Abstract
The informal group Caespitosa of Paspalum L. comprises 13–15 perennial species that are able to tolerate extreme climatic stresses, such as prolonged droughts, floods, and saltwater. Previous molecular phylogenetic studies have suggested that the Caespitosa might not be monophyletic, but they did not analyze a large enough sample of taxa for a meaningful conclusion. In this study, we evaluate the phylogeny of the genus Paspalum using parsimony, likelihood, and Bayesian inference based on four DNA regions (ETS, ndhF, rpl16, and trnH-psbA) and increasing the number of sampled species (i.e., a total of 13 taxa and 40 new accessions of the group Caespitosa). Our main objective was to analyze the positions of Caespitosa taxa, assuming a priori that they do not represent a natural group as traditionally circumscribed. Our findings showed the Caespitosa species distributed in seven morphologically distinct clades and correlated with members of the informal groups Alma, Corcovadensia, Dissecta, Lachnea, Macrophylla, Notata, Paniculata, and Rupestria. Clades containing Caespitosa taxa were characterized based on morphological, anatomical, and cytological evidence, one of which was associated with geographic isolation. A comparison with results from other studies, a brief discussion on the group Macrophylla, which our analyses showed to be polyphyletic, and comments on the need for future molecular studies in Paspalum are also included.
1. Introduction
Paspalum L. is one of the richest genera within Poaceae, comprising around 350 species [1]. Most of its species are native to the Americas, mainly distributed in tropical and subtropical regions, with a few growing in the Old World [1]. Species of Paspalum are important elements of the biodiversity of South American pasture ecosystems, and due to their forage potential, many are used for this purpose [1,2,3,4,5].
Previous authors have had differing opinions regarding the infrageneric classification of Paspalum, but as a summary, four subgenera (Paspalum, Anachyris Chase, Ceresia (Pers.) Rchb., and Harpostachys (Trin.) S. Denham) and 27 sections have been recognized [6,7,8,9,10,11,12,13,14,15,16,17,18,19,20]. In addition to these, about 30 groups without a formal taxonomic level were proposed within subgenus Paspalum, mainly based on morphological characters of inflorescences and spikelets, due to the difficulty of establishing clear limits between sections [8,9]. These informal groups, most of them without phylogenetic support [3,21], were/are used in taxonomic, cytological, and phylogenetic studies (e.g., [3,21,22,23,24,25,26,27,28,29]), as the approaches of Refs. [8,9] remain the best options to providing better knowledge of Paspalum in the absence of a fully-resolved phylogeny of the genus.
Caespitosa is one of these informal groups, and its species are distributed from the southern United States to northeastern Argentina, with six concentrated in the Caribbean and Mexico [1,8,9,30,31]. They are outstanding for their ability to tolerate extreme climatic stresses, such as prolonged droughts, floods, and saltwater [28]. When establishing the group (as Paspalum [unranked] Caespitosa), ref. [32] included a total of four species (i.e., Paspalum caespitosum Flüggé, Paspalum glabrum Poir., Paspalum helleri Nash, and Paspalum mandiocanum Trin.), characterized by the caespitose habit and paired ellipsoid spikelets glabrous or appressed-pubescent. Ref. [8], in a revision of the North American species of Paspalum, synonymized P. glabrum and P. helleri under Paspalum laxum Lam. and recognized eight species within the group Caespitosa characterized by the mostly caespitose habit, culms simple or occasionally with a single branch, inflorescences pauci to pluriracemose, and spikelets often ellipsoid. She expanded this concept in her unpublished monograph for the South American species of the genus [9], treated P. mandiocanum in the group Corcovadensia, and redefined Caespitosa to include 14 species and two varieties. Refs. [1,28] included Paspalum chacoense Parodi and Paspalum redondense Swallen within Caespitosa, bringing the number of species recognized in the group to 15. In a complete and most recent revision of the group Caespitosa, ref. [31] recognized 13 species and three varieties (i.e., Paspalum acutifolium León, Paspalum albidulum Henrard, Paspalum bakeri Hack., Paspalum blodgettii Chapm., P. caespitosum, P. chacoense, Paspalum divergens Döll, Paspalum galapageium var. galapageium Chase, Paspalum galapageium var. minoratum Chase, Paspalum galapageium var. redundans (Chase) Delfini & Zuloaga, Paspalum indecorum Mez, P. laxum, Paspalum ligulare Nees, Paspalum molle Poir., and P. redondense), characterized by including perennial plants, usually rhizomatous, with culms simple, erect, inflorescences terminal, pauci-racemose, and spikelets mostly ellipsoid to ovoid or obovoid [31].
