Abstract
The introduction of non-indigenous species (NIS) is one of the major threats to the integrity of European coastal ecosystems. DNA-based assessments have been increasingly adopted for monitoring NIS. However, the accuracy of DNA-based taxonomic assignments is largely dependent on the completion and reliability of DNA barcode reference libraries. As such, we aimed to compile and audit a DNA barcode reference library for marine invertebrate NIS occurring in Europe. To do so, we compiled a list of NIS using three databases: the European Alien Species Information Network (EASIN), the Information System on Aquatic Non-indigenous and Cryptogenic Species (AquaNIS), and the World Register of Introduced Marine Species (WRiMS). For each species, we retrieved the available cytochrome c oxidase subunit I (COI) mitochondrial gene sequences from the Barcode of Life Data System (BOLD) and used the Barcode, Audit & Grade System (BAGS) to check congruence between morphospecies names and Barcode Index Numbers (BINs). From the 1249 species compiled, approximately 42% had records on BOLD, among which 56% were discordant. We further analyzed these cases to determine the causes of the discordances and attributed additional annotation tags. Of the 622 discordant BINs, after revision, 35% were successfully solved, which increased the number of NIS detected in metabarcoding datasets from 12 to 16. However, a fair number of BINs remained discordant. Reliability of reference barcode records is particularly critical in the case of NIS, where erroneous identification may trigger action or inaction when not required.
1. Introduction
Coastal ecosystems are the source of remarkable biological productivity and an abundance of goods and services [1]. However, these ecosystems and their native biodiversity have been significantly damaged by human activities, global climate change, and biological invasions [2,3,4]. Non-indigenous species (NIS) can be introduced outside their typical distribution range naturally or by human mediation. If they survive, establish, and expand in the recipient ecosystems, they may become invasive, spreading rapidly, and displacing and out-competing native species, threatening the ecosystem’s integrity [5,6]. These introductions, which are becoming more frequent due to climate change and increasing globalization, can cause negative effects on biodiversity, human health, and welfare, as well as major economic losses [7]. In this regard, NIS introductions are now included in key legislations and directives such as the European Union Regulation 1143/2014 [8] on Invasive Alien Species and the Marine Strategy Framework Directive [9]. For these reasons, it is critical to monitor non-indigenous species for better management and protection of marine environments [10].
Most past and ongoing monitoring programs are based exclusively on morphological approaches for species identification [11,12,13]. However, this is an expertise-demanding and time-consuming procedure, also hindered by the decrease in the number of taxonomists worldwide [14,15]. With the improvement of sequencing techniques, monitoring programs have started to implement DNA-based approaches, such as DNA barcoding and metabarcoding. While DNA barcoding consists of the identification of species using standardized short DNA fragments [16], DNA metabarcoding combines it with high-throughput sequencing (HTS), allowing the identification of multiple species and taxonomic groups from complex samples, including environmental samples [17,18]. Several advantages of DNA-based approaches over the traditional methods (i.e., morphology-based identification) include the increased sensitivity and specificity in the detection of species, including cryptic taxa, as well as a greater time and cost effectiveness through the simultaneous processing of a large number of samples, ultimately improving NIS detection and monitoring in coastal and marine ecosystems [19,20,21,22,23]. The use of DNA-based tools also allows the detection of early developmental stages (e.g., gametes, propagules, planktonic larvae, eggs) or smaller organisms not amenable to morphological identification or when organisms occur at low densities [24]. NIS early detection is crucial to overcome the irreversible impacts that these species, once established, can provoke in coastal and marine ecosystems.
DNA-based species identifications are highly dependent on the quantity and quality of molecular data present in the available genetic databases [25]. The main public databases used are GenBank® (www.ncbi.nlm.nih.gov/genbank/ accessed on 25 January 2023) and the Barcode of Life Data System (BOLD) [26,27]. While GenBank possesses a much larger DNA sequence dataset, BOLD was created specifically for the acquisition, storage, analysis, and publication of DNA barcode records. The standard DNA barcode region for the animal kingdom is a ~650bp fragment of the 5′ end of the cytochrome c oxidase I (COI) mitochondrial gene, due to its ability to discriminate species in most animal taxa, including congeneric species [28]. Despite the increase in DNA barcode data availability over the years, there are still gaps in representative barcodes for a large proportion of European marine invertebrate species (e.g., [29,30,31]), reaching as much as 50 to 70% for dominant macroinvertebrate taxa [29,32], and up to 63% for NIS belonging to Animalia [33].
The Barcode Index Number (BIN) system was created in BOLD to assign a specific code (BIN) to a cluster of COI nucleotide sequences that represent Molecular Operational Taxonomic Units (MOTUs), delimited using the Refined Single Linkage (RESL) algorithm [34]. For most animal taxa, COI MOTUs match with species, hence BINs can be viewed as a proxy to species and be used as a molecular benchmark to test the taxonomic congruence of COI barcode records [34]. For a sequence to be barcode compliant, there is a set of formal criteria implemented in BOLD [27] to assess the quality of the molecular data and each record’s metadata (see also [35]). However, a number of operational errors that may occur along the production chain of the reference DNA barcodes (e.g., misidentification of specimens, cross-contamination of PCR’s DNA template, or mislabeling of records) cannot be addressed by such quality control criteria, thereby undermining the reliability of the taxonomic assignments of barcode records [32].
Indeed, a comprehensive revision of DNA barcode records of 7576 species distributed by four phyla of marine invertebrates, also revealed that overall, 39% of the species had ambiguous assignments [25]. As most DNA-based monitoring studies rely on poorly curated libraries to assign a taxonomic identification to sequences, quality control measures should be employed to ensure the taxonomic reliability of the data used, particularly in the case of NIS, where identifications of possible invasive species can trigger official government actions to prevent negative ecological and economic impacts or to manage a current invasion situation [32,36,37]. Datasets and databases have been developed with diverse curation levels and criteria depth and targeting different taxonomic groups. However, most of the quality control criteria employed focused more on the removal of sequences according to thresholds of minimum length and sequence quality and reliability parameters rather than on the accuracy of the taxonomic assignment [38], while others only flag sequences that seem discordant or with conflicting taxonomic classification [39,40,41]. There are also tools and software that examine available DNA sequences and evaluate its quality or possible errors that can lead to misidentifications [42]. Oliveira and co-authors [43] described a manual process to audit a reference library for European marine fishes by using a grading system to determine concordance or discordance between morphospecies and BINs, which was then used as a starting point to create the Barcode Audit & Grade System (BAGS), an automated tool for grading and auditing animal DNA barcode reference libraries [44]. However, most of these methodologies only detect and flag discordances but do not disentangle them. Given the fast increase in the use of DNA-based assessments in ecological studies and in the resulting molecular data, there is an urgent need to create curated DNA barcode reference libraries to improve the accuracy of DNA-based identifications [30,31,45,46].
With this work, we aimed: (i) to assess and analyze the COI barcode data available in the BOLD database, for a compiled list of non-indigenous marine invertebrate species for Europe, (ii) to further examine discordant data, i.e., when discordances between morphospecies and BINs exist, (iii) to provide a workflow to curate discordances (i.e., amending the solved discordances and signaling the unsolved ones), (iv) to create a curated DNA barcode reference library of European marine invertebrate NIS and, finally, (v) to assess the efficiency of the curated DNA barcode reference library in NIS detection, using DNA metabarcoding datasets obtained in recreational marinas in Portuguese coastal regions, known in advance to contain NIS.
