Abstract
Species inventories are a prerequisite for biodiversity monitoring and conservation, particularly in protected areas. However, the possibilities of a standardized survey of species diversity using DNA barcoding have so far hardly been implemented, especially in species-rich groups. A first-time molecular-based and nearly complete inventory of the megadiverse insect order Lepidoptera in a protected area in the Alps (Cottian Alps, Italy) was intended to test the possibilities and reliability of DNA-based identifications. From voucher material collected between 2019 and 2022, we successfully sequenced 1213 morphospecies that grouped into 1204 BINs (barcode index numbers), whereas DNA barcoding failed for another 18 species. A total of 35 species shared a BIN with one or more taxa, but a majority of 19 species could still be discriminated by divergent sequences. A total of 12 morphospecies split into two BINs. These species and a further 22 taxa with unique BINs and barcode divergences >2% to the nearest neighbor require taxonomic re-assessment. Two additional cryptic species from the study area were described recently. Finally, 16 species are newly recorded for Italy. Our study, therefore, demonstrates the importance of DNA barcoding for both faunistics and the discovery of cryptic diversity, even in apparently well-studied protected areas.
1. Introduction
Protected areas are, despite several shortcomings, regarded as important harbors of biodiversity and particularly species diversity, and their conservation is considered a major task for responsible management [1,2]. Assessment of species diversity is, therefore, an important conservational issue in nature reserves [3,4,5]. However, species inventories are, particularly for large taxonomic groups, typically surveyed by few available experts, and identification is predominantly based on morphology [6,7]. Therefore, traditional species inventories are subject to a high risk of overlooking cryptic species. In particular, large-scale comparison of morphology for most species is hardly possible due to the lack of sufficient comparative material in reference collections and often too many species to be studied in detail. Furthermore, a morphology-based approach does not consider a very important part of biodiversity, namely genetic diversity. Provided an adequate reference database is available, DNA barcoding is an excellent alternative for species identification and scanning for potential cryptic species while at the same time enabling estimation of intraspecific genetic diversity [8,9,10]. However, comprehensive DNA barcode libraries are only available for some taxonomic groups and are usually restricted to national borders or rarely to larger geographical areas [11]. Lepidoptera is one of the few diverse groups with a largely complete barcode library in Central Europe, a prerequisite for our study [12,13,14].
Most of the surveys of Lepidoptera and other diverse orders of insects within and outside nature reserves in the Alps in general and in the southwestern Alps in particular are largely based on morphological identification methods [15,16,17]. Even recent inventories only exceptionally integrate molecular identification methods, and so far, none is available for a larger protected area [18,19]. Therefore, for the first time in a protected area in the Alps, specifically in the Cottian Alps Nature Parks (Italy), we analyzed a nearly complete species inventory of Lepidoptera from genetic sequences of the mitochondrial COI gene (cytochrome c oxidase 1) (DNA barcode region) to test the effectiveness of DNA barcode reference libraries already available. The resulting DNA barcode library is placed in a supra-regional context according to available reference sequences and furthermore integrated into a final species inventory. Previously unknown intraspecific genetic diversity is considered an addition to the comprehensive Barcode of Life (BOLD) database, and in several cases, suspected potential cryptic diversity may be involved [20].
Finally, the scope of this work contributes significantly to the national and regional species inventories of Lepidoptera in Italy, adding numerous species as new records to the nationwide inventory and an uncounted number on a regional or provincial level.
2. Materials and Methods
Study area. The Cottian Alps Nature Parks, situated in the Metropolitan City of Torino (former province of Torino; Italy), were formally established in 1980. They cover an area of ca 18,000 hectares and are split into four separate entities (Figure 1): (1) Avigliana Lakes Natural Park (409 ha; ca 350–360 m; 45.048° N–45.084° N, 7.385° E–7.397° E), (2) Orsiera Rocciavrè Natural Park (10,947 ha; ca 1070–2890 m; 45.006° N–45.11° N, 7.024° E–7.241° E), (3) Gran Bosco die Salbertrand Natural Park (3775 ha; ca 1000–2700 m; 45.023° N–45.092° N, 6.857° E–6.991° E), and (4) Val Troncea Natural Park (3280 ha; ca 1680–3280 m; 44.898° N–45.044° N, 6.896° E–7.051° E) [21]. Whereas Avigliana Natural Park covers different types of relict wetland habitats, the other three parks are dominated by montane to alpine habitats, including forests, mainly dominated by softwoods such as Pinus sylvestris, Larix decidua, and Picea abies, diverse meadows and pastures, alpine grassland, and rock and scree formations, mainly on siliceous soil. The alpine parks are characterized by a temperate and rather cold climate with an average annual temperature of only 1.4 °C and annual precipitation of nearly 1400 mm in Pragelato. The climate at the Avigliana Natural Park is also temperate, rather warm, and humid, with an average annual temperature of 9.9 °C and 1259 mm annual precipitation. Sampling design. Our study is based on samples from the Nature Parks, with a special emphasis on Orsiera Rocciavrè Natural Park and Gran Bosco die Salbertrand Natural Park, whereas samples from Avigliana Lakes and Val Troncea Natural Park are underrepresented. The samples were collected between 2019 and 2022 during 16 field trips, most of which lasted several days (28.6.–1.7.2019, 23.–25.7.2019, 25.–27.8.2019, 20.–21.9.2019, 31.10.2019; 18.–20.6.2020, 10.–13.7.2020, 19.–22.7.2020, 8.–10.10.2020, 9.–11.7.2021, 10.–12.9.2021; 7.5.2022, 1.–2.6.2022, 24.–26.6.2022, 16.–19.7.2022, 19.–22.8.2022). In order to provide a species inventory as comprehensively as possible, the focus of the fieldwork was in the summer months. The material was collected mainly at night, with three to five simultaneously run gaze pyramids/towers illuminated with superactinic or black light lamps (Figure 2). Occasionally automatic light traps were also used. In addition, diurnal species were collected with a butterfly net during the day and at twilight. No attempts were made to collect immature stages. We tried to obtain a minimum of one to usually a maximum of four samples per morphospecies to enable DNA barcoding. Voucher specimens were pinned, their wings spread, and immediately dried to ensure DNA quality or alternatively later set in a classic way. Eventual genitalia preparations followed standard techniques [22].