Molecular phylogenetic studies have suggested that the group Caespitosa might not be monophyletic [4,21], but they have not included a large enough sample of species to test this rigorously (i.e., only four species were analyzed in Refs. [4,21]). Based on these previous results, our objective was to evaluate the phylogeny of the genus Paspalum by increasing the number of Caespitosa species sampled to test their positions, assuming a priori that they do not represent a natural group. To accomplish these goals, DNA sequence data from three plastid regions (ndhF, rpl16 and trnH-psbA) and one nuclear region (ETS) were used to unravel the phylogenetic relationships. We analyzed a total of 13 Caespitosa species considered to belong to this group, those treated in the taxonomic revision of ref. [31]. The relationships of Caespitosa taxa were herein discussed and related to morphological, anatomical, and ecological evidence.
2. Materials and Methods
2.1. Taxon Sampling
The DNA data matrix used in the phylogenetic analyses includes a total of 128 accessions, of which 119 are ingroup and 17 are Caespitosa [see Table S1 available in Supplementary Material here]. In total, we added to the analyses 40 new accessions of the group Caespitosa and sampled 13 taxa (i.e., Paspalum acutifolium, P. bakeri, P. blodgettii, P. caespitosum, P. chacoense, P. galapageium, P. indecorum, P. laxum, P. ligulare, P. molle, P. pleostachyum Döll, P. redondense, and P. redundans; Table S1), considering species accepted and synonyms in the taxonomic treatment in ref. [31]. The nuclear ribosomal DNA ETS matrix and chloroplast DNA (cpDNA) ndhF and rpl16 matrices previously published in ref. [21] were completed with sequences of new taxa added (13, 18, and 12, respectively). We also built a new cpDNA matrix for this study [trnH-psbA (coding region, spacer, IR-B/LSC junction), encompassing the rps19 gene [33,34] including 38 accessions, totaling 81 new sequences added. In addition, nine species belonging to eight closely related genera were selected as outgroups, based on [21,35,36,37]: Anthaenantia P. Beauv., Axonopus P. Beauv., Hildaea C. Silva & R.P. Oliveira, Hymenachne P. Beauv., Ichnanthus P. Beauv., Ocellochloa Zuloaga & Morrone, Plagiantha Renvoize, and Streptostachys Desv. Information about vouchers and accession numbers of the new sequences obtained for this study and those available in GenBank is given in Table S1.
2.2. DNA Amplification and Sequencing
Total genomic DNA was extracted from herbarium material using modified CTAB protocols from ref. [38]. For the species that failed this protocol, the DNA was isolated using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s recommendations. Each species was amplified from a single voucher specimen, but a second voucher was also included for some taxa. The four DNA regions were amplified by polymerase chain reaction (PCR) and sequenced for each taxon. The primers 18S and 26S of ref. [39] were used to amplify and sequence the external transcribed spacer (ETS); the trnH-psbA region was amplified using the primers psbA and trnHGUG proposed by ref. [40]; the rpl16 region, corresponding to the intron and partial sequences of the gene encoding ribosomal protein L16 [41,42,43], was amplified in two fragments using primers F71 [44] and R1661 [41], combined with the internal ones F584 and R584 [45]; the complete ndhF gene, coding NADH dehydrogenase subunit F, was amplified with a battery of primers in different combinations in four overlapping fragments using primer pairs specified by Refs. [46,47]: 5F–536R, 536F–972R, 972F–1666R, and 1666F–3R.