2. Materials and Methods
2.1. Compilation of the List of Non-Indigenous Invertebrate Marine Species Occurring in Europe
To compile the list of invertebrate marine NIS occurring in Europe, three databases were accessed in September 2021: the European Alien Species Information Network (EASIN) (https://easin.jrc.ec.europa.eu/easin (accessed on 15th September 2021)) [47], the Information System on Aquatic Non-indigenous and Cryptogenic Species (AquaNIS) (http://www.corpi.ku.lt/databases/index.php/aquanis/ (accessed on 15th September 2021)) [48] and the World Register of Introduced Marine Species (WRiMS) (https://www.marinespecies.org/introduced (accessed on 15th September 2021)) [49]. From the EASIN database, 1013 species records were retrieved using the following criteria—“Environment”: Marine + Oligohaline; “Species status”: Alien + Cryptogenic + Questionable; “Taxonomy”: Animalia (in “Chordata” only “Ascidiacea” was selected). From the AquaNIS database, 1439 species records were returned using as a search criterion the “Recipient region” and the following sub-criteria—“Ocean”: Atlantic + Arctic; “Ocean Region”: NE Atlantic + Arctic; “LME”: 20. Barents Sea + 21. Norwegian Sea + 22. North Sea + 23. Baltic Sea + 24. Celtic-Biscay Shelf + 25. Iberian Coastal + 26. Mediterranean Sea + 59. Iceland Shelf + 60. Faroe Plateau + 62. Black Sea + A1. Macaronesia. From the WRiMS database, 4769 species records were retrieved using the following criteria in Distributions—“Geounit”: North Atlantic + Arctic; “Rank”: is Species; “Synonyms”: only accepted names. From the resulting list, we selected only the records with the following Localities: Barents Sea, Norwegian Sea, North Sea, Baltic Sea, Celtic-Biscay Shelf, Iberian Coastal, Mediterranean Sea, Iceland Shelf, Faroe Plateau, Black Sea, and Macaronesia. Species taxonomic classification and environment were verified in the World Register of Marine Species (WoRMS) database (www.marinespecies.org (accessed on 17th September 2021)) [50]. The lists were then merged, and duplicate entries removed, resulting in a final list of 1249 species of marine and brackish invertebrate NIS occurring in Europe.
2.2. Compilation of the Genetic Data and Curation
To compile and audit the available COI barcode data for each species in the list, we used the software BAGS ([44]; https://github.com/tadeu95/BAGS (accessed on 20th September 2021)). This tool annotates species records in five grades (A–E) according to the congruency between species assignments and BINs, and the quantity of the sequences present in the BOLD database [27]: A and B for concordant morphospecies, (one BIN = only one species name), where these grades only differ in the quantity of available sequences in the library (grade A: > 10 sequences and grade B: ≤ 10 sequences), C for morphospecies assigned to multiple BINs, D for morphospecies with insufficient sequences (<3) and E for seemingly discordant morphospecies (more than one species name in the same BIN). With the graded library generated by BAGS, a workflow was designed (Figure 1) to classify the BINs and further audit graded C and E records and curate the seemingly discordant morphospecies (grade E). A system of four tags was used (adapted from [25]): AMBIG (to refer to ambiguous records), MISID (to refer to misidentifications), SYN (to refer to synonyms) or SHARE (if well-established species were aggregated in the same BIN). The final tags were set out as RELIABLE, if we successfully matched a morphospecies to a single BIN, either in case of concordance or solved BIN discordances; or UNCERTAIN, if the BIN discordance remained unsolved after auditing, or if the available sequences were insufficient to resolve the taxonomic ambiguity (criteria detailed further below).
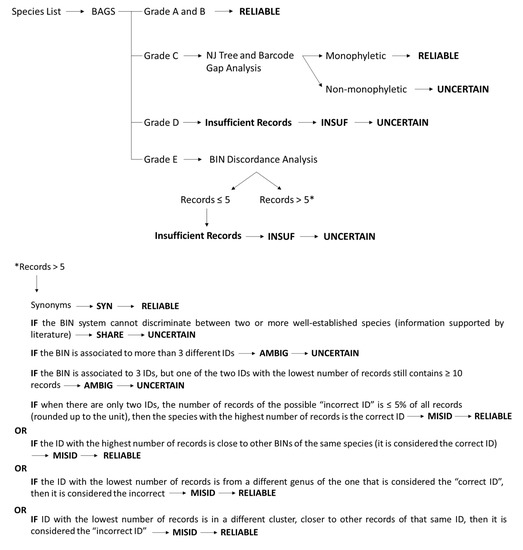
Figure 1.
Workflow designed for the curation and audit of BAGS-graded BINs. A list of marine non-indigenous invertebrates was submitted to BAGS that produced a library of graded morphospecies. A set of criteria was developed to tag the BINs associated with the morphospecies as SYN (in case of synonyms), SHARE (if the BIN system cannot discriminate between well-established species), MISID (misidentifications), or AMBIG (ambiguous records). Final RELIABLE or UNCERTAIN tags defined whether the morphospecies identification (ID) matched to each BIN was considered reliable or uncertain.* workflow used for grade E species and with more than 5 records in the BIN discordance analysis.
BINs assigned to species graded A and B were further tagged RELIABLE as they represent concordant records (i.e., the species is assigned to a single BIN). All grade D records were considered INSUF-UNCERTAIN as the data are insufficient (as we considered less than 5 records per BIN, and BAGS considers BINs with less than 3 records). For grade C morphospecies, a publicly available dataset comprising the involved BINs was created in BOLD (DS-NISEURC) and two analyses were used to determine if all BINs of a given grade C morphospecies were in a monophyletic (morphospecies considered RELIABLE) or non-monophyletic group (considered UNCERTAIN). A neighbor-joining (NJ) Tree was generated using the following criteria: BOLD Aligner and selection of Country/Ocean, and the BIN URI and GenBank Accession boxes (for the remaining, the default parameters were maintained). In the filters, nucleotide sequence length ≥300 bp was selected. A barcode gap analysis was also performed with the same settings (Table S4). This analysis flags the cases where the distance between the species and the nearest neighbor is <2% or lower than the maximum intraspecific distance. Grade E records were compiled into a different dataset (DS-NISEURE) and curated by performing a BIN discordance report on BOLD (no filters were selected) (Table S5). This analysis produces files containing concordant, discordant, and singleton records. While the BIN discordance report is a BIN-centered approach, the BAGS analysis is morphospecies-centered, meaning that in BOLD, a BIN that includes more than one species is considered discordant, while in BAGS, if a morphospecies has one discordant BIN assigned, it is automatically considered a discordant species (grade E), even if the other BINs are concordant. As such, some of the BINs of species graded E by BAGS can be considered concordant in the BIN discordance report. All BINs with ≤ 5 records were considered “Insufficient Records” (INSUF) and tagged UNCERTAIN, as 3 or fewer records (as used on BAGS) were not sufficient to solve certain discordances; BINs with more than 5 records were manually inspected using the following criteria:
- BINs were tagged SYN if the identifications (IDs) were synonyms of the accepted species name (confirmed using the WoRMS database) and were considered RELIABLE.
- BINs were tagged AMBIG and UNCERTAIN if:
- The BIN comprises more than 3 different IDs (i.e., species, genus, incomplete classification);
- The BIN comprises 3 IDs, but one of the two IDs with the lowest number of records still contains ≥ 10 records.