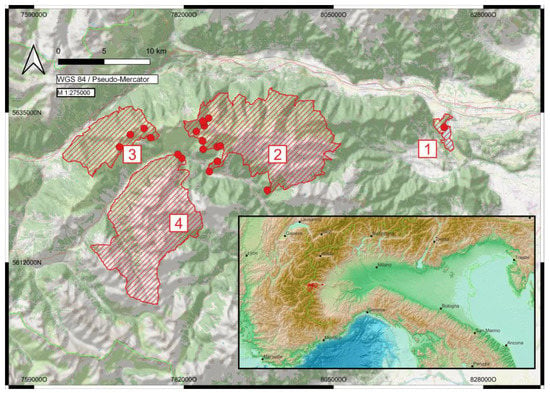
Figure 1.
Map of the study area (hatched) (1 = Avigliana Lakes Natural Park, 2 = Orsiera Rocciavrè Natural Park, 3 = Gran Bosco die Salbertrand Natural Par, 4 = Val Troncea Natural Park) with sample points in red. Sources: OpenTopoMap https://www.openstreetmap.org/; Natura2000 https://www.eea.europa.eu/; GEBCO world map https://www.gebco.net/) (all accessed on 1 February 2023).

Figure 2.
Light trapping with illuminated gaze pyramids.
DNA barcodes. DNA barcode sequences of Eumetazoa in general and Lepidoptera in particular are based on a 658 base-pair long segment of the mitochondrial COI gene (cytochrome C oxidase subunit I). DNA samples from dried legs of 1794 specimens, pre-identified to species level from external morphology, were prepared according to the prescribed standards [23]. The material was processed at the Canadian Centre for DNA Barcoding (CCDB, Biodiversity Institute of Ontario, University of Guelph) using standard high-throughput protocols to obtain DNA barcodes. Primer sets employed in the amplification of COI barcode region (658 bp) are mainly LepF1 (ATTCAACCAATCATAAAGATATTGG) and LepR1 (TAAACTTCTGGATGTCCAAAAAATCA), for shorter amplicons of 307 bp, 407 bp or 189 bp LepF1 (ATTCAACCAATCATAAAGATATTGG), MLepR2 (GTTCAWCCWGTWCCWGCYCCATTTTC), MLepF1 (GCTTTCCCACGAATAAATAATA), LepR1 (TAAACTTCTGATGTCCAAAAAATCA), MLepR2 (GTTCAWCCWGTWCCWGCYCC-ATTTTC) (https://boldsystems.org/index.php/Public_Primer_PrimerSearch) (accessed on 21 January 2023). Details of sequenced specimens, including complete voucher data, images, and sequences, can be accessed in the public dataset “Lepidoptera of Cottian Alps” (DS-LEPICOTT) dx.doi.org/10.5883/DS-LEPICOTT in the Barcode of Life Data Systems (BOLD) [20]. Sequences were finally submitted to GenBank. All sequences were assigned to barcode index numbers (BINs), algorithm-based operational taxonomic units that provide an accurate proxy for the true species [24]. BINs were automatically calculated for records in BOLD that are compliant with the DNA barcode standard. A total of 12 records not yet attached to a BIN in BOLD but with a full match to available BINs are attached here. In the case of BINs covering more than one taxon in BOLD (BIN sharing, misidentifications, contaminations), identification was based on external morphology, in critical cases, and also on genital morphology. Degrees of intra- and interspecific variation of DNA barcode fragments were calculated under the Kimura 2 parameter model of nucleotide substitution using analytical tools of BOLD systems v. 4.0. (http:// www.boldsystems.org) (accessed on 30 December 2022).
