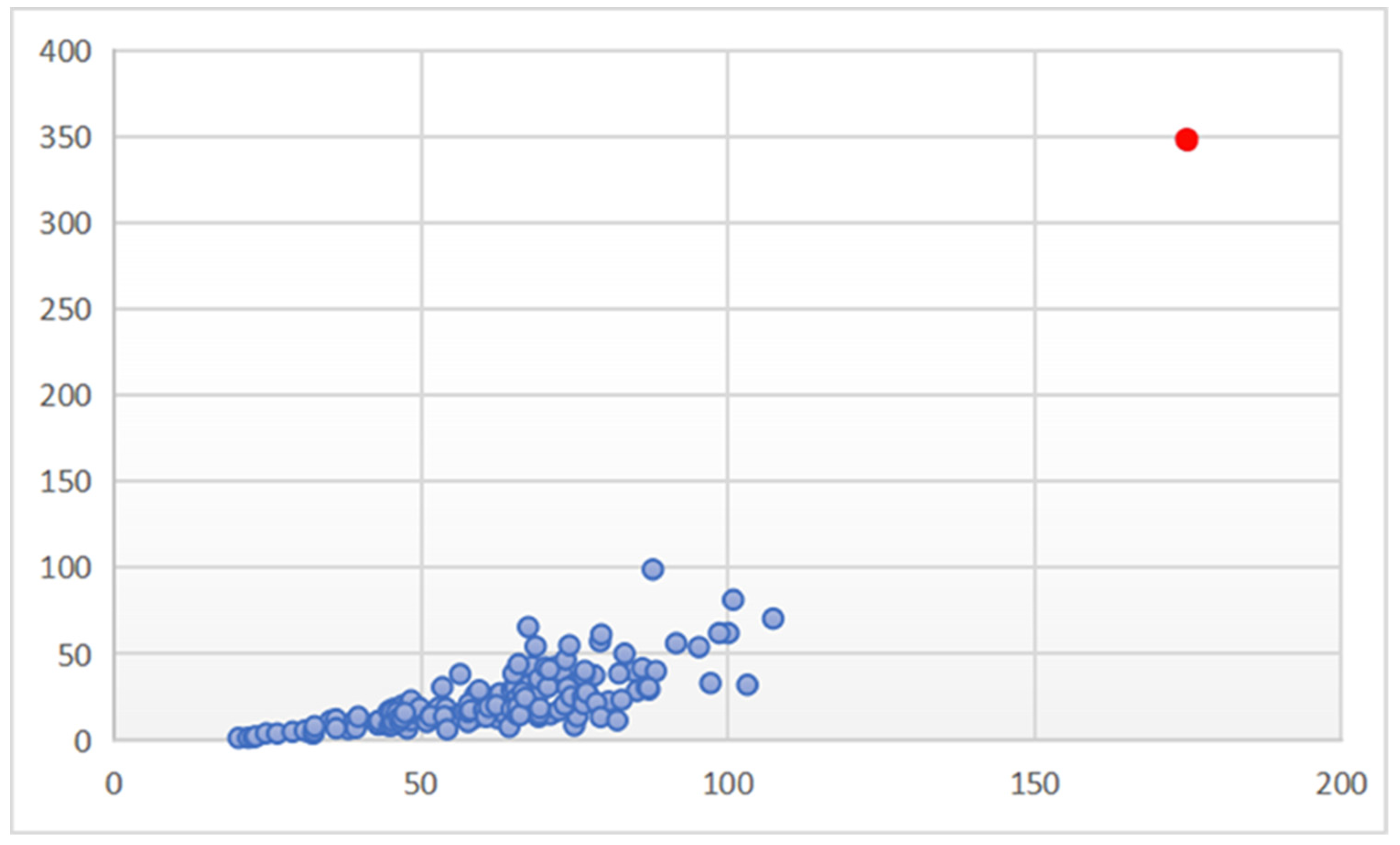
3.1.3. Review of the Aurelia Species with Widespread and Disjunct Biogeographic Distributions
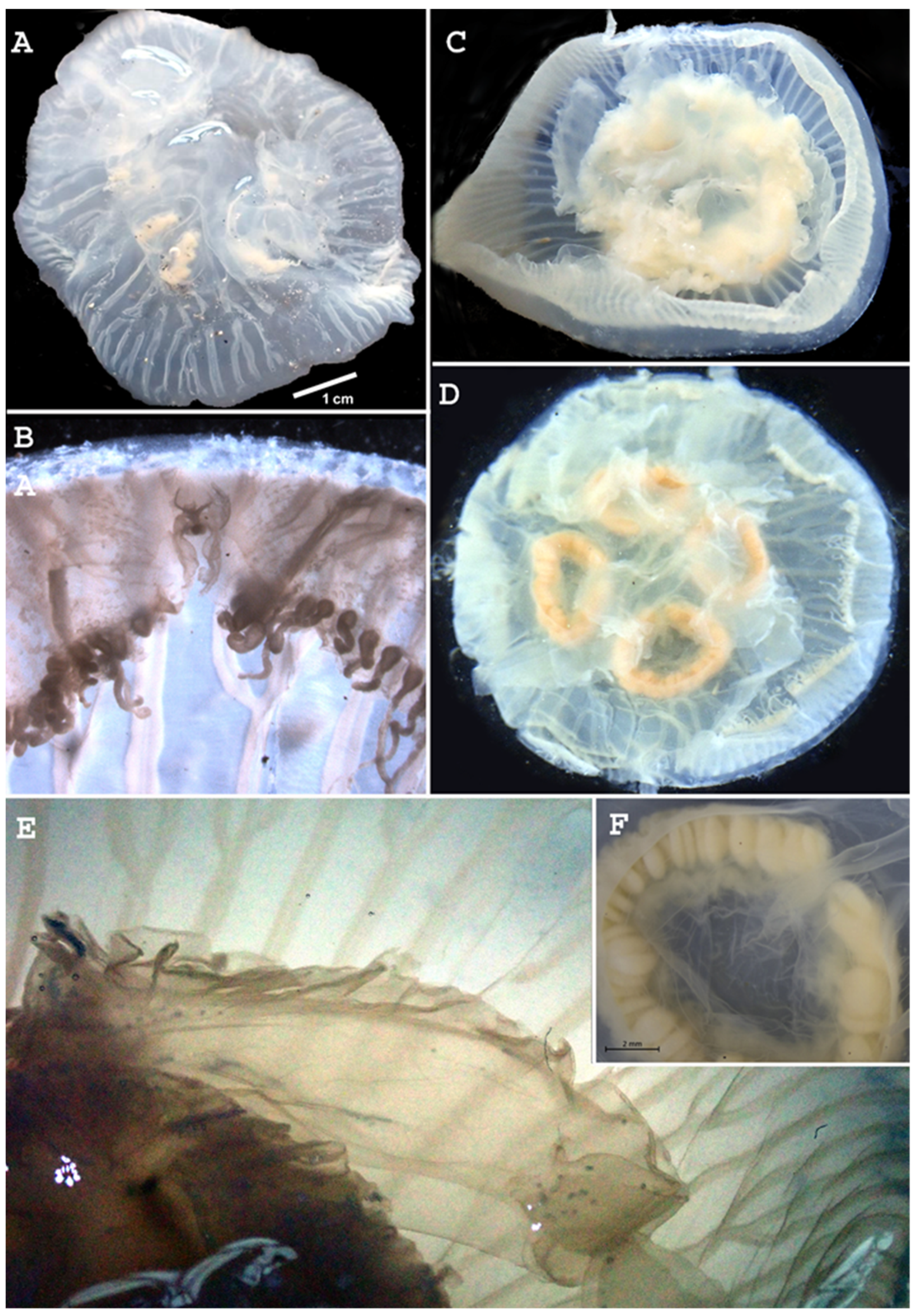
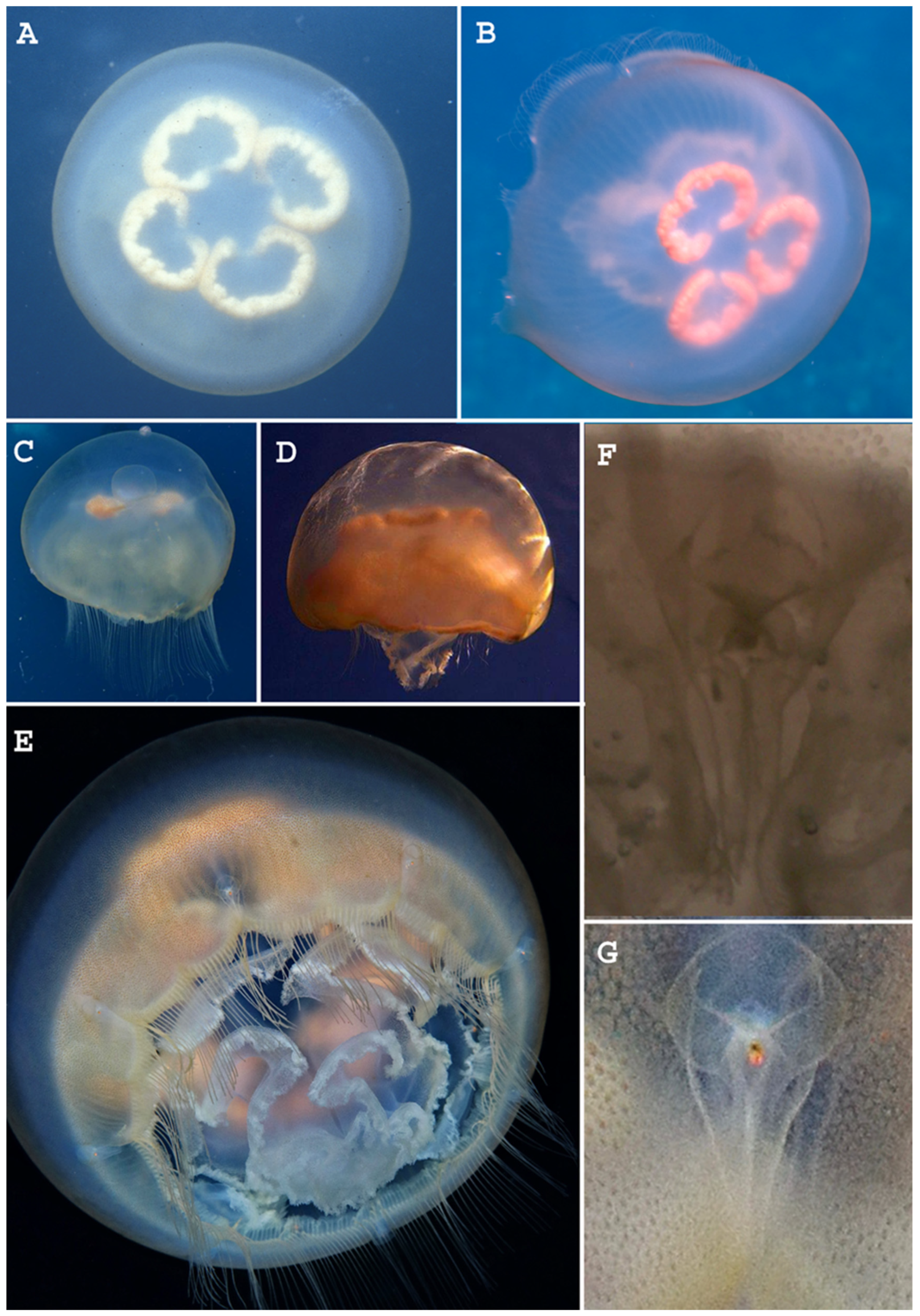
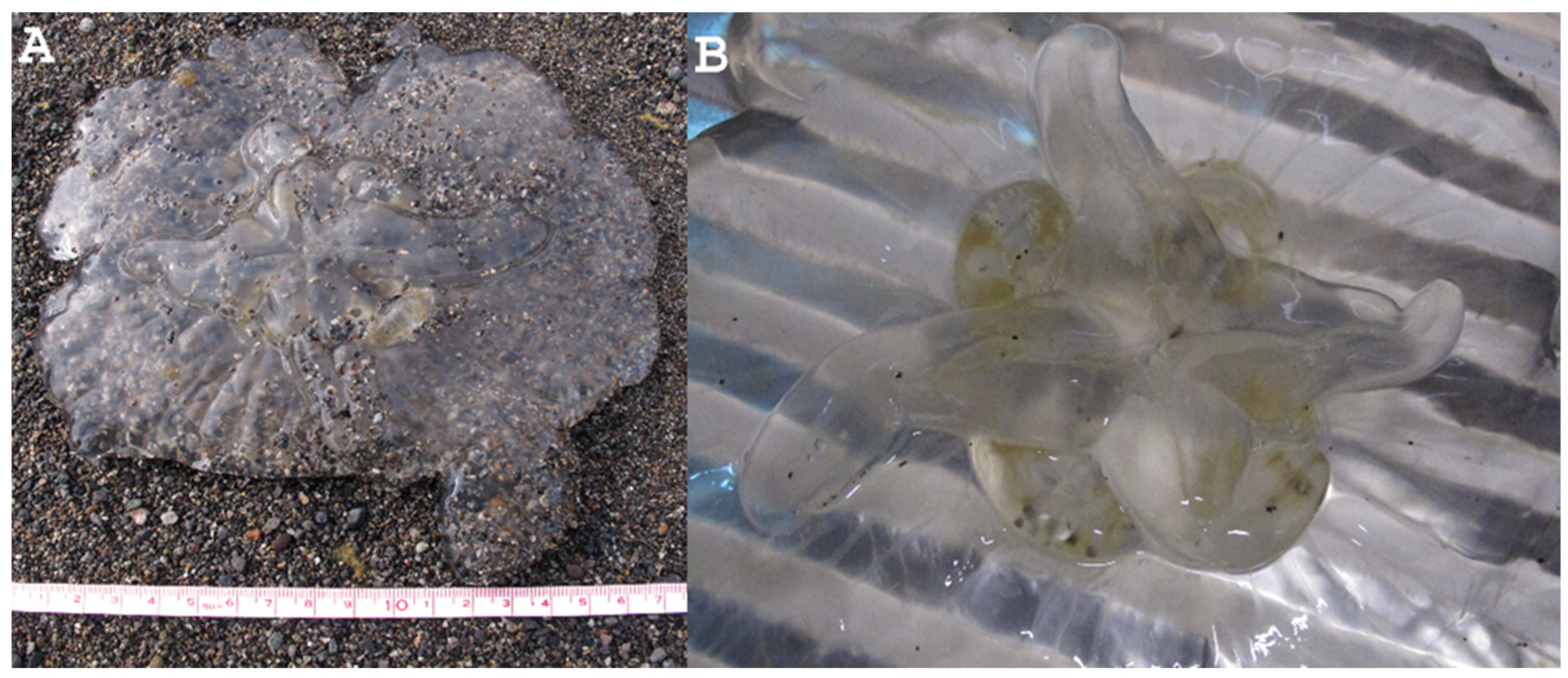
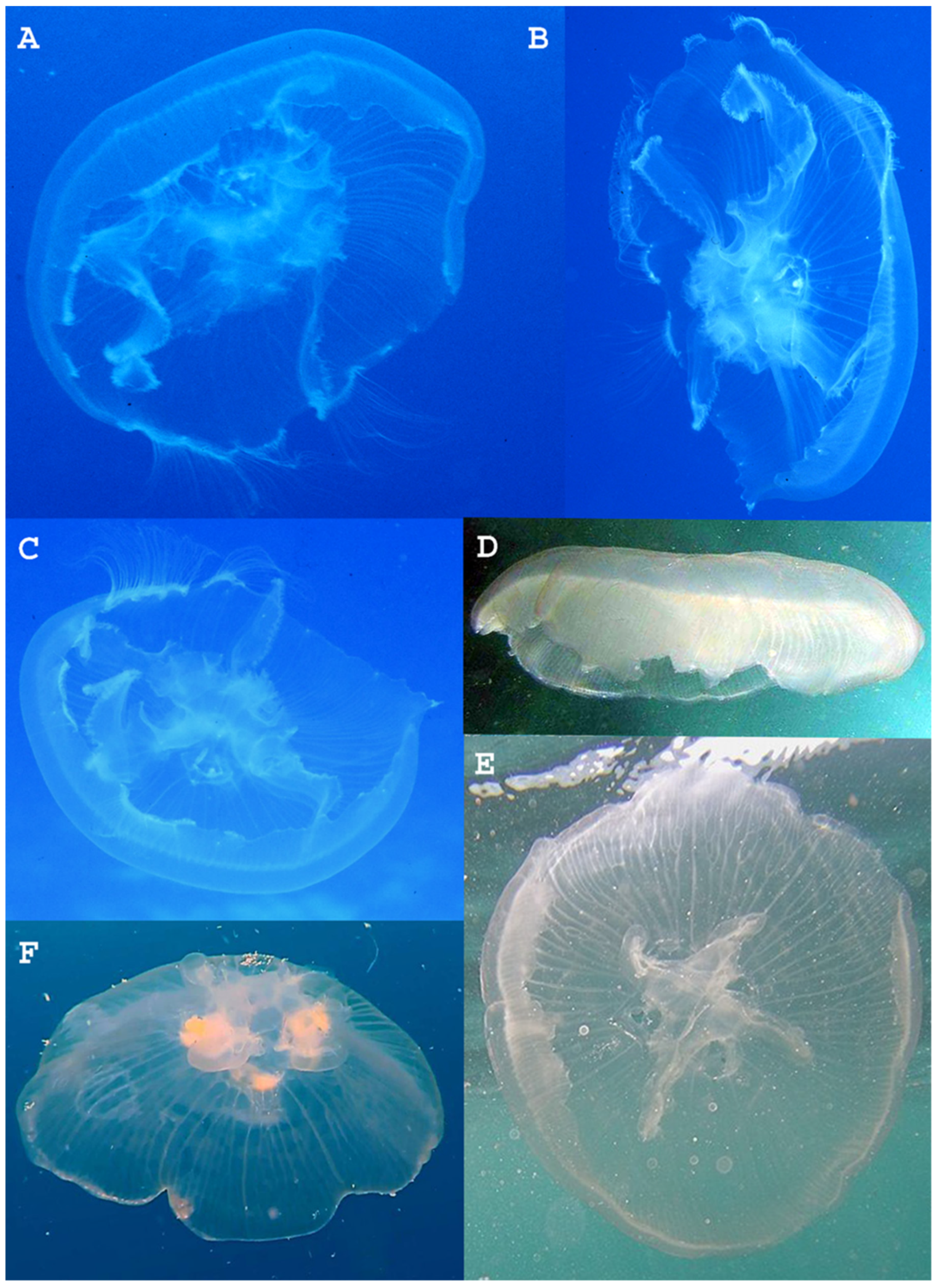
3.2. Systematic Account
3.3. Moon Jellies Reports in the Azores
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mayer, A.G. The Medusae of the World. Volume III. The Scyphomedusae; Carnegie Institution of Washington: Washington, DC, USA, 1910; Volume 109, pp. 499–735. [Google Scholar]
- Purcell, J.E.; Brown, E.D.; Stokesbury, K.; Haldorson, L.H.; Shirley, T.C. Aggregations of the jellyfish Aurelia labiata: Abundance, distribution, association with age-0 walleye pollock, and behaviors promoting aggregation in Prince William Sound, Alaska, USA. Mar. Ecol. Prog. Ser. 2000, 195, 145–158. [Google Scholar] [CrossRef]
- Dawson, M.N.; Martin, L.E. Geographic variation and ecological adaptation in Aurelia (Scyphozoa: Semaeostomeae): Some implications from molecular phylogenetics. Hydrobiologia 2001, 451, 259–273. [Google Scholar] [CrossRef]
- Lucas, C.H. Reproduction and life history strategies of the common jellyfish, Aurelia aurita, in relation to its ambient environment. Hydrobiologia 2001, 451, 229–246. [Google Scholar] [CrossRef]