- BINs were tagged MISID, and the discordance was solved if:
- When there are only two IDs, and the number of records of the putative “incorrect ID” is ≤ 5% of all records in the BIN (rounded up to the unit), the species with the highest number of records is considered the correct ID;
- The ID with the highest number of records is close to other BINs of the same species (when analyzing the NJ tree for the genus), then it is considered the correct ID;
- The ID with the lowest number of records is from a different genus than the one that is considered the “correct ID”, then it is considered the incorrect ID;
- When analyzing the NJ tree for the genus, the ID with the lowest number of records is in a different cluster, containing only records of that same ID, then it is considered the incorrect ID.
For the analysis of points 3.b, 3.c, and 3.d, NJ trees were generated for the genus, family, or order (depending on the discordance level), to determine the correct ID according to each criterion.
A reference library composed of three DNA sequences for each BIN was constructed using all the RELIABLE BINs. A script was developed to select only the curated BIN records associated with morphospecies from the graded library (resultant from the BAGS analysis), with the curated identification, and then to select three random DNA sequences, prioritized according to size and availability of country data for each record. Nine size classes were created to select the sequences: (1) sequences with 658 bp; (2) ≥650 and <658 bp; (3) ≥625 and <650 bp; (4) ≥600 and <625 bp; (5) ≥575 and <600 bp; (6) ≥550 and <575 bp; (7) ≥525 and <550 bp; (8) ≥500 and <525 bp; (9) <500 and >658 bp. The script developed to create the DNA sequence reference library is available at https://github.com/tadeu95/Curated-BINs-Reference-Library (accessed on 25 January 2023).
With this curated library, a new dataset was created on the BOLD database using the reference library process IDs (DS-NISEUREF).
2.3. Testing the Impact of the Curated Reference Library on the Accuracy and Amount of NIS Detected through DNA Metabarcoding of Natural Communities
High-throughput sequencing data were generated from three types of samples collected in a recreational marina (Costa Nova, Portugal: 40°37′11.6″ N, 8°44′55.7″ W): zooplankton collected from the water column (with a plankton net with 55 µm mesh size), water eDNA (1 L collected at 1 m depth), and a sample of marine invertebrates scraped from hard substrates (e.g., pontoons, cables, buoys), to conduct this analysis (considering only the marine or oligohaline invertebrates detected). Briefly, zooplankton and eDNA samples were vacuum filtered through 0.45 μm, 47 mm nitrocellulose membranes (Millipore, Corp., Bedford, MA, USA) and DNA from half of the filters was extracted using the DNeasy® PowerSoil® Kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. Marine invertebrate samples were preserved in absolute ethanol, and DNA extraction was carried out using the protocol described in [51]. High-throughput sequencing (HTS) was carried out at Genoinseq (Biocant, Cantanhede, Portugal) in an Illumina MiSeq® platform using the primer pair mlCOIintF (5′-GGWACWGGWTGAACWGTWTAYCCYCC-3′) [52] and LoboR1 (5′-AAACYTCWGGRTGWCCRAARAAYCA-3′) [53] to amplify a 313 bp region of the COI gene.
MiSeq data were analyzed on the Multiplex Barcode Research And Visualization Environment (mBRAVE; http://mbrave.net/ (accessed on 15th June 2022)) [54], and three tests were performed, which differed mainly in the reference sequences libraries used to conduct the taxonomic assignments: (1) BOLD system reference libraries, namely SYS-CRLCHORDATA, SYS-CRLINSECTA, SYS-CRLNONARTHINVERT, and SYS-CRLNONINSECTARTH; (2) a dataset containing only records from grade A, B, and C (audited with BAGS); and (3) our curated reference library. mBRAVE settings were as follows: Trimming—Trim front: 0 bp, Trim end: 0 bp, Trim length: 313 bp, Primer masking: off; Filtering—Min QV: 10, Min length: 150 bp, Max bases with low QV: 25%, Max bases with ultralow QV: 25%; No pre-clustering, ID distance threshold: 3%, Exclude from OTU threshold: 3%, Minimum OTU size: 1, OTU threshold: 3%; Paired End Merging: Pool, Assembler min overlap: 20 bp, Assembler max substitutions: 5 bp. This allowed us to compare results on the accuracy and number of identifications using non-curated libraries (BOLD system libraries), an audited library, and a fully curated library. For the analysis of the results using the non-curated library, we eliminated all species that were not marine or brackish invertebrates, records with hits to taxonomic ranks higher than species level (as for NIS, information at the species level is mandatory), and records with hits with more than one species/taxon.
3. Results
3.1. List of Non-Indigenous Invertebrate Marine Species Occurring in Europe
After the removal of duplicated records and records with taxonomic ranks higher than species level, our final list contained 1249 non-indigenous species (Table S1), of which 38% were found in both EASIN and AquaNIS, 21% were retrieved exclusively from AquaNIS, 40% from EASIN and 1% from WRiMS. The species were distributed into 16 phyla, with a dominance of Arthropoda (28%), Mollusca (28%), and Annelida (18%) (Figure 2).
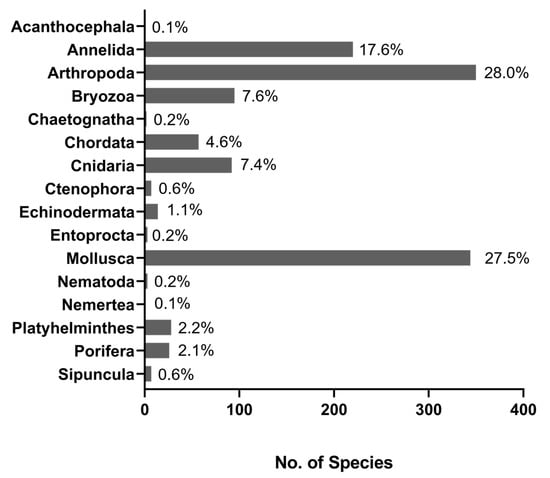
Figure 2.
Taxonomic composition of the 1249 invertebrate marine non-indigenous species compiled from the three databases: EASIN, AquaNIS, and WRiMS. Chordata only includes Ascidiacea.
3.2. Curation of the Taxonomic Assignments of Barcode Records
Of the 1249 NIS, only 42% (530 species) had COI barcode sequences in BOLD (Table S2), and the most well-represented groups with sequence data were the same that dominated the species list (Arthropoda, Mollusca, and Annelida). For these 530 species, BAGS retrieved 25,291 records belonging to 1105 BINs (Table S3). Only 11.4% of these BINs were assigned to morphospecies graded concordant (A + B), 23% to morphospecies graded C (more than 1 BIN per species, with the species being the exclusive member of the BINs), 9% to morphospecies graded D (insufficient data), and the majority (56.3%) to morphospecies graded E (discordant) (Figure 3). Arthropoda and Mollusca were the most well-represented phyla in all grades (ranging from 20% to 45% of the BINs in each grade). European NIS belonging to Acanthocephala, Chaetognatha, Nemertea, Platyhelminthes, and Porifera do not have records with concordant BINs (A and/or B); however, these BINs only represent 3% of the total number of BINs in the dataset.
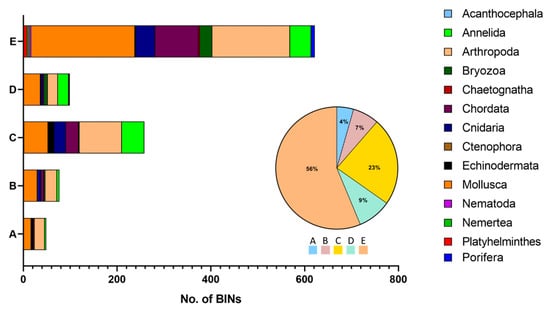
Figure 3.