Faunistic assessment. Suspected potential new records for the Italian fauna were initially assessed from existing catalogs on the national fauna, in particular, the checklists of the fauna of Italy, Fauna Europaea, and Lepiforum [25,26,27]. Finally, regional experts were contacted (specifically: Dr. Giorgio Baldizzone, Dr. Graziano Bassi, and Dr. Manuela Pinzari). Despite all these efforts, it cannot be ruled out that records published in unreferenced journals have been overlooked.
Assessment of potential cryptic diversity. Unique BINs with a minimum 2% divergence to the nearest neighbor BIN are considered potential cryptic species. All these taxa require further integrative studies.
Specimen repositories. LMK = kärnten.museum, Klagenfurt, Austria; TLMF = Tiroler Landesmuseum Ferdinandeum, Innsbruck, Austria.
3. Results
3.1. General Overview—BIN System versus Linnean Taxa
DNA barcoding of 1794 specimens resulted in 1743 sequences, with only three short sequences <400 bp. The remaining 1740 DNA barcodes could be attributed to a BIN. The barcode library includes 1,204 BINs belonging to 1213 Linnean species (including cases of potential cryptic species) (Table S1). A total of 923 Linnean species are represented by a single sequence and a unique BIN from the study area. The remaining 281 species cover between two and 16 sequences. Only 12 species have more than one BIN (Infurcitinea atrifasciella, Infurcitinea roesslerella, Coleophora gryphipennella, Elachista atricomella, Agnoea langohri, Aproaerema anthyllidella, Aproaerema incognitana, Phalonidia manniana, Cydia succedana, Agriphila tristella, Eupithecia icterata, and Caradrina selini), but intraspecific DNA barcode variation is insufficiently known due to the underlying limitations of sampling concept.
A barcode gap analysis of all taxa showed a mean distance of 7.11% to the nearest neighbor species (min. dist. 0%, max. dist. 16.37%).
3.2. BIN-Sharing Morphospecies
A total of 35 morphospecies from the study area cannot be separated from one or more related sympatric species by their BIN (Table 1). However, 19 of these species still show diagnostic characters in DNA barcode sequences. The DNA barcodes of the remaining 16 species overlap or species share barcodes with one or two taxa and, therefore, cannot be separated by COI sequences (Table 2).

Table 1.
BIN-sharing species in the study area. BIN-sp = number of Linnean species in BIN.

Table 2.
Species with barcode sharing or barcode overlap in the study area.
A particularly high intra-BIN variation is documented for BIN BOLD: AAC0206, covering 48 taxa and an average distance of 3.95% and a maximum distance of 7.33%, whereas the distance to the nearest BIN is only 2.23%. Six members of this BIN have been found in the study areas, but all of them, with the exception of Agonopterix angelicella, can be identified from unique barcodes. A. angelicella shows a barcode overlap with A. paraselini, a species unknown from the study area, but Central European specimens cluster separately.
The three BIN-sharing species separated in Euxoa, namely E. tritici, E. vitta, and E. obelisca, primarily differ in weak phenotypic characters. Other BIN-sharing taxa in the families Noctuidae and Lycaenidae are easily separated by their appearance or genitalia morphology.
3.3. Cryptic Diversity
By Central European standards, a significant number of 22 morphospecies could not be assigned to any known BIN in BOLD. According to superficial morphological criteria, the majority of these species largely correspond to well-known taxa from the alpine region (Table 3). In addition, two local endemic species, viz. Caryocolum lamai and Megacraspedus cottiensis (Gelechiidae) were described from material collected in the framework of our study [28,29]. In contrast, Lymphia chalybella (Pyralidae), only a single named species without a reference sequence in BOLD, was successfully sequenced for the first time and added to the BOLD database.

Table 3.
Potential cryptic species, min. divergence >2% to nearest neighbor (NN). Taxon * = DNA barcode cluster endemic to Cottian Alps; Taxon ** = DNA barcode cluster also in other regions/countries; BM = total number of BIN members; BMC = total number of BIN members from study area; d NN = distance to nearest neighbor; BIN NN = BIN of nearest neighbor; NN = nearest neighbor.
3.4. New Faunistic Records for Italy
The Lepidoptera fauna of Italy is documented in voluminous checklists with several additions published during the last two decades [25,26,27]. A faunal list for the region of Piemont is available for “Macrolepidoptera” but not for the majority of other taxonomic groups [30]. Therefore, a regional faunistic analysis has not been carried out. For the “Macrolepidoptera”, we found a single species new to Piemont, viz. Standfussiana wiskotti. However, the unexpected number of 16 new records for Italy published earlier and herein (Table 4) also indicates a large number of regional or provincial novelties. Detailed label data of hitherto unpublished records are given below. Those already published species can be found in the dataset as well as in [31].

Table 4.
New faunistic records from the study area for Italy (species published earlier [31] are marked with *).
