Investigating the Potential Utility of Environmental DNA to Provide a Relative Abundance Index for the Depleted Teleost, Mulloway, Argyrosomus japonicus
Abstract
1. Introduction
2. Materials and Methods
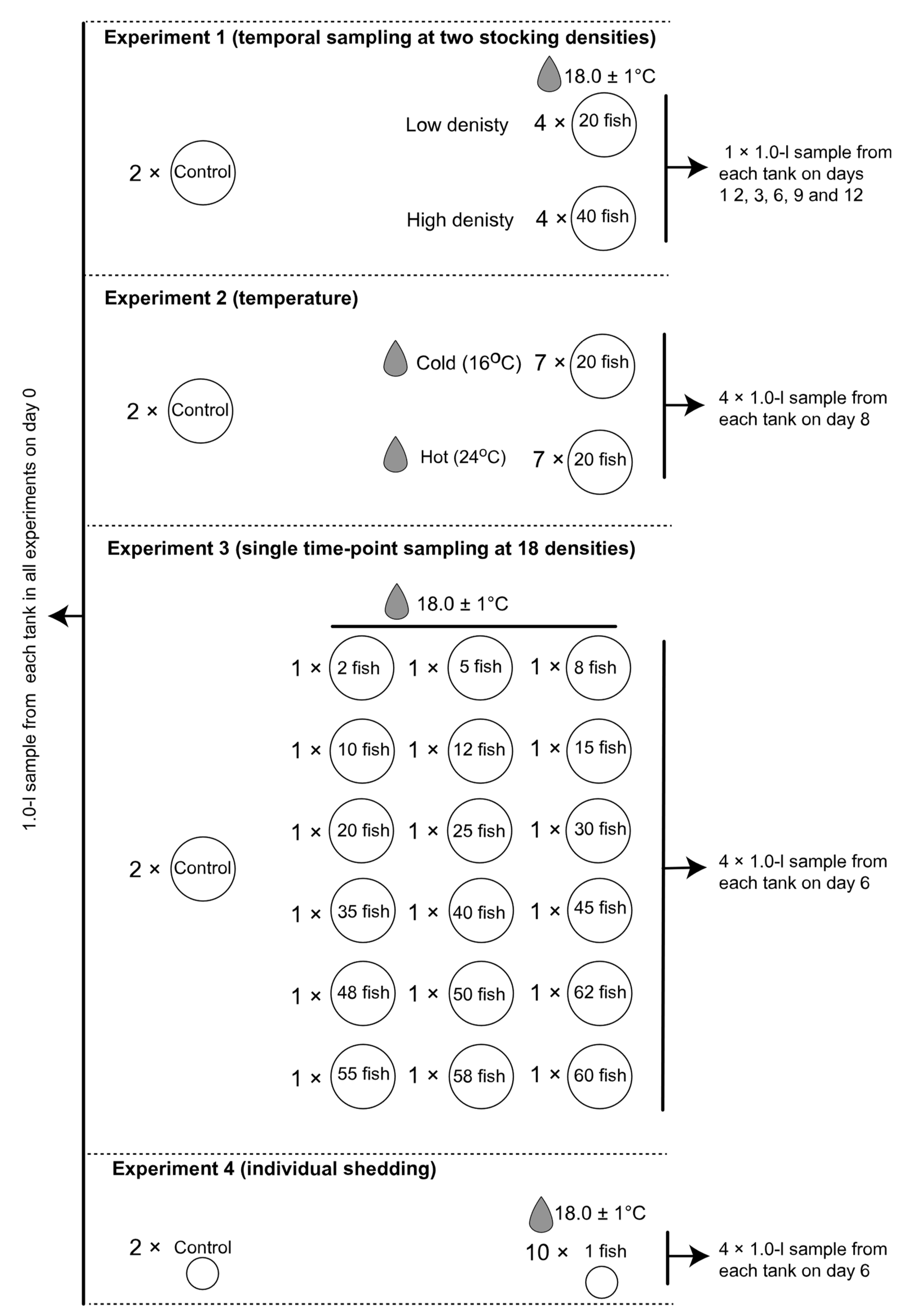
2.1. Aquaria Configuration and Fish Collection
2.2. Experimental Procedure
2.3. Filters, DNA Extraction, and Quantitative PCR
2.4. Statistical Analyses
3. Results
3.1. Experimental Fish
3.2. Quality Control
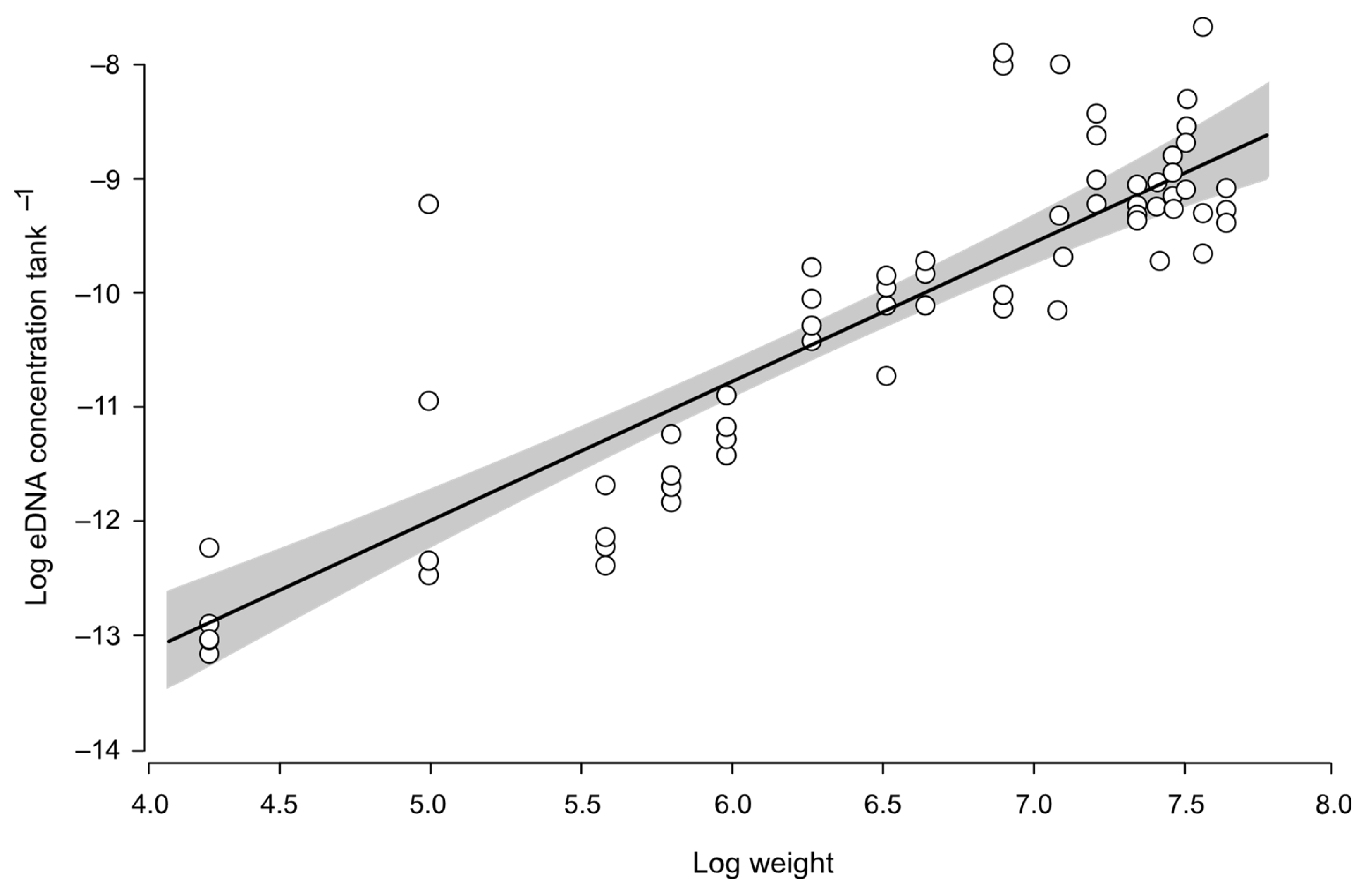
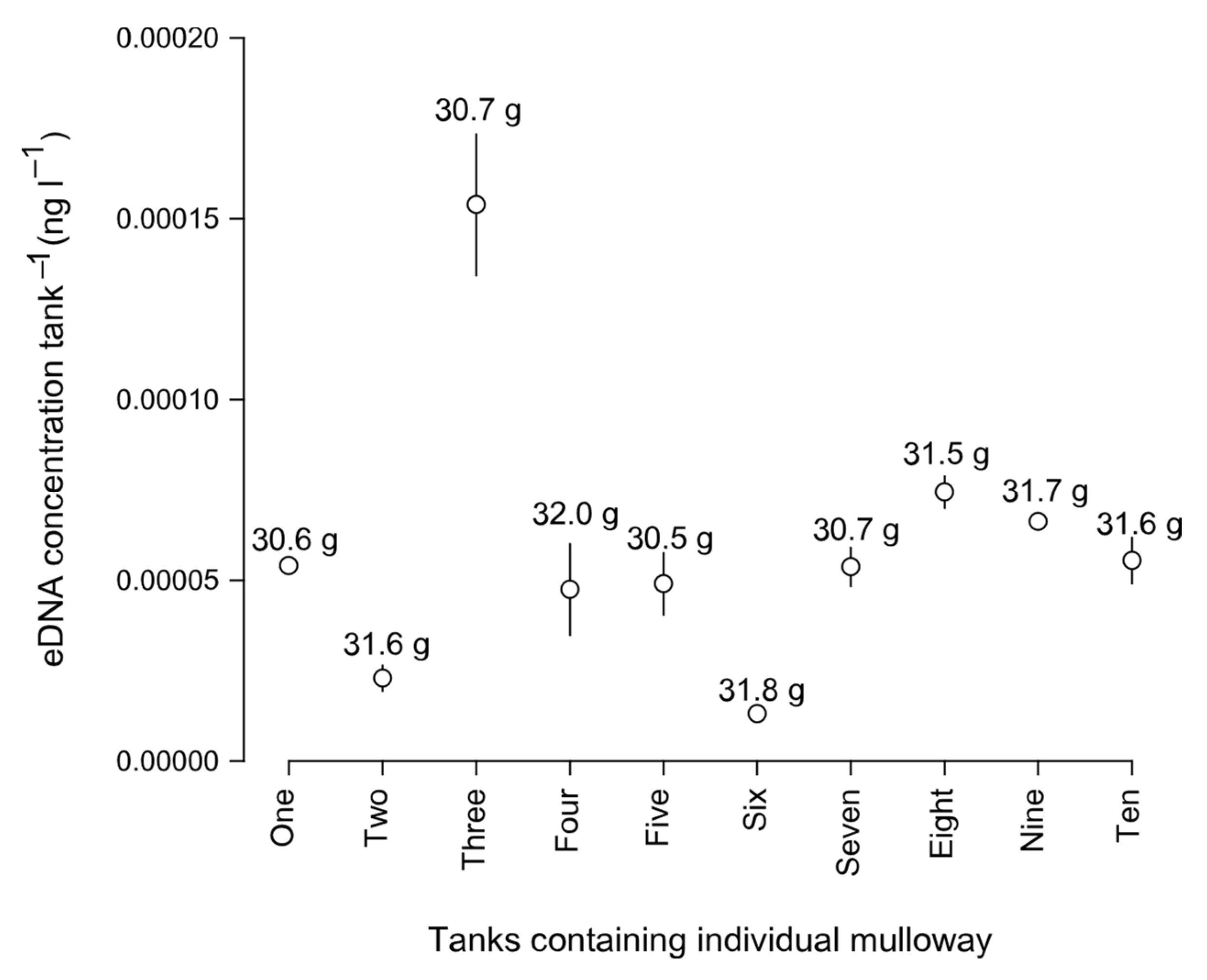
3.3. Environmental DNA Variability within and among Experiments
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petit-Marty, N.; Liu, M.; Tan, I.; Chung, A.; Terrasa, B.; Guijarro, B.; Ordines, F.; Ramırez-Amaro, S.; Massutı, E.; Schunter, C. Declining Population Sizes and Loss of Genetic Diversity in Commercial Fishes: A Simple Method for a First Diagnostic. Front. Mar. Sci. 2022, 9, 1–11. [Google Scholar] [CrossRef]
- Attard, C.R.M.; Möller, L.M.; Sasaki, M.; Hammer, M.P.; Bice, C.M.; Brauer, C.J.; Carvalho, D.C.; Harris, J.O.; Beheregaray, L.B. A novel holistic framework for genetic-based captive-breeding and reintroduction programs. Conserv. Biol. 2016, 30, 1060–1069. [Google Scholar] [CrossRef]
- Hilborn, R.; Quinn, T.P.; Schindler, D.E.; Rogers, D.E. Biocomplexity and fisheries sustainability. Proc. Natl. Acad. Sci. USA 2003, 100, 6564–6568. [Google Scholar] [CrossRef]
- King, M. Fisheries Biology, Assessment and Management. Fishing News Books; Blackwell Science: London, UK, 1995; p. 342. [Google Scholar]
- Tilzey, R.D.J.; Rowling, K.R. History of Australia’s South East Fishery: A scientist’s perspective. Mar. Freshw. Res. 2001, 52, 361–375. [Google Scholar] [CrossRef]