Taxonomic composition of the graded BINs resulting from the BAGS analysis. A and B represent concordant records, C records with multiple BINs for the same species, D records with insufficient data, and E discordant records (more than one species in the same BIN). Chordata only includes Ascidiacea.
All BINs corresponding to grade C morphospecies were considered RELIABLE as all groups were monophyletic (Figure S1). The BIN discordance report that was created for grade E records on BOLD separates the BINs into three categories: singletons (BINs with a single record), concordant (BINs with no taxonomic discordance), and discordant (BINs with taxonomic discordance). For the 622 graded E BINs, this report indicated that 20% were singletons, 31% were concordant, and 49% were discordant (Figure 4). Singletons in graded E BINs are likely the result of private records (records that researchers have deposited on BOLD but are not publicly available); these were immediately tagged UNCERTAIN since they hold insufficient records. Concordant and discordant records were analyzed case by case following the established workflow (Figure 1). Of the 194 concordant BINs, 99 (51%) were considered UNCERTAIN (91 INSUF). Three BINs (BOLD:AAA2185, BOLD:AAA4734, and BOLD:ACQ2249) assigned to species of the Mytilus genus were considered UNCERTAIN as the Mytilus edulis species complex is composed of three closely related species: M. edulis Linné, 1758, M. galloprovincialis Lamarck, 1819 and M. trossulus Gould, 1850 [55]. Of the 302 discordant, 174 were tagged AMBIG, 92 MISID, 31 SYN, and 5 SHARE (Figure 4). Most BINs tagged SYN, SHARE and INSUF belonged to Mollusca (77%, 88%, and 38%, respectively), while Arthropoda and Mollusca dominated the MISID (42% and 30%, respectively) and AMBIG records (28 and 26%, respectively) (Figure 5). After the curation, 216 BINs (35%) were solved and considered RELIABLE. A total of 70% of all AMBIG records were tagged INSUF, which represents 34% of all BINs (378 in the total 1105 BINs) (Figure 4).
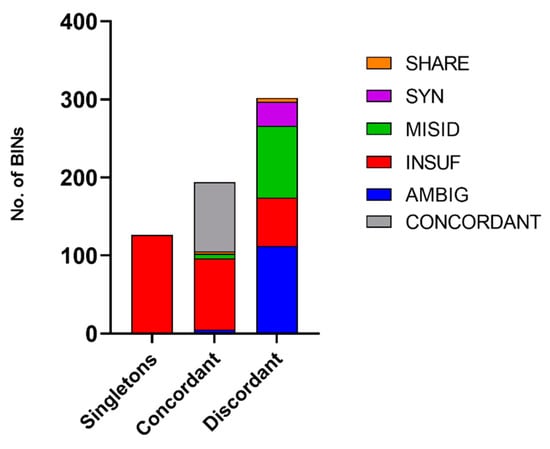
Figure 4.
Results from the BIN discordance report performed on the BOLD database. This analysis separates the BINs into three categories: singletons (BINs with only one record), concordant (BINs with no taxonomic discordance), and discordant (BINs with taxonomic discordance). The BINs were then tagged according to the designed workflow into: AMBIG (ambiguous records), INSUF (insufficient records), MISID (misidentifications), SYN (synonyms), or SHARE (if the BIN system was not able to discriminate well-established species).
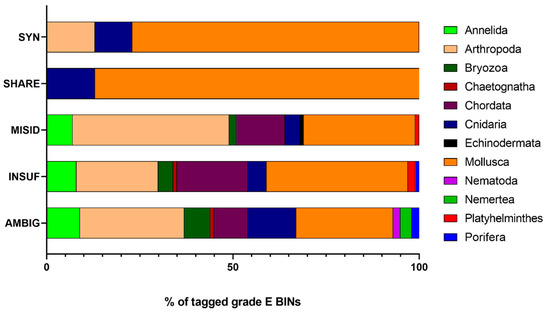
Figure 5.
Percentual taxonomic composition of the tagged graded E BINs. AMBIG: ambiguous records, INSUF: insufficient records, MISID: misidentifications, SYN: synonyms, and SHARE: when the BIN system was not able to discriminate well-established species. Chordata only includes Ascidiacea.
3.3. Impact of the Curated Reference Library on the Accuracy and Amount of NIS Detected through DNA Metabarcoding of Natural Communities
Only RELIABLE BINs were included in the final curated reference library (Table S6). This library included 597 BINs (54% of the initial number) corresponding to 356 species, of which 21% were to grade A + B morphospecies, 43% to grade C, and 36% corresponded to the original graded E that were deemed RELIABLE after applying the curation workflow (Table S8). If the post-audit library was assembled only with records graded A, B, and C, it would only consist of 384 BINs (36% fewer BINs than the curated reference library created using our grade E-curation workflow) (Table S7).
High-throughput sequencing reads analysis of the three samples (eDNA, zooplankton, and fouling marine invertebrate samples) against the non-curated libraries was assigned to a total of 29 marine invertebrate species (Table 1) out of a total of 44 species. The highest number of NIS was detected using our curated library (16 NIS), followed by the non-curated libraries (12 NIS) and the audited library (9 NIS) (Table 1). Haliclystus tenuis, Scruparia ambigua, Syllidia armata, and Terebella lapidaria were exclusively detected with the non-curated libraries. Upon further investigation, we determined that the records from Haliclystus tenuis and Terebella lapidaria belonged to BINs corresponding to grade D morphospecies (insufficient records), and the BINs of the other two species were not present in the BOLD database (probably private records)—rendering these results as uncertain detections. Eight NIS were not detected using the non-curated libraries but were present in either one or both the audited and curated libraries. Amphibalanus improvisus, Paracalanus indicus, and Ruditapes philippinarum were exclusively detected with our curated library.

Table 1.
Number of sequences (sum of reads of all the samples) per species detected with each of the three libraries: non-curated, audited, and curated, using the mBRAVE engine.
4. Discussion
By analyzing the availability and quality of the COI records belonging to non-indigenous invertebrate marine species occurring in Europe, our study brings to the forefront six main conclusions: (1) most species still lack genetic information for this marker (58%); (2) the majority of the existent records were graded discordant (56%); (3) data curation can increase the number of concordant records (36%); (4) the use of a curated database can increase the chances of detecting NIS (33%); (5) some NIS can be categorized as possible cryptic species complexes (23%); and (6) many BINs were represented by singletons (i.e., only one sequence record, 20% of all graded E morphospecies BINs).
DNA metabarcoding has proven to be a powerful tool in biomonitoring and species detection [56]. In particular, the ability to detect premature life stages, such as propagules, larvae or small juveniles, or recently introduced species that are still at a low density and confined to a small area, maximizes the early detection of NIS before harming ecosystems [57]. However, the use of DNA metabarcoding, in particular at the taxonomic assignment step, is dependent on the availability of taxonomically accurate sequence data in public databases. In this study, we verified that 58% of the species analyzed did not have publicly available COI sequence data. Gaps of up to 90% in DNA barcodes have been reported earlier for different taxonomic groups of marine invertebrates [29,30,32,58]. These gaps can probably be explained by the difficulties inherent in generating reference libraries for this phylogenetically very diverse set of taxa, for example, in the initial identification of specimens through morphology-based approaches [59] or the lower PCR amplification success rates when using universal primers [53], and also a lower number of studies and commitment to produce reference libraries for marine invertebrates, in comparison to other groups such as fishes or freshwater invertebrates [32]. This was also noticeable in the current study by the significant number of UNCERTAIN records that were tagged “INSUF” due to insufficient records in the database (70%).