Euhyponomeutoides albithoracellus Gaj, 1954 (Yponomeutidae)
Prov. Torino, Salbertrand, 2 km SE Colle dell Assieta, 2240 m, 45.0606° N, 6.97889° E, 25.7.2019, leg. Huemer, DNA Barcode ID TLMF Lep 27687.
Mercantouria neli Huemer, Lopez-Vaamonde, and Triberti, 2016 (Gracillariidae)
Prov. Torino, Salbertrand, 1.5 km SE Colle dell Assieta, 2350 m, 45.061° N, 6.973° E, 18.7.2022, leg. Huemer, DNA Barcode ID TLMF_Lep_33416; Prov. Torino, Meana di Susa, PN Orsiera - Rocciavre, Colle delle Finestre N, 2090 m, 45.076° N, 7.052° E, 16.7.2022, leg. Huemer, TLMF_Lep_33437.
Coleophora tricolor Walsingham, 1899 (Coleophoridae)
Prov. Torino, Fenestrelle E, Depot, 1090 m, 45.028° N, 7.052° E, 11.7.2021, leg. Wieser, DNA Barcode ID KLM Lep 15810.
Depressaria gallicella Chrétien, 1908 (Depressariidae)
Prov. Torino, Pequerel NE, Via Colle delle Finestre, 1800 m, 45.053° N, 7.075° E, 29.6.2019, leg. Wieser, DNA Barcode ID KLM Lep 14956.
Athrips pruinosella (Lienig and Zeller, 1846) (Gelechiidae)
Prov. Torino, Colle delle Finestre N, 2180 m, 45.072° N, 7.053° E, 24.7.2019, leg. Huemer, DNA Barcode ID TLMF Lep 27656.
Tila capsophilella (Chrétien, 1900) (Gelechiidae)
Prov. Torino, Salbertrand, 1.5 km SE Colle dell Assieta, 2350 m, 45.061° N, 6.973° E, 22.8.2022, leg. Huemer, DNA Barcode IDs TLMF_Lep_33545, TLMF_Lep_33546.
Stomopteryx pyrenaella Varenne and Nel, 2020 (Gelechiidae)
Prov. Torino, Fenestrelle, Chambons-Depot NWN, 1080 m, 45.028° N, 7.06° E, 22.7.2020, leg. Huemer, DNA Barcode IDs TLMF Lep 29425, TLMF Lep 29426; Prov. Torino, Fenestrelle, Pracatinat, Forte delle Valli, 17000 m, 45.038° N, 7.071° E, 19.7.2022, leg. Huemer, DNA Barcode IDs TLMF_Lep_33391, TLMF_Lep_33392. First record for Italy and the Alps!
Remarks: Two further barcoded specimens from Cottian Alps (Prov. Cuneo, Val Varaita, Sampeyre W, N Villar, 1450 m, 44.5889° N, 7.14111° E, 18.7.2001, leg. Huemer, DNA Barcode IDs TLMF Lep 03796, TLMF Lep 03797) so far remain unidentified.
Sphinx maurorum (Jordan, 1931) (Sphingidae)
Prov. Torino, Pequerel NE, Via Colle delle Finestre, 1800 m, 45.053° N, 7.075° E, 11.7.2020, leg. Wieser, DNA Barcode ID KLM Lep 15265.
Remarks: Examination of the male genitalia of this unique specimen did not fully support the identification by the barcode, indicating potential introgression between S. maurorum and S. pinastri.
4. Discussion
DNA barcodes are a valuable and efficient tool for species identification, provided that a carefully validated and comprehensive reference library of DNA barcodes is available [10,32,33]. Lepidoptera has been one of the focal taxonomic groups in DNA barcoding, and several initiatives have helped build DNA barcode libraries for the diverse insect order from regional to continental scales [12,13,32,33,34,35,36,37,38]. However, a reasonably complete reference library is still lacking, and an estimated 20% of the European fauna of Lepidoptera have not yet been sequenced. These shortcomings cause major problems for reliable species identification, particularly in the Mediterranean area [39]. In contrast to southern regions, the coverage with DNA reference sequences in Central Europe and especially in the alpine region seems to have progressed much further [13,14]. As well as unresolved cases of intra- or interspecific genetic variation, only a single Linnean species from our study was an addition to the BOLD database. In almost all taxa, our comprehensive species inventory finally led to an undisputed assignment to a BIN, corresponding with a Linnean name. Such a close congruence of formal species to the BIN system was already documented for several larger families of Lepidoptera in Europe [37,38,40,41,42], and following our study seems to be relevant for the vast majority of Lepidoptera in the Alps. In our dataset, only 35 out of 1213 morphospecies are inseparable by their BIN from other species occurring in the study area. The group of BIN-sharing species includes unresolved taxonomic problems, i.e., in the disputed species group of Euxoa tritici. Taxa in this group are virtually indistinguishable in the genitalia morphology but also in DNA. In some taxa, BIN sharing is probably overestimated due to the misidentification of specimens boosting the number of taxa within one BIN. Such misidentifications are also likely for the BIN, including Lacanobia oleracea and L. splendens, with two alleged L. suasa in this BIN and also the genus Mesapamea. Finally, Conistra ligula shares its BIN and DNA barcode sequence with C. alicia, a species not present in Italy but clusters separately from the sympatric C. vaccinii. However, the majority of BIN-sharing species are divergent from the nearest neighbor, even by diagnostic DNA barcode sequences. The few observed cases of DNA barcode sharing require further assessment and may be due to both incomplete lineage sorting and hybridization, as proved for the genus Lysandra [43]. Other taxa, such as the above-mentioned Euxoa, require further studies involving additional nuclear markers [44].