- Maunder, M.N.; Sibert, J.R.; Fonteneau, A.; Hampton, J.; Kleiber, P.; Harley, S.J. Interpreting catch per unit effort data to assess the status of individual stocks and communities. ICES J. Mar. Sci. 2006, 63, 1373–1385. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Rourke, M.L.; Fowler, A.M.; Hughes, J.M.; Broadhurst, M.K.; DiBattista, J.D.; Fielder, S.; Wilkes Walburn, J.; Furlan, E.M. Environmental DNA (eDNA) as a tool for assessing fish biomass: A review of approaches and future considerations for resource surveys. Environ. DNA 2022, 4, 9–33. [Google Scholar] [CrossRef]
- Yao, M.; Zhang, S.; Lu, Q.; Chen, X.; Zhang, S.-Y.; Kong, Y.; Zhao, J. Fishing for fish environmental DNA: Ecological applications, methodological considerations, surveying designs, and ways forward. Mol. Ecol. 2022, 31, 5132–5164. [Google Scholar] [CrossRef]
- Lamb, P.D.; Fonseca, V.G.; Maxwell, D.L.; Nnanatu, C.C. Systematic review and meta-analysis: Water type and temperature affect environmental DNA decay. Mol. Ecol. Resour. 2022, 22, 2494–2505. [Google Scholar] [CrossRef]
- Xing, Y.; Gao, W.; Shen, Z.; Zhang, Y.; Bai, J.; Cai, X.; Ouyang, J.; Zhao, Y. A Review of Environmental DNA Field and Laboratory Protocols Applied in Fish Ecology and Environmental Health. Front. Environ. Sci. 2022, 10, 1–17. [Google Scholar] [CrossRef]
- Taylor, M.D.; van der Meulen, D.E.; Ives, M.C.; Walsh, C.T.; Reinfelds, I.V.; Gray, C.A. Shock, Stress or Signal? Implications of Freshwater Flows for a Top-Level Estuarine Predator. PLoS ONE 2014, 9, e95680. [Google Scholar] [CrossRef]
- Hughes, J.M.; Meadows, N.M.; Stewart, J.; Booth, D.J.; Fowler, A.M. Movement patterns of an iconic recreational fish species, mulloway (Argyrosomus japonicus), revealed by cooperative citizen-science tagging programs in coastal eastern Australia. Fish. Res. 2022, 247, 106179. [Google Scholar] [CrossRef]