Although there are fewer studies aiming at completing and auditing DNA barcode reference libraries of marine invertebrates, recently, there have been some initiatives worth mentioning. During the 8th iBOL conference hackathon in 2019, a group of researchers reviewed 83,712 DNA barcode records belonging to four major taxonomic groups of marine invertebrates (Echinodermata, Mollusca: Bivalvia and Gastropoda, Annelida: Polychaeta and Arthropoda: Crustacea), which resulted in more than 2700 records flagged and removed from BOLD [25]. In addition, the project GEANS aims to verify taxonomically over 90% of the macroinvertebrate species of the North Sea [60]. For this reference library, quality control will be performed by a taxonomic curator. By 2021, this reference library covered over 30% of North Sea species, but morphological identification can be a time-consuming process that becomes more difficult at a wider European scale. In contrast, we concur that taxonomic expertise is crucial to guarantee accurate identifications of the specimens used to generate the DNA barcodes for records flagged as discordant already deposited in genetic databases. A taxonomic congruency check, such as the one performed in the current study, may be the most straightforward and fast approach or even the only one available for records lacking vouchers [25]. However, in the cases where the discordances are not solvable (and in our study, were 65% of grade E BINs), the taxonomic revision of the original specimens (if stored properly, and for that repositories would remain crucial) is the only solution. For instance, Ciona intestinalis and C. robusta (Chordata: Tunicata) were recently reclassified since there were many sequences of C. robusta on GenBank falsely attributed to C. intestinalis [61,62]. In addition, sequences of Botrylloides diegensis (Chordata: Tunicata) (a global marine invader) were found to be erroneously assigned to B. leachii (a putatively native species in Europe) [63], and sequences from Acartia tonsa (Arthropoda: Crustacea), an invasive species recorded in many ecoregions of the world, including in several European countries, formed several distinct clades, some of which clustered with A. hudsonica [64].
Our analysis highlighted that in addition to the lack of COI barcode data for these species, the majority of the BINs were graded discordant. This has also been found for Macaronesian non-indigenous marine invertebrates, where approximately 50% of the species were graded discordant [31], which can represent a major constraint for the use of DNA metabarcoding in studies targeting marine invertebrate communities. In the particular case of NIS, species misidentification can be even more problematic since it can trigger both action or inaction when not required.
Comprehensive and curated reference libraries are imperative for accurate DNA-based taxonomic identifications [65]. However, a standard procedure for the curation of BINs is still lacking. Guidelines for the curation of specimen vouchers and barcode data and metadata, as recently published by Rimet and collaborators [35], are an important component of quality control, but a further taxonomy congruency check is still necessary at the end of the reference library production chain [66]. With our curation workflow based on an automated system (BAGS), we first detected the discordant records, and by inspecting each, we were able to increase the number of concordant records using a conservative approach. We also strived for a simple workflow that can be easily employed, at least for moderately sized sets of species. However, our curation protocol also has its own limitations because it is based only on the congruence between morphospecies and BINs. Moreover, for this purpose, we considered it important to use all data available. The workflow did not evaluate the associated metadata. In addition, occasionally, records that were previously considered reliable or morphospecies concordant can be later considered uncertain, or vice-versa, due to the frequent update of BIN information and taxonomic revision. This is also one of the reasons, in case of doubt or lack of representativity, why we opted for a conservative approach in the current study, where most curated BINs had explicit ambiguities of simple resolution. In addition, as previously mentioned, the fact that this workflow involves manual curation (albeit initially supported by BAGS) makes it challenging for bigger datasets. However, we believe that the criteria can be automated, which would facilitate the work of researchers, as well as any time databases are updated. Moreover, finally, even if some criteria do not guarantee the most accurate identification (indeed, some records remained uncertain and unsolvable, at least until more data becomes available), we still believe that this curation workflow is a quick and reliable approach, as some records can, in fact, be considered reliable and definitely solved (e.g., records tagged SYN, MISID).
In our study, we also detected almost twice the number of BINs compared to the number of morphospecies, flagging the potential presence of hidden diversity for several NIS occurring in European waters. Indeed, species complexes have been uncovered for many invasive species (e.g., Bryozoa: Bugula neritina, Watersipora sp. [67,68]) and marine invertebrates in general [69,70,71]. This highlights that reference libraries should cover a balanced representation of specimens across each species’ distributional range, including native and recipient locations, to account for possible genetic variability among different geographic regions. In addition, we found a large fraction of NIS represented by singletons, preventing their use as reliable records and the detection of possible intraspecific variability or unrecognized diversity.
As a result of our analysis, we were able to assemble a reliable reference library for NIS using only concordant records (597 BINs) belonging to 356 species with confirmed occurrence in Europe. When the trial metabarcoding datasets were matched against this curated library, the number of NIS detected increased by a third. Furthermore, the comparative tests using the non-curated library, an audited library, and the final curated reference library revealed the possible existence of four false detections. Two of the species that were exclusively detected using the non-curated libraries were associated with BINs that were not publicly available on BOLD. In our opinion, these could be either private records, which can represent a major issue since researchers cannot verify the detailed information, and, thus, audit and guarantee its credibility, or records that, in the meantime, were removed from BOLD but not immediately updated on BOLD imported databases on mBRAVE. Weigand and collaborators [32] showed that for some groups, such as Annelida and Sipuncula, the number of private records was even higher than that of public records. The other two species detected using the non-curated libraries had insufficient records in the database, which renders them uncertain using our defined standards for this study. Without proper curation, these four “false detections” could lead to false conclusions during a biomonitoring study, which highlights the importance of proper curation of taxonomic assignments of barcode records. The use of a curated dataset also revealed four NIS that were not initially detected using a non-curated library. Testing our metabarcoding reads against these three datasets showed that data curation is crucial to improve species detection, as previously reported [72]. Importantly, it also revealed that the effort to curate discordant data should be employed instead of just removing it from the datasets, as the audited library recovered the lowest number of NIS.
5. Conclusions
Although completing the gaps in reference libraries is essential for making the most of the potential of the DNA metabarcoding in NIS surveillance in European marine and coastal ecosystems [23,33], a careful compilation, verification, and annotation of available sequences is also crucial to support rigorous species identifications, as demonstrated in our study. This can have major implications as introduced species can be misidentified as putative native species or vice-versa when employing DNA-based tools and conducting taxonomic assignments against non-curated databases. Unfortunately, and as also flagged by our study, these database errors are frequent, and thus, auditing existing records and building/compiling curated reference libraries with reliable taxonomic assignments is equally important as generating new reference sequences for NIS.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15020174/s1, Figure S1: Neighbor-joining (NJ) tree generated on BOLD using all the records from grade C morphospecies, Table S1: List of the non-indigenous invertebrate marine species occurring in Europe retrieved in September of 2021 from the databases EASIN, AquaNIS, and WRiMS, Table S2: List of the non-indigenous invertebrate marine species occurring in Europe without available COI barcode data in the BOLD database, Table S3: Graded library generated by BAGS using the list of the non-indigenous invertebrate marine species occurring in Europe containing available COI barcode data, Table S4: Barcode gap analysis performed on BOLD for grade C morphospecies using the dataset DS-NISEURC, Table S5: Analysis of graded E records and assignment of TAGS using the BIN discordance report performed on BOLD with the dataset of grade E records (DS-NISEURE), Table S6: List of curated RELIABLE BINs, Table S7: Audited library composed of reference sequences for each BIN (using BINs graded A, B, and C by BAGS), compiled using the developed script, and Table S8: Curated library composed of reference sequences for each RELIABLE BIN, compiled using the developed script.