However, despite such examples, the extremely high level of congruence between our DNA barcode library and previously published studies demonstrate the usefulness of the COI barcode sequence region for species identification. 22 morphospecies showed a p-distance of >2% to the nearest neighbor in BOLD, and all these taxa were finally grouped into new BINs. This particularly interesting segment of unique, unidentified BINs in our species inventory gives evidence of unexpected intraspecific genetic diversity but probably also covers cryptic species, which would have been overlooked with morphologically based identification methods. All of these taxa require integrative studies to check for eventual cryptic species but can be immediately used as unique intraspecific units for conservation, irrespective of the future assessment of taxonomic status. Exceptionally, however, these genetic lineages are obviously new species and, therefore, strictly speaking, not reflecting cryptic species. However, the p-distances alone are not sufficiently meaningful for the delimitation of species, even if there is a certain consensus for species demarcation of Lepidoptera. In this insect order, a threshold value of 2–3% divergence is often considered as a guideline to assess cryptic diversity [12]. However, unfortunately, no objective threshold is established to reliably distinguish between interspecific and intraspecific divergence. This restriction is, in extreme cases, reflected by well-supported species that share a DNA barcode or, alternatively, in cases of extraordinary intraspecific variation up to 14% in the genus Megacraspedus [45]. Summing up, voucher specimens from unidentified BINs need careful integrative taxonomic re-assessment to check inter- or intraspecific status [46,47]. In addition to a meticulous investigation of potentially diagnostic morphological characters, the consideration of other molecular markers, especially nuclear genes, will be useful and necessary for the ultimate clarification of the taxonomic status of these critical taxa [48]. Ultimately, such an integrative taxonomic analysis should also include deviating sequences below the threshold of 2%, such as for two sequences that were currently assigned to Eudonia vallesialis, though in a separate BIN, and which indicate at least a geographical structure of the DNA barcodes.
Extensive genetic surveys of megadiverse groups of animals in protected areas have so far been rare, although the advantages of such inventories for Lepidoptera were demonstrated early, i.e., in a protected tropical rainforest of Costa Rica [49]. The resulting inventory of more than 11,000 genetic units of Lepidoptera, mostly representative of named and unnamed species, is extraordinary in terms of European diversity, which covers about the same number of species for the whole continent [27]. Despite the obvious advantages of DNA barcoding for increasingly objective identification of species and the possibility of recognizing cryptic diversity, there are no comprehensive, DNA-based inventories for protected areas in Europe available for Lepidoptera. Even large programs such as All Taxa Biodiversity Inventories (ATBI) in Europe, inspired by earlier projects in Central and North America, largely relied on classical taxonomic expertise [50]. A nearly complete species inventory in protected areas of the French/Italian Alps resulted in more than 12,640 species, including 50 new to science, but with few exceptions, i.e., for Mollusca, did not integrate molecular methods [50]. However, our preliminary results on the megadiverse Lepidoptera demonstrate the value of molecular data for the assessment of local fauna since they ultimately lead to greater completeness in the species inventory and a more objective and comprehensive assessment work by the experts involved.
DNA barcoding should therefore be integrated as an important method for creating inventories in protected areas, regardless of whether complete reference libraries are already available or not. In the best case, as in many regions of Europe, this will achieve completeness in the recording of species and genetic diversity. In the worst case, at least a serious estimate of genetic diversity is possible.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15020214/s1, Table S1: List of sequenced taxa from the study area, BINs, number of BIN members in BOLD, and no. of BIN members from the study area.
Author Contributions
Conceptualization, P.H.; Data curation, P.H. and C.W.; Formal analysis, P.H. and C.W.; Methodology, P.H. and C.W.; Writing—original draft, P.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
All 1743 COI sequences are available in DS-LEPICOTT on BOLD (dx.doi.org/10.5883/DS-LEPICOTT) at https://www.boldsystems.org/ (accessed on 7 January 2022) and in the NCBI GenBank under acc. nrs. MN549781, MN549792, MN549808, MN804381, MN805171, MN85764, MT048462, MT048467, MT048469, MT456708, MT456716, MT456734, MW211194, MW212076, MW212396, MW212680, MW212717, MW212995, MW213197, MW214289, OP443315–OP443316, OQ181468–OQ183188.