- Stewart, J.; Hughes, J.M.; Stanley, C.; Fowler, A.M. The influence of rainfall on recruitment success and commercial catch for the large sciaenid, Argyrosomus japonicus, in eastern Australia. Mar. Environ. Res. 2020, 157, 104924. [Google Scholar] [CrossRef]
- Silberschneider, V.; Gray, C.A.; Stewart, J. Age, growth, maturity and the overfishing of the iconic sciaenid, Argyrosomus japonicus, in south-eastern, Australia. Fish. Res. 2009, 95, 220–229. [Google Scholar] [CrossRef]
- Earl, J.; Fairclough, D.; Fisher, E.; Hughes, J.; Roelofs, A. Mulloway Argyrosomus Japonicus; Fisheries Research and Development Corporation: Canberra, NSW, Austria, 2021. [Google Scholar]
- Fielder, S.; Heasman, M.P. Hatchery Manual for the Production of Australian Bass, Mulloway and Yellowtail Kingfish; Industry and Investment NSW and Fisheries Research and Development Corporation: Orange, NSW, Australia, 2011. [Google Scholar]
- Furlan, E.M.; Gleeson, D.; Hardy, C.M.; Duncan, R.P. A framework for estimating the sensitivity of eDNA surveys. Mol. Ecol. Resour. 2016, 16, 641–654. [Google Scholar] [CrossRef]
- Wilkes Walburn, J.; Rourke, M.L.; Furlan, E.; DiBattista, J.D.; Broadhurst, M.K.; Fowler, A.M.; Hughes, J.M.; Fielder, S. Robust environmental DNA assay development and validation: A case study with two vulnerable Australian fish. Aquat. Conserv. Mar. Freshwat. Ecosyst. 2022, 32, 1225–1231. [Google Scholar] [CrossRef]
- Harrison, J.B.; Sunday, J.M.; Rogers, S.M. Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc. R. Soc. B 2019, 286, 20191409. [Google Scholar] [CrossRef]
- Klymus, K.E.; Richter, C.A.; Chapman, D.C.; Paukert, C. Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 2015, 183, 77–84. [Google Scholar] [CrossRef]
- Eichmiller, J.J.; Miller, L.M.; Sorensen, P.W. Optimizing techniques to capture and extract environmental DNA for detection and quantification of fish. Mol. Ecol. Resour. 2016, 16, 56–68. [Google Scholar] [CrossRef]