Author Contributions
Conceptualization, A.S.L., F.O.C., P.E.V. and S.D.; methodology, A.S.L., F.O.C., J.T.F., P.E.V. and S.D.; software, A.S.L., J.T.F. and P.E.V.; validation, A.S.L. and J.T.F.; formal analysis, A.S.L. and J.T.F.; investigation, A.S.L., F.O.C., J.T.F., P.E.V. and S.D.; resources, F.O.C. and S.D.; data curation, A.S.L. and J.T.F.; writing—A.S.L. and S.D.; writing—review and editing, A.S.L., F.O.C., J.T.F., P.E.V. and S.D.; visualization, A.S.L.; supervision, F.O.C. and S.D.; project administration, F.O.C. and S.D.; funding acquisition, F.O.C. and S.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by national funds through the Foundation for Science and Technology (FCT I.P.), grant number PTDC/BIA-BMA/29754/2017 (NIS-DNA: Early detection and monitoring of non-indigenous species (NIS) in coastal ecosystems based on high-throughput sequencing tools) and by the “Contrato-Programa” UIDB/04050/2020. Financial support granted by the FCT I.P. to S.D. (CEECIND/00667/2017) is also acknowledged. A.S.L. (UI/BD/150871/2021) and J.T.F. (UI/BD/150910/2021) are supported by the Collaboration Protocol for Financing the Multiannual Research Grants Plan for Doctoral Students with financial support from FCT I.P. and the European Social Fund under the Northern Regional Operational Program—Norte2020.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The script developed to create the DNA sequence reference library is available at https://github.com/tadeu95/Curated-BINs-Reference-Library (accessed on 25 January 2023). Metabarcoding datasets will be made available upon request since they belong to a study that is not published yet.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Costanza, R.; d’Arge, R.; de Groot, R.; Farber, S.; Grasso, M.; Hannon, B.; Limburg, K.; Naeem, S.; O’Neill, R.V.; Paruelo, J.; et al. The value of the world’s ecosystem services and natural capital. Nature 1997, 387, 253–260. [Google Scholar] [CrossRef]
- Jackson, J.B.C.; Kirby, M.X.; Berger, W.H.; Bjorndal, K.A.; Botsford, L.W.; Bourque, B.J.; Bradbury, R.H.; Cooke, R.; Erlandson, J.; Estes, J.A.; et al. Historical Overfishing and the Recent Collapse of Coastal Ecosystems. Science 2001, 293, 629–637. [Google Scholar] [CrossRef]
- Turner, R.E.; Rabalais, N.N. Coastal eutrophication near the Mississipi river Delta. Nature 1994, 368, 619–621. [Google Scholar] [CrossRef]
- Rilov, G.; Crooks, J.A. Biological Invasions in Marine Ecosystems: Ecological, Management and Geographic Perspectives; Springer: Berlin/Heidelberg, Germany, 2009; p. 30. [Google Scholar] [CrossRef]
- Bax, N.; Williamson, A.; Aguero, M.; Gonzalez, E.; Geeves, W. Marine invasive alien species: A threat to global biodiversity. Mar. Policy 2003, 27, 313–323. [Google Scholar] [CrossRef]
- Katsanevakis, S.; Wallentinus, I.; Zenetos, A.; Leppäkoski, E.; Çinar, M.E.; Oztürk, B.; Grabowski, M.; Golani, D.; Cardoso, A.C. Impacts of invasive alien marine species on ecosystem services and biodiversity: A pan-European review. Aquat. Invasions 2014, 9, 391–423. [Google Scholar] [CrossRef]
- Diagne, C.; Leroy, B.; Vaissière, A.-C.; Gozlan, R.E.; Roiz, D.; Jarić, I.; Salles, J.-M.; Bradshaw, C.J.A.; Courchamp, F. High and rising economic costs of biological invasions worldwide. Nature 2021, 592, 571–576. [Google Scholar] [CrossRef] [PubMed]
- EU Regulation 1143/2014; European Union Regulation (EU) No 1143/2014 of the European Parliament and the Council of 22 October 2014 on the Prevention and Management of the Introduction and Spread of Invasive Alien Species. European Commission: Brussel, Belgium, 2014; Volume 317, pp. 35–55.
- Directive 2008/56/EC; European Comission Directive of the European Parliament and the Council Establishing a Framework for Community Action in the Field of Marine Environmental Policy (Marine Strategy Framework Directive). European Parliament and of the Council: Brussel, Belgium, 2008.
- Lehtiniemi, M.; Ojaveer, H.; David, M.; Galil, B.; Gollasch, S.; McKenzie, C.; Minchin, D.; Occhipinti-Ambrogi, A.; Olenin, S.; Pederson, J. Dose of truth—Monitoring marine non-indigenous species to serve legislative requirements. Mar. Policy 2015, 54, 26–35. [Google Scholar] [CrossRef]
- Afonso, I.; Berecibar, E.; Castro, N.; Costa, J.; Frias, P.; Henriques, F.; Moreira, P.; Oliveira, P.; Silva, G.; Chainho, P. Assessment of the colonization and dispersal success of non-indigenous species introduced in recreational marinas along the estuarine gradient. Ecol. Indic. 2020, 113, 106147. [Google Scholar] [CrossRef]
- Chainho, P.; Fernandes, A.; Amorim, A.; Ávila, S.P.; Canning-Clode, J.; Castro, J.J.; Costa, A.C.; Costa, J.L.; Cruz, T.; Gollasch, S.; et al. Non-indigenous species in Portuguese coastal areas, coastal lagoons, estuaries and islands. Estuar. Coast. Shelf Sci. 2015, 167, 199–211. [Google Scholar] [CrossRef]
- Mancinelli, G.; Chainho, P.; Cilenti, L.; Falco, S.; Kapiris, K.; Katselis, G.; Ribeiro, F. The Atlantic blue crab Callinectes sapidus in southern European coastal waters: Distribution, impact and prospective invasion management strategies. Mar. Pollut. Bull. 2017, 119, 5–11. [Google Scholar] [CrossRef]
- Hopkins, G.W.; Freckleton, R.P. Declines in the numbers of amateur and professional taxonomists: Implications for conservation. Anim. Conserv. 2002, 5, 245–249. [Google Scholar] [CrossRef]
- Kim, K.C.; Byrne, L.B. Biodiversity loss and the taxonomic bottleneck: Emerging biodiversity science. Ecol. Res. 2006, 21, 794–810. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, M.E. From barcoding single individuals to metabarcoding biological communities: Towards an integrative approach to the study of global biodiversity. Trends Ecol. Evol. 2014, 29, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Hajibabaei, M. The golden age of DNA metasystematics. Trends Genet. 2012, 28, 535–537. [Google Scholar] [CrossRef] [PubMed]
- Holman, L.E.; de Bruyn, M.; Creer, S.; Carvalho, G.; Robidart, J.; Rius, M. Detection of introduced and resident marine species using environmental DNA metabarcoding of sediment and water. Sci. Rep. 2019, 9, 11559. [Google Scholar] [CrossRef]
- Pochon, X.; Zaiko, A.; Hopkins, G.A.; Banks, J.C.; Wood, S.A. Early detection of eukaryotic communities from marine biofilm using high-throughput sequencing: An assessment of different sampling devices. Biofouling 2015, 31, 241–251. [Google Scholar] [CrossRef]