PCR reactions were performed in a 25 µL final volume with 50–100 ng of template DNA, 2.5 µL PCR buffer (10×), 1.5 µL MgCl2 (25 mM), 1 µL dNTP (10 mM), 0.5 µL of each primer (10 pM), and 0.3 µL of Taq polymerase (5 u/µL) provided by Promega (Madison, Wisconsin, U.S.A.). Variations in dNTP (1–1.25 µL), primers (0.5–1 µL), and total DNA dilutions (1:5, 1:10 and 1:50) were used. The reactions were carried out using the following parameters: (1) for the nuclear ETS: one cycle of 94 °C for 5 min, 39 cycles of 94 °C for 30 s, 52 °C for 1 min, and 72 °C for 1 min, and a final extension cycle of 72 °C for 10 min; and (2) for the chloroplast regions of ndhF, rpl16 and trnH-psbA: 95 °C for 2 min, 39 cycles of 95 °C for 30 s, 48 °C for 30 s, 72 °C for 1.5 min, and a final extension cycle of 72 °C for 10 min. PCR products were run out on a 1% TBE (Tris-Borate-EDTA) agarose gel stained with SYBR Safe DNA gel stain (Invitrogen Life Technologies) and visualized in a blue-light transilluminator. Automated sequencing was performed by Macrogen, Inc. (Seoul, South Korea). Forward and reverse strands were sequenced for all fragments, with a minimum overlap of 80%.
2.3. Phylogenetic Analyses
Sequence editing and assembly were performed with MEGA v. 7.0 [48]. The accuracy of the sequences was assessed through the visual inspection of the chromatograms. Alignments were generated with Clustal X v. 2 [49] under the default settings and were trimmed to remove part of the 3′ end, for which many sequences were incomplete. The alignments obtained were then checked and improved manually, when necessary, by visual refinement using the program MEGA v. 7.0 [48]. The phylogenetic reconstruction was based on parsimony (MP) [50], maximum likelihood (ML) [51,52], and Bayesian inference (BI) [53] methods. In all analyses, gaps were considered missing data. The best-fitting nucleotide substitution models for each region were selected by the Akaike information criterion (AIC), as implemented in jModeltest 2.1.1 [54]: SYM+I+G (ETS), TIM1+G (ndhF), TVM+I+G (rpl16), and F81+I+G (trnH-psbA).
Molecular analyses of nuclear ETS and three cpDNA regions were performed separately and combined. Two combined matrices were constructed: the “plastid matrix”, combining the three plastid regions (ndhF, rpl16 and trnH-psbA), and the “total dataset matrix,” combining the plastid and the nuclear ETS matrices. Separate analyses are useful to investigate the possibility of reticulation or hybridization events, while combined analyses better maximize cladistic parsimony, allowing secondary signals to emerge and produce the best-supported hypotheses [55]. Congruence between the combined nuclear and plastid datasets was assessed using the partition homogeneity (incongruence length difference [ILD]) test [56] implemented in PAUP* v. 4.0b10 [57], with 1000 replicates, MaxTrees set at 1000, and TBR branch swapping.
Separate and combined parsimony analyses were performed using TNT ver. 1.1 [58] with Fitch parsimony [50] as the optimality criterion. All characters were equally weighted and treated as unordered. A heuristic search was conducted using 1000 random taxon-addition replicates with the tree-bisection-reconnection (TBR) algorithm, saving up to 15 trees per replicate to prevent extensive swapping on islands with many trees. The resulting trees were then used as starting trees for a second-round search using TBR branch swapping with an upper limit of 10,000 trees. Nonparametric bootstrap support (BS) was estimated using 10,000 pseudo-replicates, and the same parameters were used in our MP analyses [59]. Bootstrap percentages of 50 to 80 were considered weak, 81 to 90 moderate, and >90 strong.