- Russell, A.; Taylor, M.D.; Barnes, T.C.; Johnson, D.D.; Gillanders, B.M. Habitat transitions by a large coastal sciaenid across life history stages, resolved using otolith chemistry. Mar. Environ. Res. 2022, 176, 105614. [Google Scholar] [CrossRef]
- Yates, M.C.; Glaser, D.M.; Post, J.R.; Cristescu, M.E.; Fraser, D.J.; Derry, A.M. The relationship between eDNA particle concentration and organism abundance in nature is strengthened by allometric scaling. Mol. Ecol. 2021, 30, 3068–3082. [Google Scholar] [CrossRef]
- Takeuchi, A.; Iijima, T.; Kakuzen, W.; Watanabe, S.; Yamada, Y.; Okamura, A.; Horie, N.; Mikawa, N.; Miller, M.J.; Kojima, T.; et al. Release of eDNA by different life history stages and during spawning activities of laboratory-reared Japanese eels for interpretation of oceanic survey data. Sci. Rep. 2019, 9, 6074. [Google Scholar] [CrossRef]
- Rourke, M.L.; Walburn, J.W.; Broadhurst, M.K.; Fowler, A.M.; Hughes, J.M.; Fielder, D.S.; DiBattista, J.D.; Furlan, E.M. Poor utility of environmental DNA for estimating the biomass of a threatened freshwater teleost; but clear direction for future candidate assessments. Fish. Res. 2023, 258, 106545. [Google Scholar] [CrossRef]
- Metcalfe, N.B.; Van Leeuwen, T.E.; Killen, S.S. Does individual variation in metabolic phenotype predict fish behaviour and performance? J. Fish Biol. 2016, 88, 298–321. [Google Scholar] [CrossRef]
- Ghosal, R.; Eichmiller, J.J.; Witthuhn, B.A.; Sorensen, P.W. Attracting Common Carp to a bait site with food reveals strong positive relationships between fish density, feeding activity, environmental DNA, and sex pheromone release that could be used in invasive fish management. Ecol. Evol. 2018, 8, 6714–6727. [Google Scholar] [CrossRef]
- Raoult, V.; Brown, C.; Zuberi, A.; Williamson, J.E. Blood cortisol concentrations predict boldness in juvenile mulloway (Argyosomus japonicus). J. Ethol. 2012, 30, 225–232. [Google Scholar] [CrossRef]