- Rey, A.; Basurko, O.C.; Rodriguez-Ezpeleta, N. Considerations for metabarcoding-based port biological baseline surveys aimed at marine nonindigenous species monitoring and risk assessments. Ecol. Evol. 2020, 10, 2452–2465. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Hajibabaei, M.; Rieseberg, L.H. Environmental DNA. Mol. Ecol. 2012, 21, 1789–1793. [Google Scholar] [CrossRef]
- Duarte, S.; Vieira, P.E.; Lavrador, A.S.; Costa, F.O. Status and prospects of marine NIS detection and monitoring through (e)DNA metabarcoding. Sci. Total Environ. 2021, 751, 141729. [Google Scholar] [CrossRef]
- Knowlton, N. Sibling species in the sea. Annu. Rev. Ecol. Syst. 1993, 24, 189–216. [Google Scholar] [CrossRef]
- Radulovici, A.E.; Vieira, P.E.; Duarte, S.; Teixeira, M.A.L.; Borges, L.M.S.; Deagle, B.E.; Majaneva, S.; Redmond, N.; Schultz, J.A.; Costa, F.O. Revision and annotation of DNA barcode records for marine invertebrates: Report of the 8th iBOL conference hackathon. Metabarcoding Metagenomics 2021, 5, e67862. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Ratnasingham, S.; De Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef]
- Paz, G.; Rinkevich, B. Gap analysis of DNA barcoding in ERMS reference libraries for ascidians and cnidarians. Environ. Sci. Eur. 2021, 33, 4. [Google Scholar] [CrossRef]
- Leite, B.R.; Vieira, P.E.; Teixeira, M.A.L.; Lobo-Arteaga, J.; Hollatz, C.; Borges, L.M.S.; Duarte, S.; Troncoso, J.S.; Costa, F.O. Gap-analysis and annotated reference library for supporting macroinvertebrate metabarcoding in Atlantic Iberia. Reg. Stud. Mar. Sci. 2020, 36, 101307. [Google Scholar] [CrossRef]
- Vieira, P.E.; Lavrador, A.S.; Parente, M.I.; Parretti, P.; Costa, A.C.; Costa, F.O.; Duarte, S. Gaps in DNA sequence libraries for Macaronesian marine macroinvertebrates imply decades till completion and robust monitoring. Divers. Distrib. 2021, 27, 2003–2015. [Google Scholar] [CrossRef]
- Weigand, H.; Beermann, A.J.; Čiampor, F.; Costa, F.O.; Csabai, Z.; Duarte, S.; Geigerg, M.F.; Grabowski, M.; Rimet, F.; Rulik, B.; et al. DNA barcode reference libraries for the monitoring of aquatic biota in Europe: Gap-analysis and recommendations for future work. Sci. Total Environ. 2019, 678, 499–524. [Google Scholar] [CrossRef]
- Duarte, S.; Vieira, P.E.; Costa, F.O. Assessment of species gaps in DNA barcode libraries of nonindigenous species (NIS) occurring in European coastal regions. Metabarcoding Metagenomics 2020, 4, 35–46. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef] [PubMed]
- Rimet, F.; Aylagas, E.; Borja, A.; Bouchez, A.; Canino, A.; Chauvin, C.; Chonova, T.; Jr, F.C.; Costa, F.O.; Ferrari, B.J.D.; et al. Metadata standards and practical guidelines for specimen and DNA curation when building barcode reference libraries for aquatic life. Metabarcoding Metagenomics 2021, 5, e58056. [Google Scholar] [CrossRef]
- Leese, F.; Altermatt, F.; Bouchez, A.; Ekrem, T.; Hering, D.; Meissner, K.; Mergen, P.; Pawlowski, J.; Piggott, J.; Rimet, F.; et al. DNAqua-Net: Developing new genetic tools for bioassessment and monitoring of aquatic ecosystems in Europe. Res. Ideas Outcomes 2016, 2, e11321. [Google Scholar] [CrossRef]
- Zaiko, A.; Pochon, X.; Garcia-Vazquez, E.; Olenin, S.; Wood, S.A. Advantages and Limitations of Environmental DNA/RNA Tools for Marine Biosecurity: Management and Surveillance of Non-Indigenous Species. Front. Mar. Sci. 2018, 5. [Google Scholar] [CrossRef]
- Machida, R.J.; Leray, M.; Ho, S.-L.; Knowlton, N. Metazoan mitochondrial gene sequence reference datasets for taxonomic assignment of environmental samples. Sci. Data 2017, 4, 170027. [Google Scholar] [CrossRef]
- Neto, L.; Pinto, N.; Proença, A.; Amorim, A.; Conde-Sousa, E. 4SpecID: Reference DNA Libraries Auditing and Annotation System for Forensic Applications. Genes 2021, 12, 61. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Bucklin, A.; Peijnenburg, K.T.C.A.; Kosobokova, K.N.; O’Brien, T.D.; Blanco-Bercial, L.; Cornils, A.; Falkenhaug, T.; Hopcroft, R.R.; Hosia, A.; Laakmann, S.; et al. Toward a global reference database of COI barcodes for marine zooplankton. Mar. Biol. 2021, 168, 78. [Google Scholar] [CrossRef]
- Nugent, C.M.; Elliott, T.A.; Ratnasingham, S.; Adamowicz, S.J. Coil: An R package for cytochrome c oxidase I (COI) DNA barcode data cleaning, translation, and error evaluation. Genome 2020, 63, 291–305. [Google Scholar] [CrossRef]
- Oliveira, L.M.; Knebelsberger, T.; Landi, M.; Soares, P.; Raupach, M.J.; Costa, F.O. Assembling and auditing a comprehensive DNA barcode reference library for European marine fishes. J. Fish Biol. 2016, 89, 2741–2754. [Google Scholar] [CrossRef]
- Fontes, J.T.; Vieira, P.E.; Ekrem, T.; Soares, P.; Costa, F.O. BAGS: An automated Barcode, Audit & Grade System for DNA barcode reference libraries. Mol. Ecol. Resour. 2021, 21, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.; Dimech, M.; Arculeo, M.; Biondo, G.; Martins, R.; Carneiro, M.; Carvalho, G.R.; Brutto, S.L.; Costa, F. DNA Barcoding for Species Assignment: The Case of Mediterranean Marine Fishes. PLoS ONE 2014, 9, e106135. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.M.; Brodnicke, O.B.; Evankow, A.M.; Ferreira, A.O.; Fontes, J.T.; Hansen, A.K.; Jensen, M.R.; Kalaycı, T.E.; Leeper, A.; Patil, S.K.; et al. The Future of DNA Barcoding: Reflections from Early Career Researchers. Diversity 2021, 13, 313. [Google Scholar] [CrossRef]
- Katsanevakis, S.; Bogucarskis, K.; Gatto, F.; Vandekerkhove, J.; Deriu, I.; Cardoso, A.C. Building the European Alien Species Information Network (EASIN): A Novel Approach for the Exploration of Distributed Alien Species Data. BioInvasions Rec. 2012, 1, 235–245. [Google Scholar] [CrossRef]
- Olenin, S.; Narščius, A.; Minchin, D.; David, M.; Galil, B.; Gollasch, S.; Marchini, A.; Occhipinti-Ambrogi, A.; Ojaveer, H.; Zaiko, A. Making non-indigenous species information systems practical for management and useful for research: An aquatic perspective. Biol. Conserv. 2014, 173, 98–107. [Google Scholar] [CrossRef]
- Rius, M.; Ahyong, S.; Bieler, R.; Boudouresque, C.; Costello, M.J.; Downey, R.; Galil, B.S.; Gollasch, S.; Hutchings, P.; Kamburska, L.; et al. World Register of introduced Marine Species (WRiMS). Available online: https://www.marinespecies.org/introduced (accessed on 15 September 2021).