ML analyses were conducted using RAxML-HPC2 on XSEDE (v. 8.2.12) [60] in the Cyberinfrastructure for Phylogenetic Research (CIPRES) Portal v. 3.3 [61]. For these analyses, we used the implemented algorithm, which allows one to perform optimal tree searches and obtain bootstrap support [59] in one single analysis [62]. To this end, we performed 1000 bootstrap replicates with a subsequent search of the maximum likelihood tree, using the GTRGAMMA nucleotide substitution model [60], individual per-site substitution rates (-c), and the default setting of likelihood acceptance (-e), 25 and 0.1, respectively. Bootstrap percentages of 50 to 80 were considered weak, 81 to 90 moderate, and >90 strong.
Individual and combined BI analyses were performed using MrBayes v. 3.2.7a [63] in the CIPRES Portal [61] with nst = 6 and rates = invgamma (ETS and rpl16), nst = 6 and rates = gamma (ndhF), and nst = 1 and rates = invgamma (trnH-psbA), unlinking models across loci for combined analyses. The datasets were analyzed in two independent runs of 10 million generations, each with four Markov chains (one cold chain and three heated chains), sampling every 1000 generations. The convergence and effective sample size (ESS) of the runs were assessed in Tracer v.1.7 [64], checking that ESS > 200 for all parameters. After discarding the initial 2500 trees of each run as burn-in (25%), the remaining trees (15,002) were used to generate a 50% majority-rule consensus tree. The cutoff for strong support in the Bayesian analyses was 0.95 (roughly equal to p < 0.05) posterior probabilities, and values below 0.8 were considered not supported.
3. Results
General features and descriptive statistics of the DNA datasets used in MP analyses are given in Table S2 [available in Supplementary Material here]. Nuclear ETS and plastid ndhF had the highest sequencing success of the Caespitosa taxa recovered in the datasets, followed by trnH-psbA and rpl16; however, all four regions failed to amplify for P. albidulum and P. divergens. The nuclear ETS region was the most variable marker, with 54.07% phylogenetically informative sites, at least more than four times as variable as the plastid regions, which had 12.83% (rpl16), 11.25% (ndhF), and 2.13% (trnH-psbA) phylogenetically informative sites. The ILD test of the plastid and total datasets combined did not indicate incongruence between partitions (ndhF + rpl16 + trnH-psbA, ILD: P = 0.75; ETS + ndhF + rpl16 + trnH-psbA, ILD: P = 0.32). Strict consensus tree from MP, 50% majority-rule consensus tree from BI, and ML tree recovered similar topologies showing the same strongly supported clades; therefore, only the BI tree of the combined four-region DNA dataset is presented here (Figure 1) along with branch support obtained under MP and ML analyses for clades that include Caespitosa taxa. All aligned data matrices and trees of separate and combined datasets from the three methods of analysis are available in the Repositorio Institucional CONICET Digital under the following link: http://hdl.handle.net/11336/179173, accessed on 23 December 2022.
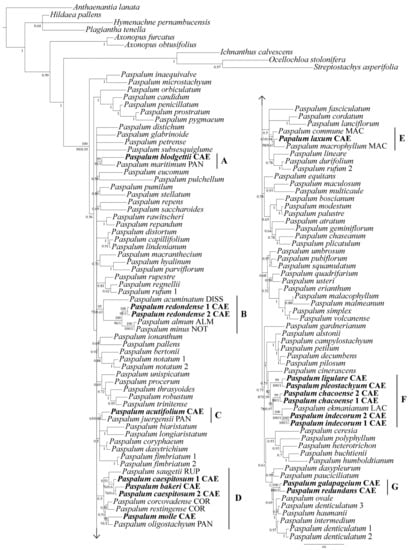
Figure 1.