- Pirozzi, I.; Booth, M.A.; Pankhurst, P.M. The effect of stocking density and repeated handling on the growth of juvenile mulloway, Argyrosomus japonicus (Temminck & Schlegel 1843). Aquac. Int. 2008, 17, 199. [Google Scholar] [CrossRef]
- Person-Le Ruyet, J.; Mahé, K.; Le Bayon, N.; Le Delliou, H. Effects of temperature on growth and metabolism in a Mediterranean population of European sea bass, Dicentrarchus labrax. Aquaculture 2004, 237, 269–280. [Google Scholar] [CrossRef]
- Takahara, T.; Yamanaka, H.; Suzuki, A.A.; Honjo, M.N.; Minamoto, T.; Yonekura, R.; Itayama, T.; Kohmatsu, Y.; Ito, T.; Kawabata, Z.I. Stress response to daily temperature fluctuations in common carp, Cyprinus carpio L. Hydrobiologia 2011, 675, 65. [Google Scholar] [CrossRef]
- Jo, T.; Murakami, H.; Yamamoto, S.; Masuda, R.; Minamoto, T. Effect of water temperature and fish biomass on environmental DNA shedding, degradation, and size distribution. Ecol. Evol. 2019, 9, 1135–1146. [Google Scholar] [CrossRef]
- Thalinger, B.; Rieder, A.; Teuffenbach, A.; Pütz, Y.; Schwerte, T.; Wanzenböck, J.; Traugott, M. The Effect of Activity, Energy Use, and Species Identity on Environmental DNA Shedding of Freshwater Fish. Front. Ecol. Evol. 2021, 9, 1–13. [Google Scholar] [CrossRef]
- Takahara, T.; Minamoto, T.; Yamanaka, H.; Doi, H.; Kawabata, Z.i. Estimation of Fish Biomass Using Environmental DNA. PLoS ONE 2012, 7, e35868. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2016, 17, 1–17. [Google Scholar] [CrossRef]
- Eichmiller, J.J.; Best, S.E.; Sorensen, P.W. Effects of Temperature and Trophic State on Degradation of Environmental DNA in Lake Water. Environ. Sci. Technol. 2016, 50, 1859–1867. [Google Scholar] [CrossRef]
- Tsuji, S.; Ushio, M.; Sakurai, S.; Minamoto, T.; Yamanaka, H. Water temperature-dependent degradation of environmental DNA and its relation to bacterial abundance. PLoS ONE 2017, 12, e0176608. [Google Scholar] [CrossRef]