Acknowledgments
The authors are grateful to Michele Ottino for issuing the necessary permits. Furthermore, the continuous support by BOLD staff under the leadership of Paul D.N. Hebert is gratefully acknowledged. The map was produced by Romed Unterasinger. Robert (Bob) Heckford and Stella Beavan kindly helped with the improvement of the manuscript. Finally, we are most grateful to the guest editors and two anonymous reviewers for their careful work and valuable input.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rodrigues, A.S.L.; Andelman, S.J.; Bakarr, M.I.; Boitani, L.; Brooks, T.M.; Cowling, R.M.; Fishpool, L.D.C.; da Fonseca, G.A.B.; Gaston, K.J.; Hoffmann, M.; et al. Effectiveness of the global protected area network in representing species diversity. Nature 2004, 428, 640–643. [Google Scholar] [CrossRef] [PubMed]
- Bruner, A.G.; Gullison, R.E.; Rice, R.E.; Fonseca, G.A.B. Effectiveness of parks in protecting tropical biodiversity. Science 2001, 291, 125–128. [Google Scholar] [CrossRef]
- Engelbrecht, I. Invertebrate species inventories in protected area management: Are they useful? Afr. Entomol. 2010, 18, 235–245. [Google Scholar] [CrossRef]
- Encyclopaedia Britannica. Saving Earth. Species Inventories and Biodiversity Protection. Available online: https://www.britannica.com/explore/savingearth/species-inventories-and-biodiversity-protection (accessed on 29 December 2022).
- Pascher, K.; Švara, V.; Jungmeier, M. Environmental DNA-Based Methods in Biodiversity Monitoring of Protected Areas: Application Range, Limitations, and Needs. Diversity 2022, 14, 463. [Google Scholar] [CrossRef]
- De Biaggi, M.; Leccia, M.-F.; Kroupa, A.; Monje, J.C. Creating a biodiversity inventory in protected areas to increase knowledge of their natural heritage and to improve land management. J. Prot. Mt. Areas Res. Manag. 2010, 2, 49–52. [Google Scholar] [CrossRef]
- Ichter, J.; Leccia, M.-F.; Touroult, J.; Blandin, P.; Aberlenc, H.-P.; Holtof, J.-F.; Foret, J.; Bonet, R.; Pascal, O.; Dusoulier, F.; et al. Les inventaires généraux de la biodiversité en France et dans le monde. In Revue des All Taxa Biodiversity Inventories; UMS PatriNat/Parc national du Mercantour: Paris, France, 2010; 51p, Available online: http://www.patrinat.fr/sites/patrinat/files/atoms/files/2018/11/patrinat_2018_-_108_-_ichter_et_al_2018_les_inventaires_generaux_de_la_biodiversite_atbi.pdf (accessed on 2 January 2023).
- Hobern, D. BIOSCAN: DNA barcoding to accelerate taxonomy and biogeography for conservation and sustainability. Genome 2020, 64, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Mutanen, M.; Kivelä, S.M.; Vos, R.A.; Doorenweerd, C.; Ratnasingham, S.; Hausmann, A.; Huemer, P.; Dincă, V.; van Nieukerken, E.J.; Lopez-Vaamonde, C.; et al. Species-Level Para-and Polyphyly in DNA Barcode Gene Trees: Strong Operational Bias in European Lepidoptera. Syst. Biol. 2016, 65, 1024–1040. [Google Scholar] [CrossRef]
- Roslin, T.; Somervuo, P.; Pentinsaari, M.; Hebert, P.D.N.; Agda, J.; Ahlroth, P.; Anttonen, P.; Aspi, J.; Blagoev, G.; Blanco, S.; et al. A molecular-based identification resource for the arthropods of Finland. Mol. Ecol. Res. 2021, 22, 803–822. [Google Scholar] [CrossRef]
- Bergsten, J.; Bilton, D.T.; Fujisawa, T.; Elliott, M.; Monaghan, M.T.; Balke, M.; Hendrich, L.; Geijer, J.; Herrmann, J.; Foster, G.N.; et al. The effect of geographical scale of sampling on DNA barcoding. Syst. Biol. 2012, 61, 851–869. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, A.; Haszprunar, G.; Segerer, A.H.; Speidel, W.; Behounek, G.; Hebert, P.D.N. Now DNA barcoded: The butterflies and larger moths of Germany (Lepidoptera: Rhopalocera, Macroheterocera). Spixiana 2011, 34, 47–58. [Google Scholar]
- Huemer, P.; Mutanen, M.; Sefc, K.M.; Hebert, P.D.N. Testing DNA Barcode Performance in 1000 Species of European Lepidoptera: Large Geographic Distances Have Small Genetic Impacts. PLoS ONE 2014, 9, e115774. [Google Scholar] [CrossRef] [PubMed]