50% majority-rule consensus tree from the Bayesian inference analysis of the total combined dataset (ETS + ndhF + rpl16 + trnH-psbA). Posterior probabilities from BI are listed below the branches and bootstrap supports from ML (below the branches) and from MP (above the branches) are also indicated for clades that contain Caespitosa taxa. Nodes with “–” have bootstrap supports <50. Clades (A–G) correspond to the Caespitosa taxa discussed in the text. Group abbreviations: ALM = Alma, CAE = Caespitosa, COR = Corcovadensia, DISS = Dissecta, LAC = Lachnea, MAC = Macrophylla, NOT = Notata, PAN = Paniculata, RUP = Rupestria.
For four Caespitosa taxa, we were able to include two accessions (i.e., Paspalum caespitosum, P. chacoense, P. indecorum, and P. redondense) [see Table S1 available in Supplementary Material here]. In most cases, the two accessions of the same species had identical or nearly identical sequences and were placed together by the three analyses. In the case of P. caespitosum, mutations in the sequences led to distinct placements, but the accessions still formed a clade with P. bakeri (Figure 1), although in parsimony these relationships are not clearly defined. With a single exception of clade D, the remaining clades that contain Caespitosa taxa received some support in the three analyses.
The topology of the four-region DNA dataset tree (Figure 1) grouped Paspalum species in a strongly supported clade (Bayesian posterior probability (BPP) 0.95/ML bootstrap (MLB) 99/parsimony bootstrap (PB) 100). Although the relationships among clades remain largely unresolved, the three combined analyses placed the 13 Caespitosa taxa in seven clades (Figure 1A–G) related to members of the informal groups Alma, Corcovadensia, Dissecta, Lachnea, Macrophylla, Notata, Paniculata, and Rupestria, as detailed next.
Clade A is a strongly supported clade (BPP 1/MLB 91/PB 99), including P. blodgettii (group Caespitosa) as sister to Paspalum maritimum Trin. (group Paniculata), two morphologically quite distinct species.
Clade B, strongly (MP) supported (BPP 0.63/MLB 75/PB 95), consists of members of groups Alma (Paspalum almum Chase), Dissecta (Paspalum acuminatum Raddi), Notata (Paspalum minus E. Fourn.), and P. redondense (group Caespitosa). Paspalum minus is sister to P. almum (BPP 1/MLB 100/PB 100), and both are related to P. redondense (BPP 1/MLB 96/PB 100); Paspalum acuminatum was resolved as the sister group to all clade B species with strong branch support in the MP analysis. Due to the ploidy variation within P. redondense [see Table S3 available in Supplementary Material here], we included two accessions for this species, and both were placed together (BPP 1/MLB 95/PB 100).
Within clade C, P. acutifolium (group Caespitosa) was resolved as closely related to Paspalum juergensii Hack. (group Paniculata) with moderate branch support in the BI analysis (BPP 0.84/MLB 65/PB < 50).
Paspalum caespitosum is a member of clade D, a clade with almost no support in BI (BPP 0.50) and not recovered in ML and MP. Clade D is divided into two subclades: the first one includes two accessions of P. caespitosum related to P. bakeri (group Caespitosa) (BPP 0.60/MLB 76/PB < 50), and Paspalum saugetii Chase (group Rupestria) as its sister group (BPP 0.60); within the second subclade, P. molle (group Caespitosa) is sister to Paspalum oligostachyum Salzm. ex Steud. (group Paniculata) (BPP 1/MLB 91/PB 98), and both were resolved closely related to Paspalum corcovadense Raddi (group Corcovadensia) and Paspalum restingense Renvoize (group Corcovadensia), although without support (BPP 0.50).