- Arai, M.N. A Functional Biology of Scyphozoa; Chapman & Hall: London, UK, 1997; Volume 6. [Google Scholar] [CrossRef]
- Gershwin, L. Systematics and Biogeography of the Jellyfish Aurelia Labiata (Cnidaria: Scyphozoa). Biol. Bull. 2001, 201, 104–119. [Google Scholar] [CrossRef]
- Ishii, H.; Takagi, A. Development time of planulae larvae on the oral arms of the scyphomedusa Aurelia aurita. J. Plankton Res. 2003, 25, 1447–1450. [Google Scholar] [CrossRef]
- Schiariti, A.; Morandini, A.C.; Jarms, G.; Paes, R.V.G.; Franke, S.; Mianzan, H. Asexual reproduction strategies and blooming potential in Scyphozoa. Mar. Ecol. Prog. Ser. 2014, 510, 241–253. [Google Scholar] [CrossRef]
- Hansson, L. Effect of temperature on growth rate of Aurelia aurita (Cnidaria, Scyphozoa) from Gullmarsfjorden, Sweden. Mar. Ecol. Prog. Ser. 1997, 161, 145–153. Available online: https://www.jstor.org/stable/24859021 (accessed on 15 February 2023). [CrossRef]
- Gröndahl, F. A comparative ecological study on the scyphozoans Aurelia aurita, Cyanea capillata and C. lamarckii in the Gullmar Fjord, western Sweden, 1982 to 1986. Mar. Biol. 1988, 97, 541–550. [Google Scholar] [CrossRef]
- Graham, W.M. Numerical increases and distributional shifts of Chrysaora quinquecirrha (Desor) and Aurelia aurita (Linne) (Cnidaria: Scyphozoa) in the northern Gulf of Mexico. Hydrobiologia 2001, 451, 97–111. [Google Scholar] [CrossRef]