- Huemer, P.; Hebert, P.D.N. DNA Barcode Bibliothek der Schmetterlinge Südtirols und Tirols (Italien, Österreich)–Impetus für integrative Artdifferenzierung im 21. Jahrhundert. Gredleriana 2016, 16, 141–164. [Google Scholar]
- Baldizzone, G. I microlepidotteri del Parco Naturale del Mont Avic e zone limitrofe (Valle d’Aosta-Val Chalamy-Alpi Graie orientali). Rev. Valdotaine Hist. Nat. 1997, 50, 55–141. [Google Scholar]
- Baldizzone, G. I Microlepidotteri del Parco Naturale Alpi Marittime (Italia, Piemonte) (Lepidoptera). Boll. Museo Reg. Sci. Nat. Torino 2004, 22, 1–318. [Google Scholar]
- Hellmann, E.; Bertaccini, E. I Macrolepidotteri della Valle di Susa Italia Nord-occidentale (Alpi Cozie-Graie). Monogr. Museo Reg. Sci. Nat. Torino 2004, 40, 392. [Google Scholar]
- Huemer, P.; Hebert, P.D.N. DNA-Barcoding von Schmetterlingen (Lepidoptera) in Waldstandorten Südtirols (IT01 Ritten und IT02 Montiggl). For. Obs. 2012, 6, 75–98. [Google Scholar]
- Piccini, I.; Depetris, M.; Paradiso, F.; Cochis, F.; Audisio, M.; Artioli, P.; Smargiassi, S.; Bonifacino, M.; Giuliano, D.; La Cava, S.; et al. Macro-moth (Lepidoptera) Diversity of a Newly Shaped Ecological Corridor and the Surrounding Forest Area in the Western Italian Alps. Diversity 2023, 15, 95. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. Available online: www.barcodinglife.org (accessed on 15 December 2022). [CrossRef]
- Ente di Gestione Dell Aree Protette Delle Alpi Cozie. Available online: https://www.parchialpicozie.it/ (accessed on 29 December 2022).
- Robinson, G.S. The preparation of slides of Lepidoptera genitalia with special reference to the Microlepidoptera. Entomol. S Gaz. 1976, 27, 127–132. [Google Scholar]
- deWaard, J.R.; Ivanova, N.V.; Hajibabaei, M.; Hebert, P.D.N. Assembling DNA Barcodes: Analytical Protocols. In Methods in Molecular Biology: Environmental Genomics; Martin, C.C., Ed.; Humana Press Inc.: Totowa, NJ, USA, 2008; pp. 275–293. [Google Scholar]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-based registry for all animal species: The Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef]
- Italian Ministry of the Environment. Checklist of the Italian Fauna On-Line. 2003. Available online: https://www.faunaitalia.it/checklist/ (accessed on 15 December 2022).
- Karsholt, O.; van Nieukerken, E.J. Lepidoptera, Moths. Fauna Europaea 2013, Version 2021.12. Available online: https://fauna-eu.org (accessed on 15 December 2022).
- Lepiforum: Website zur Bestimmung von Schmetterlingen (Lepidoptera) und Ihren Präimaginalstadien. Available online: http://www.lepiforum.org (accessed on 15 December 2022).
- Huemer, P.; Karsholt, O.; Wieser, C. Megacraspedus cottiensis sp. nov. (Lepidoptera, Gelechiidae) from northern Italy–a case of taxonomic confusion. ZooKeys 2020, 963, 141–152. [Google Scholar] [CrossRef]
- Huemer, P. Integrative revision of the Caryocolum schleichi species group–A striking example of a temporally changing species concept (Lepidoptera, Gelechiidae). Alp. Entomol. 2020, 4, 39–63. [Google Scholar] [CrossRef]
- Hellmann, F.; Parenzan, P. I Macrolepidotteri del Piemonte. Museo Regionale die Scienze Naturali; Monografie XLVI: Torino, Italy, 2010; pp. 1–1058. [Google Scholar]
- Huemer, P.; Wieser, C. Bemerkenswerte Neufunde von Schmetterlingen (Lepidoptera) für Italien (Cottische Alpen). Carinth. II 2020, 210, 457–470. [Google Scholar]
- Ratnasingham, S.; Hebert, P.D.N. BOLD’s role in barcode data management and analysis: A response. Mol. Ecol. Notes 2011, 11, 941–942. [Google Scholar] [CrossRef]
- deWaard, J.R.; Ratnasingham, S.; Zakharov, E.V.; Borisenko, A.V.; Steinke, D.; Telfer, A.C.; Perez, K.H.J.; Sones, J.E.; Young, M.R.; Levesque-Beaudin, V.; et al. A reference library for the identification of Canadian invertebrates: 1.5 million DNA barcodes, voucher specimens, and genomic samples. Sci. Data 2019, 6, 308. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; deWaard, J.R.; Zakharov, E.V.; Prosser, S.W.J.; Sones, J.E.; McKeown, J.T.A.; Mantle, B.; La Salle, J. A DNA ‘barcode blitz’: Rapid digitization and sequencing of a natural history collection. PLoS ONE 2013, 10, e68535. [Google Scholar] [CrossRef] [PubMed]