Clade E is strongly (BI) supported (BPP 0.94/MLB 63/PB < 50) and includes two members of the group Macrophylla (Paspalum commune Lillo and Paspalum macrophyllum Trin.), plus P. laxum (group Caespitosa). Paspalum laxum is sister to P. macrophyllum (BPP 0.63/MLB 58/PB < 50) and both were resolved as sister clades to P. commune.
Within clade F, strongly (BI) supported (BPP 0.99/MLB 87/PB 82), the two accessions of P. indecorum (group Caespitosa) (BPP 1/MLB 100/PB 100) form a strongly supported clade with Paspalum ekmanianum Henrard (group Lachnea) (BPP 1/MLB 100/PB 100) and the two accessions of P. chacoense (group Caespitosa) (BPP 1/MLB 88/PB 99) as its sister group. Paspalum ligulare (group Caespitosa) is related to P. pleostachyum (group Caespitosa) (BPP 1/MLB 100/PB 99) and was resolved as a sister group to all other clade F species.
Clade G groups the two Caespitosa species restricted to the Galapagos archipelago (i.e., Paspalum galapageium and P. redundans) in a strongly supported clade by the three analyses (BPP 1/MLB 100/PB 100).
4. Discussion
4.1. Major Results and Comparison with Previous Molecular Studies
Relationships among species were similar to the total evidence tree from MP analyses presented in ref. [21] with differences in the compositions of some previously proposed clades (e.g., Paniculata and Macrophylla clades) or placements of some species (e.g., Paspalum regnellii Mez nested with one accession of Paspalum rufum Nees ex Steud. and P. juergensii as sister to P. acutifolium), possibly due to our larger sampling of species and accessions.
The polyphyly of the group Caespitosa was also indicated by previous molecular phylogenetic analyses [4,21], but these studies included too few Caespitosa accessions for a meaningful conclusion. Ref. [4] considered as Caespitosa P. chacoense, P. indecorum, and Paspalum trichostomum Hack.; however, the latter species belongs to the Barbinodia group [9], and therefore, it has not been analyzed here. Ref. [21] analyzed P. caespitosum, P. chacoense, P. indecorum, and P. redondense, considering the latter as an ungrouped species since it was described after the classification from Refs. [8,9]. As we follow ref. [31], P. redondense was considered here as a member of the group Caespitosa.
Data presented here greatly increased the sample of Caespitosa species analyzed and confirmed that this informal group as traditionally circumscribed (sensu [8,9,31]) is not monophyletic. An analysis of the different clades and relationships among species is discussed next.
4.2. Relationships of Caespitosa Taxa
Our phylogeny showed the Caespitosa species distributed in seven morphologically distinct clades (Figure 1A–G). As most previously infrageneric categories recognized in Paspalum (i.e., subgenera, sections, and informal groups) failed to define monophyletic groups [6,7,8,9,10,11,12,13,14,15,16,17,18,19,22,23,24,25], here we presented a limited discussion about relationships of Caespitosa taxa, since the current tree is not strongly supported enough to warrant taxonomic revision of the recovered clades.
Clade A, strongly supported by the three analyses, is newly identified in this study and links P. blodgettii with P. maritimum (Figure 1). These species are distinct in general aspect, ploidy level (Table S3; [65]), and geographic distribution but share papillose-scabrous spikelets and the habitat (Figure 2A). Paspalum blodgettii is a caespitose plant that lacks conspicuous rhizomes and is distributed from the southern United States, Mexico, and the Caribbean islands to Panama [31], while P. maritimum is a long-rhizomatous [66] and exclusively polyploid species [65], more widely distributed (i.e., in Brazil, the Caribbean islands, Colombia, Guyanas, Paraguay, Suriname, and Venezuela [30]). Despite their divergent distributions, both species are frequently found in sandy and humid soils along the seacoast [31,67]. In P. maritimum, the basal cell of microhairs is sunken into the epidermis (Figure 3A–B) and probably functions as salt glands because, when in contact with the mesophyll cells, it acts as a collecting cell and the upper cell as an excreting one [67,68]. There are no similar ecological anatomical data for P. blodgettii that confirm the presence of such secretory structures; however, as both species tolerate a level of salinity, they can be defined as halophytes. Ref. [66] also highlighted the morphological affinity of P. maritimum with P. pleostachyum, but our analyses suggest that they are not related; the latter species is placed in clade F as a sister species to P. ligulare (Figure 1).