- Muller, W.; Leitz, T. Metamorphosis in the Cnidaria. Can. J. Zool. 2002, 80, 1755–1771. [Google Scholar] [CrossRef]
- Purcell, J.E. Environmental effects on asexual reproduction rates of the scyphozoan Aurelia labiata. Mar. Ecol. Prog. Ser. 2007, 348, 183–196. [Google Scholar] [CrossRef]
- Widmer, C.L.; Fox, C.J.; Brierley, A.S. Effects of temperature and salinity on four species of north-eastern Atlantic scyphistomae (Cnidaria: Scyphozoa). Mar. Ecol. Prog. Ser. 2016, 559, 73–88. [Google Scholar] [CrossRef]
- Yongze, X.; Qian, L.; Mei, Z.; Yu, Z.; Tiezhu, M.; Zhigang, Y. Effects of temperature and salinity on the asexual reproduction of Aurelia coerulea polyps. J. Oceanol. Limnol. 2020, 38, 133–142. [Google Scholar] [CrossRef]
- Schäfer, S.; Gueroun, S.K.M.; Andrade, C.; Canning-Clode, J. Combined Effects of Temperature and Salinity on Polyps and Ephyrae of Aurelia solida (Cnidaria: Scyphozoa). Diversity 2021, 13, 573. [Google Scholar] [CrossRef]
- Hamner, W.M.; Gilmer, R.W.; Hamner, P.P. The physical, chemical, and biological characteristics of a stratified, saline, sulfide lake in Palau. Limnol. Oceanogr. 1982, 27, 896–909. [Google Scholar] [CrossRef]
- Malej, A.; Kogovšek, T.; Ramšak, A.; Catenacci, L. Blooms and population dynamics of moon jellyfish in the Northern Adriatic. Cah. Biol. Mar. 2012, 53, 3. [Google Scholar]