- Zahiri, R.; Lafontaine, J.D.; Schmidt, B.C.; deWaard, J.R.; Zakharov, E.V.; Hebert, P.D.N. A transcontinental challenge—A test of DNA barcode performance for 1,541 species of Canadian Noctuoidea (Lepidoptera). PLoS ONE 2014, 9, e92797. [Google Scholar] [CrossRef]
- Dinca, V.; Dapporto, L.; Somervuo, P.; Vodă, R.; Cuvelier, S.; Gascoigne-Pees, M.; Huemer, P.; Mutanen, M.; Hebert, P.D.N.; Vila, R. High resolution DNA barcode library for European butterflies reveals continental patterns of mitochondrial genetic diversity. Commun. Biol. 2021, 4, 315. [Google Scholar] [CrossRef]
- Huemer, P.; Karsholt, O.; Aarvik, L.; Berggren, K.; Bidzilya, O.; Junnilainen, J.; Landry, J.-F.; Mutanen, M.; Nupponen, K.; Segerer, A.; et al. DNA barcode library for European Gelechiidae (Lepidoptera) suggests greatly underestimated species diversity. ZooKeys 2020, 921, 141–157. [Google Scholar] [CrossRef]
- Lopez-Vaamonde, C.; Kirichenko, N.; Cama, A.; Doorenweerd, C.; Godfray, H.C.J.; Guiguet, A.; Gomboc, S.; Huemer, P.; Landry, J.-F.; Laštůvka, A.; et al. Evaluating DNA Barcoding for Species Identification and Discovery in European Gracillariid Moths. Front. Ecol. Evol. 2021, 9, 626752. [Google Scholar] [CrossRef]
- Huemer, P.; Mutanen, M. An Incomplete European Barcode Library Has a Strong Impact on the Identification Success of Lepidoptera from Greece. Diversity 2022, 14, 118. [Google Scholar] [CrossRef]
- Hausmann, A.; Godfray, H.C.J.; Huemer, P.; Mutanen, M.; Rougerie, R.; van Nieukerken, E.J.; Ratnasingham, S.; Hebert, P.D.N. Genetic Patterns in European Geometrid Moths Revealed by the Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e84518. [Google Scholar] [CrossRef]
- Huemer, P.; Wieser, C.; Stark, W.; Hebert, P.D.N.; Wiesmair, B. DNA barcode library of megadiverse Austrian Noctuoidea (Lepidoptera)—A nearly perfect match of Linnean taxonomy. Biodivers. Data J. 2019, 7, e37734. [Google Scholar] [CrossRef]
- Ortiz, A.; Rubio, R.; Guerrero, J.; Garre, M.; Serrano, J.; Hebert, P.D.N.; Hausmann, A. Close congruence between Barcode Index Numbers (bins) and species boundaries in the Erebidae (Lepidoptera: Noctuoidea) of the Iberian Peninsula. Biodivers. Data J. 2017, 5, e19840. [Google Scholar] [CrossRef]
- Talavera, G.; Lukhtanov, V.A.; Rieppel, L.; Pirece, N.E. In the shadow of phylogenetic uncertainty: The recent diversification of Lysandra butterflies through chromosomal change. Mol. Phylogenet. Evol. 2013, 69, 449–478. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, J.R.; Roe, A.D.; Sperling, F.A.H. Multi-locus species delimitation in closely related animal and fungi: One marker is not enough. Mol. Ecol. 2012, 21, 4422–4436. [Google Scholar] [CrossRef]
- Huemer, P.; Karsholt, O. Revision of the genus Megacraspedus Zeller, 1839, a challenging taxonomic tightrope of species delimitation (Lepidoptera, Gelechiidae). ZooKeys 2018, 800, 1–278. [Google Scholar] [CrossRef]
- Dayrat, B. Towards integrative taxonomy. Biol. J. Linn. Soc. 2005, 85, 407–417. [Google Scholar] [CrossRef]
- De Queiroz, K. Species concepts and species delimitation. Syst. Biol. 2007, 56, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Janzen, D.H.; Hallwachs, W. DNA barcoding the Lepidoptera inventory of a large complex tropical conserved wildland, Area de Conservacion Guanacaste, northwestern Costa Rica. Genome 2016, 59, 641–660. [Google Scholar] [CrossRef] [PubMed]
- Deharveng, L.; Bedos, A.; Daugeron, C.; Villemant, C.; Judson, M.L.I. Organization, usefulness and limitations of an ATBI (All Taxa Biodiversity Inventory): The inventory in the Mercantour National Park. Zoosystema 2015, 37, 9–30. [Google Scholar] [CrossRef]
- Ichter, J.; Gargominy, O.; Leccia, M.-F.; Robert, S.; Poncet, L. The first large-scale All Taxa Biodiversity Inventory in Europe: Description of the Mercantour National Park ATBI datasets. Biodivers. Data J. 2022, 10, e85901. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).