- Lacoursière-Roussel, A.; Rosabal, M.; Bernatchez, L. Estimating fish abundance and biomass from eDNA concentrations: Variability among capture methods and environmental conditions. Mol. Ecol. Resour. 2016, 16, 1401–1414. [Google Scholar] [CrossRef]
- Pirozzi, I.; Booth, M.A. The effect of temperature and body weight on the routine metabolic rate and postprandial metabolic response in mulloway, Argyrosomus japonicus. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 2009, 154, 110–118. [Google Scholar] [CrossRef]
- Shogren, A.J.; Tank, J.L.; Andruszkiewicz, E.; Olds, B.; Mahon, A.R.; Jerde, C.L.; Bolster, D. Controls on eDNA movement in streams: Transport, Retention, and Resuspension. Sci. Rep. 2017, 7, 5065. [Google Scholar] [CrossRef]
- Hallam, J.; Clare, E.L.; Jones, J.I.; Day, J.J. Biodiversity assessment across a dynamic riverine system: A comparison of eDNA metabarcoding versus traditional fish surveying methods. Environ. DNA 2021, 3, 1247–1266. [Google Scholar] [CrossRef]
- Sanches, T.M.; Schreier, A.D. Optimizing an eDNA protocol for estuarine environments: Balancing sensitivity, cost and time. PLoS ONE 2020, 15, e0233522. [Google Scholar] [CrossRef]
- Pochardt, M.; Allen, J.M.; Hart, T.; Miller, S.D.L.; Yu, D.W.; Levi, T. Environmental DNA facilitates accurate, inexpensive, and multiyear population estimates of millions of anadromous fish. Mol. Ecol. Resour. 2020, 20, 457–467. [Google Scholar] [CrossRef]
- Lafferty, K.D.; Garcia-Vedrenne, A.E.; McLaughlin, J.P.; Childress, J.N.; Morse, M.F.; Jerde, C.L. At Palmyra Atoll, the fish-community environmental DNA signal changes across habitats but not with tides. J. Fish Biol. 2021, 98, 415–425. [Google Scholar] [CrossRef]