- Hamner, W.M.; Hamner, P.P.; Strand, S.W. Sun-compass migration by Aurelia aurita (Scyphozoa): Population retention and reproduction in Saanich Inlet, British Columbia. Mar. Biol. 1994, 119, 347–356. [Google Scholar] [CrossRef]
- Greenberg, N.; Garthwaite, R.L.; Potts, D.C. Allozyme and morphological evidence for a newly introduced species of Aurelia in San Francisco Bay, California. Mar. Biol. 1996, 125, 401–410. [Google Scholar] [CrossRef]
- Dawson, M.N.; Jacobs, D.K. Molecular Evidence for Cryptic Species of Aurelia Aurita (Cnidaria, Scyphozoa). Biol. Bull. 2001, 200, 92–96. [Google Scholar] [CrossRef]
- Schroth, W.; Jarms, G.; Streit, B.; Schierwater, B. Speciation and Phylogeography in the Cosmopolitan Marine Moon Jelly, Aurelia Sp. BMC Evol. Biol. 2002, 2, 1. [Google Scholar] [CrossRef]
- Dawson, M.N.; Gupta, A.S.; England, M.H. Coupled Biophysical Global Ocean Model and Molecular Genetic Analyses Identify Multiple Introductions of Cryptogenic Species. Proc. Natl. Acad. Sci. USA 2005, 102, 11968. [Google Scholar] [CrossRef]
- Scorrano, S.; Aglieri, G.; Boero, F.; Dawson, M.N.; Piraino, S. Unmasking Aurelia Species in the Mediterranean Sea: An Integrative Morphometric and Molecular Approach. Zool. J. Linn. Soc. 2017, 180, 243–267. [Google Scholar] [CrossRef]
- Holland, B.S. Genetics of marine bioinvasions. Hydrobiologia 2000, 420, 63–71. [Google Scholar] [CrossRef]