- Ahyong, S.; Boyko, C.B.; Bailly, N.; Bernot, J.; Bieler, R.; Brandão, S.N.; Daly, M.; De Grave, S.; Gofas, S.; Hernandez, F.; et al. World Register of Marine Species (WoRMS). Available online: https://www.marinespecies.org (accessed on 17 September 2021).
- Duarte, S.; Vieira, P.E.; Leite, B.R.; Teixeira, M.A.L.; Neto, J.M.; Costa, F.O. Comparing DNA metabarcoding with morphology in the assessment of macrozoobenthos in Portuguese transitional waters in the scope of the water framework directive monitoring. Biorxiv 2022, 1–38. [Google Scholar] [CrossRef]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef]
- Lobo, J.; Costa, P.M.; Al Teixeira, M.; Ferreira, M.S.; Costa, M.H.; O Costa, F. Enhanced primers for amplification of DNA barcodes from a broad range of marine metazoans. BMC Ecol. 2013, 13, 34. [Google Scholar] [CrossRef]
- Ratnasingham, S. MBRAVE: The Multiplex Barcode Research and Visualization Environment. Biodivers. Inf. Sci. Stand. 2019, 3, e37986. [Google Scholar] [CrossRef]
- Zouros, E. The exceptional mitochondrial DNA system of the mussel family Mytilidae. Genes Genet. Syst. 2000, 75, 313–318. [Google Scholar] [CrossRef]
- Leese, F.; Bouchez, A.; Abarenkov, K.; Altermatt, F.; Borja, A.; Bruce, K.; Ekrem, T.; Čiampor, F.; Čiamporová-Zaťovičová, Z.; Costa, F.O.; et al. Why We Need Sustainable Networks Bridging Countries, Disciplines, Cultures and Generations for Aquatic Biomonitoring 2.0: A Perspective Derived from the DNAqua-Net COST Action. Adv. Ecol. Res. 2018, 58, 63–99. [Google Scholar] [CrossRef]
- Duarte, S.; Leite, B.R.; Feio, M.J.; Costa, F.O.; Filipe, A.F. Integration of DNA-Based Approaches in Aquatic Ecological Assessment Using Benthic Macroinvertebrates. Water 2021, 13, 331. [Google Scholar] [CrossRef]
- Aylagas, E.; Borja, Á.; Rodríguez-Ezpeleta, N. Environmental Status Assessment Using DNA Metabarcoding: Towards a Genetics Based Marine Biotic Index (gAMBI). PLoS ONE 2014, 9, e90529. [Google Scholar] [CrossRef] [PubMed]
- Coleman, C.O. Taxonomy in times of the taxonomic impediment—Examples from the community of experts on amphipod crustaceans. J. Crustac. Biol. 2015, 35, 729–740. [Google Scholar] [CrossRef]
- Christodoulou, M.; van der Hoorn, B.; Van den Bulcke, L.; Derycke, S.; De Backer, A.; Kröncke, I.; Arbizu, P.M. A reliable DNA barcode reference library for the identification of benthic invertebrates: Essential for biomonitoring of the North Sea. ARPHA Conf. Abstr. 2021, 4, 10–11. [Google Scholar] [CrossRef]
- Gissi, C.; Hastings, K.E.M.; Gasparini, F.; Stach, T.; Pennati, R.; Manni, L. An unprecedented taxonomic revision of a model organism: The paradigmatic case of Ciona robusta and Ciona intestinalis. Zool. Scr. 2017, 46, 521–522. [Google Scholar] [CrossRef]
- Couton, M.; Comtet, T.; Le Cam, S.; Corre, E.; Viard, F. Metabarcoding on planktonic larval stages: An efficient approach for detecting and investigating life cycle dynamics of benthic aliens. Manag. Biol. Invasions 2019, 10, 657–689. [Google Scholar] [CrossRef]
- Viard, F.; Roby, C.; Turon, X.; Bouchemousse, S.; Bishop, J. Cryptic Diversity and Database Errors Challenge Non-Indigenous Species Surveys: An Illustration with Botrylloides spp. in the English Channel and Mediterranean Sea. Front. Mar. Sci. 2019, 6, 615. [Google Scholar] [CrossRef]
- Lacoursière-Roussel, A.; Howland, K.; Normandeau, E.; Grey, E.K.; Archambault, P.; Deiner, K.; Lodge, D.M.; Hernandez, C.; LeDuc, N.; Bernatchez, L. eDNA metabarcoding as a new surveillance approach for coastal Arctic biodiversity. Ecol. Evol. 2018, 8, 7763–7777. [Google Scholar] [CrossRef]
- Costa, F.O.; Antunes, P.M. The contribution of the barcode of life initiative to the discovery and monitoring of biodiversity. In Natural Resources, Sustainability and Humanity; Mendonca, A., Cunha, A., Chakrabarti, R., Eds.; Springer: Dordrecht, The Netherlands, 2012; ISBN 9789400713215. [Google Scholar]
- Rimet, F.; Chaumeil, P.; Keck, F.; Kermarrec, L.; Vasselon, V.; Kahlert, M.; Franc, A.; Bouchez, A. R-Syst: Diatom: An open-access and curated barcode database for diatoms and freshwater monitoring. Database 2016, 2016, baw016. [Google Scholar] [CrossRef]
- Mackie, J.A.; Darling, J.A.; Geller, J.B. Ecology of cryptic invasions: Latitudinal segregation among Watersipora (Bryozoa) species. Sci. Rep. 2012, 2, srep00871. [Google Scholar] [CrossRef] [PubMed]
- Fehlauer-Ale, K.H.; Mackie, J.A.; Lim-Fong, G.E.; Ale, E.; Pie, M.R.; Waeschenbach, A. Cryptic species in the cosmopolitan Bugula neritina complex (Bryozoa, Cheilostomata). Zool. Scr. 2014, 43, 193–205. [Google Scholar] [CrossRef]
- Lobo-Arteaga, J.; Ferreira, M.S.; Antunes, I.C.; Teixeira, M.A.L.; Borges, L.M.S.; Sousa, R.; Gomes, P.A.; Costa, M.H.; Cunha, M.R.; Costa, F. Contrasting morphological and DNA barcode-suggested species boundaries among shallow-water amphipod fauna from the southern European Atlantic coast. Genome 2017, 60, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Vieira, P.E.; Desiderato, A.; Azevedo, S.L.; Esquete, P.; Costa, F.O.; Queiroga, H. Molecular evidence for extensive discontinuity between peracarid (Crustacea) fauna of Macaronesian islands and nearby continental coasts: Over fifty candidate endemic species. Mar. Biol. 2022, 169, 64. [Google Scholar] [CrossRef]
- Teixeira, M.A.L.; E Vieira, P.; Ravara, A.; O Costa, F.; Nygren, A. From 13 to 22 in a second stroke: Revisiting the European Eumida sanguinea (Phyllodocidae: Annelida) species complex. Zool. J. Linn. Soc. 2022, 196, 169–197. [Google Scholar] [CrossRef]
- Stoeckle, M.Y.; Das Mishu, M.; Charlop-Powers, Z. Improved Environmental DNA Reference Library Detects Overlooked Marine Fishes in New Jersey, United States. Front. Mar. Sci. 2020, 7, 226. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).