- Kelly, R.P.; Gallego, R.; Jacobs-Palmer, E. The effect of tides on nearshore environmental DNA. PeerJ 2018, 6, e4521. [Google Scholar] [CrossRef]
- Saenz-Agudelo, P.; Delrieu-Trottin, E.; DiBattista, J.D.; Martínez-Rincon, D.; Morales-González, S.; Pontigo, F.; Ramírez, P.; Silva, A.; Soto, M.; Correa, C. Monitoring vertebrate biodiversity of a protected coastal wetland using eDNA metabarcoding. Environ. DNA 2022, 4, 77–92. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rourke, M.L.; Broadhurst, M.K.; Fowler, A.M.; Wilkes Walburn, J.; Hughes, J.M.; Fielder, D.S.; DiBattista, J.D.; Furlan, E.M. Investigating the Potential Utility of Environmental DNA to Provide a Relative Abundance Index for the Depleted Teleost, Mulloway, Argyrosomus japonicus. Diversity 2023, 15, 322. https://doi.org/10.3390/d15030322
Rourke ML, Broadhurst MK, Fowler AM, Wilkes Walburn J, Hughes JM, Fielder DS, DiBattista JD, Furlan EM. Investigating the Potential Utility of Environmental DNA to Provide a Relative Abundance Index for the Depleted Teleost, Mulloway, Argyrosomus japonicus. Diversity. 2023; 15(3):322. https://doi.org/10.3390/d15030322
Chicago/Turabian StyleRourke, Meaghan L., Matt K. Broadhurst, Ashley M. Fowler, Jackson Wilkes Walburn, Julian M. Hughes, Donald Stewart Fielder, Joseph D. DiBattista, and Elise M. Furlan. 2023. "Investigating the Potential Utility of Environmental DNA to Provide a Relative Abundance Index for the Depleted Teleost, Mulloway, Argyrosomus japonicus" Diversity 15, no. 3: 322. https://doi.org/10.3390/d15030322
APA StyleRourke, M. L., Broadhurst, M. K., Fowler, A. M., Wilkes Walburn, J., Hughes, J. M., Fielder, D. S., DiBattista, J. D., & Furlan, E. M. (2023). Investigating the Potential Utility of Environmental DNA to Provide a Relative Abundance Index for the Depleted Teleost, Mulloway, Argyrosomus japonicus. Diversity, 15(3), 322. https://doi.org/10.3390/d15030322