- Grosholz, E.D. Ecological and evolutionary consequences of coastal invasions. Trends Ecol. Evol. 2002, 17, 22–27. [Google Scholar] [CrossRef]
- Stiasny, G. Die scyphomedusen. Dana Rep. 1940, 8, 1–27. [Google Scholar]
- WoRMS Editorial Board. World Register of Marine Species. Available online: https://www.marinespecies.org (accessed on 11 March 2022).
- Lawley, J.W.; Gamero-Mora, E.; Maronna, M.M.; Chiaverano, L.M.; Stampar, S.N.; Hopcroft, R.R.; Collins, A.G.; Morandini, A.C. The Importance of Molecular Characters When Morphological Variability Hinders Diagnosability: Systematics of the Moon Jellyfish Genus Aurelia (Cnidaria: Scyphozoa). PeerJ 2021, 9, e11954. [Google Scholar] [CrossRef]
- Linnaeus, C. Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, Cum Characteribus, Differentiis, Synonymis, Locis. Editio Decima, Reformata, 10th ed.; Laurentius Salvius: Holmiae, Turkey, 1758; Volume 1, pp. 1–824. [Google Scholar] [CrossRef]
- Kramp, P. Synopsis of the medusae of the world. J. Mar. Biol. Assoc. UK 1961, 40, 7–382. [Google Scholar] [CrossRef]
- Russell, F.S. The Medusae of the British Isles, Vol II: Pelagic scyphozoa, with supplement to Vol I on Hydromedusae. C. U. P. 1970, 56, 686. [Google Scholar] [CrossRef]
- Larson, R.J. Pelagic scyphomedusae (Scyphozoa: Coronatae and Semaeostomeae) of the Southern Ocean. Antarct. Res. Ser. 1986, 41, 59–165. [Google Scholar] [CrossRef]
- Larson, R.J. Scyphomedusae and Cubomedusae from the Eastern Pacific. Bull. Mar. Sci. 1990, 47, 546–556. [Google Scholar]
- Brown, M.; Scorrano, S.; Kuplik, Z.; Kuyper, D.; Ras, V.; Thibault, D.; Engelbrecht, A.; Gibbons, M.J. A New Macromedusa from the Coast of Mozambique: Aurelia Mozambica sp. nov. (Scyphozoa: Ulmaridae). Zootaxa 2021, 4933, 263–276. [Google Scholar] [CrossRef]
- Garic, R.; Batistic, M. Description of Aurelia pseudosolida sp. nov. (Scyphozoa, Ulmaridae) from the Adriatic Sea. Water 2022, 14, 135. [Google Scholar] [CrossRef]
- Chiaverano, L.M.; Bayha, K.M.; Graham, W.M. Local versus generalized phenotypes in two sympatric Aurelia species: Understanding jellyfish ecology using genetics and morphometrics. PLoS ONE 2016, 11, 1–24. [Google Scholar] [CrossRef]
- Chiaverano, L.M.; Graham, W.M. Morphological plasticity in Aurelia polyps, with subsequent effects on asexual fecundity and morphology of young medusae. Mar. Ecol. Prog. Ser. 2017, 582, 79–92. [Google Scholar] [CrossRef]
- Freitas, R.; Romeiras, M.; Silva, L.; Cordeiro, R.; Madeira, P.; González, J.A.; Wirtz, P.; Falcón, J.M.; Brito, A.; Floeter, S.R.; et al. Restructuring of the ‘Macaronesia’ biogeographic unit: A marine multi-taxon biogeographical approach. Sci. Rep. 2019, 9, 15792. [Google Scholar] [CrossRef]
- Maas, O. Méduses provenant des campagnes des yachts Hirondelle et princesse Alice. (1886–1903). Résultats des campagnes scientifiques accomplies sur son yacht par Albert Ier. Prince Souver. De Monaco 1904, 28, 1–71. [Google Scholar]
- Broch, H. Scyphomedusae from the “Michael Sars” North Atlantic Deep-Sea Expedition 1910. In Reports of the Scientific Results of the “Michael Sars” North Atlantic Deep-Sea Expedition; Bergen Museum: Bergen, Norway, 1910; Volume 1913, pp. 4–20. [Google Scholar] [CrossRef]
- Ranson, G. Scyphoméduses provenant des Campagne du Prince Albert 1er de Monaco. Richard, M.J., Monaco. 1945. Available online: https://bibnum.explore.psl.eu/s/psl/ark:/18469/1k380 (accessed on 15 February 2023).
- Frick, M.G.; Williams, K.L.; Bolten, A.B.; Bjorndal, K.A.; Martins, H.R. Foraging ecology of oceanic-stage loggerhead turtles Caretta caretta. Endanger. Species Res. 2009, 9, 91–97. [Google Scholar] [CrossRef]
- Raposo, N.; do Azul, A. Underwater Photography Exhibition; Municipality of Lagoa: São Miguel, Azores, 2018; 13p. [Google Scholar]
- Moura, C.J.; Ropa, N.; Magalhães, B.I.; Gonçalves, J.M. Insight into the cryptic diversity and phylogeography of the peculiar fried egg jellyfish Phacellophora (Cnidaria, Scyphozoa, Ulmaridae). PeerJ 2022, 10, e13125. [Google Scholar] [CrossRef]
- Gawinski, C.; Jaspers, C.; Reusch, T.H.B. Direct GenBank Submission. 2021. Available online: https://www.ncbi.nlm.nih.gov/nucleotide/ (accessed on 15 February 2023).
- Rekstad, M.E.; Majaneva, S.; Borgersen, Å.L.; Aberle, N. Occurrence and Habitat Characteristics of Aurelia sp. Polyps in a High-Latitude Fjord. Front. Mar. Sci. 2021, 8, 12. [Google Scholar] [CrossRef]
- Seo, Y.; Muhammad, B.L.; Chae, J.; Ki, J.-S. Genetic Structure and Diversity of the Moon Jellyfish Aurelia coerulea Polyp Population in Jaran Bay, Korea, Revealed by Mitochondrial COI and 16S rRNA Genes. Ocean Sci. J. 2021, 56, 99–105. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart model selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hütter, T.; Ganser, M.H.; Kocher, M.; Halkic, M.; Agatha, S.; Augsten, N. DeSignate: Detecting signature characters in gene sequence alignments for taxon diagnoses. BMC Bioinform. 2020, 21, 151. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Cunningham, C.W.; Buss, L.W. Molecular Evidence for Multiple Episodes of Paedomorphosis in the Family Hydractiniidae. Biochem. Syst. Ecol. 1993, 21, 57–69. Available online: https://linkinghub.elsevier.com/retrieve/pii/030519789390009G (accessed on 15 February 2023). [CrossRef]
- Mizrahi, G. Phylogenetic Analysis of Gelatinous Marine Fauna in the Eastern Mediterranean Basin—An Ecosystem under Anthropogenic Stress. Ph.D. Thesis, University of Haifa, Haifa, Israel, 2014. [Google Scholar]
- Frolova, A.; Miglietta, M.P. Insights on Bloom Forming Jellyfish (Class: Scyphozoa) in the Gulf of Mexico: Environmental Tolerance Ranges and Limits Suggest Differences in Habitat Preference and Resistance to Climate Change Among Congeners. Front. Mar. Sci. 2020, 7, 93. [Google Scholar] [CrossRef]
- Lucas, C.; Gelcich, S.; Uye, S.-i. Living with Jellyfish: Management and adaptation strategies. In Jellyfish Blooms; Pitt, K., Lucas, C., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 129–150. [Google Scholar] [CrossRef]
- Schneider, G.; Behrends, G. Top-down control in a neritic plankton system by Aurelia aurita medusae—A summary. Ophelia 1998, 48, 71–82. [Google Scholar] [CrossRef]
- Hansson, L.J.; Moeslund, O.; Kiørboe, T.; Riisgård, H.U. Clearance Rates of Jellyfish and Their Potential Predation Impact on Zooplankton and Fish Larvae in a Neritic Ecosystem (Limfjorden, Denmark). Mar. Ecol. Prog. Ser. 2005, 304, 117–131. Available online: https://www.jstor.org/stable/24869846 (accessed on 15 February 2023). [CrossRef]
- Natural History Museum. Natural History Museum (London) Collection Specimens. Occurrence Dataset. 2022. Available online: https://www.gbif.org/occurrence/1056282115 (accessed on 3 May 2022).
- Orrell, T.; Informatics Office. NMNH Extant Specimen Records (USNM, US). Version 1.54. National Museum of Natural History, Smithsonian Institution. Occurrence Dataset. 2022. Available online: https://www.gbif.org/occurrence/1321383749 (accessed on 3 May 2022).
- Orrell, T.; Informatics Office. NMNH Extant Specimen Records (USNM, US). Version 1.54. National Museum of Natural History, Smithsonian Institution. Occurrence Dataset. 2022. Available online: https://www.gbif.org/occurrence/1319477001 (accessed on 3 May 2022).
- Furtado ECV. O mistério dos ilhéus das Formigas: Uma viagem pelo “paraíso” do Atlântico, 1st ed.; Nova Gráfica: Ponta Delgada, Portugal, 2007; 374p, ISBN 989-20-0354-3. [Google Scholar]
- Haeckel, E. Das System der Acraspeden. 2te Hälfte des Systems der Medusen. Acht Nachträge zur Vervollständigung des Systems. Denkschr. Der Med.-Nat. Ges. Zu Jena 1880, 2, 361–672. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moura, C.J.; Magalhães, B.I.; Gonçalves, J.M. DNA Barcoding of Moon Jellyfish (Cnidaria, Scyphozoa, Ulmaridae, Aurelia): Two Cryptic Species from the Azores (NE Atlantic, Macaronesia), and Evaluation of the Non-Indigenous Species (NIS). Diversity 2023, 15, 323. https://doi.org/10.3390/d15030323
Moura CJ, Magalhães BI, Gonçalves JM. DNA Barcoding of Moon Jellyfish (Cnidaria, Scyphozoa, Ulmaridae, Aurelia): Two Cryptic Species from the Azores (NE Atlantic, Macaronesia), and Evaluation of the Non-Indigenous Species (NIS). Diversity. 2023; 15(3):323. https://doi.org/10.3390/d15030323
Chicago/Turabian StyleMoura, Carlos J., Bruno I. Magalhães, and João M. Gonçalves. 2023. "DNA Barcoding of Moon Jellyfish (Cnidaria, Scyphozoa, Ulmaridae, Aurelia): Two Cryptic Species from the Azores (NE Atlantic, Macaronesia), and Evaluation of the Non-Indigenous Species (NIS)" Diversity 15, no. 3: 323. https://doi.org/10.3390/d15030323
APA StyleMoura, C. J., Magalhães, B. I., & Gonçalves, J. M. (2023). DNA Barcoding of Moon Jellyfish (Cnidaria, Scyphozoa, Ulmaridae, Aurelia): Two Cryptic Species from the Azores (NE Atlantic, Macaronesia), and Evaluation of the Non-Indigenous Species (NIS). Diversity, 15(3), 323. https://doi.org/10.3390/d15030323