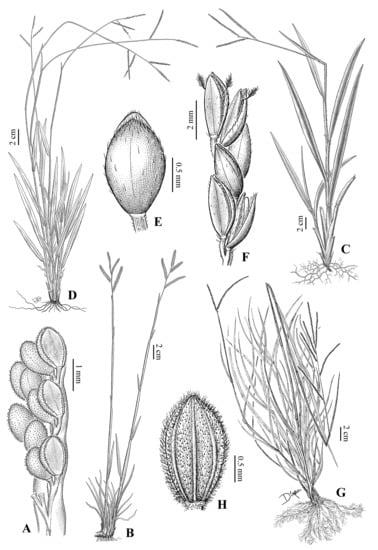
Figure 2.
Morphological variation in species of the Caespitosa group: (A) Paspalum blodgettii, raceme detail; (B) Paspalum redondense, habit; (C) Paspalum acutifolium, habit; (D,E) Paspalum caespitosum: (D) Habit; (E) Spikelet, upper glume view; (F) Paspalum molle, raceme detail; (G,H) Paspalum laxum: (G) Habit; (H) Spikelet, upper glume view [(A): N.L. Britton et al. 15790 (NY); (B): G. Hatschbach & A. Manosso 50346 (NY); (C): Bro. León & Father M. Roca 8164 (US); (D,E): H. Pittier 258 (US); (F): O. Morrone et al. 4730 (SI); (G,H): Fre. León 12305 (US)].
As presented by ref. [21], P. acuminatum, P. almum, P. minus, and P. redondense formed a clade (clade B), but our analyses support that the relationships are stronger than those retrieved previously. Clade B groups species of wet/aquatic habitats with rhizomes or stolons ([1]; Figure 2B). Paspalum acuminatum, the only species with rachis foliaceous and spongy tissue in the medulla [1,69], is confirmed by the three analyses as sister to all other clade B taxa, whereas in a previous study its position was uncertain. The spongy tissue in the rachis of P. acuminatum (Figure 4, type II) is a characteristic of species that live in aquatic or humid places; however, not all hydrophytic taxa of the genus have the same structures [69], which is the case for P. almum and P. minus. These last two species have the most common anatomical type of rachis within the genus (i.e., a triangular solid keel without spongy tissue; Figure 4, type I) [69].
There are no similar anatomical data available for P. redondense indicating the type of inflorescences; nevertheless, as this species also lacks the spongy pith, it will probably share the same rachis type as P. almum and P. minus. Paspalum redondense is morphologically similar to P. indecorum, mainly in terms of general aspect [1], but a close relationship between them was not supported by our findings (Figure 1).
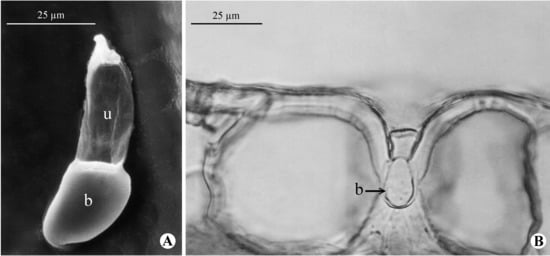
Figure 3.
Paspalum maritimum: (A) Detail of a michrohair in adaxial surface of the leaf blade view with scanning electron micrographs (b = basal cell; u = upper cell); (B) Light micrograph of leaf blade in cross section showing a microhair with the basal cell (b) sunken into the epidermis [(A): A. Chase 9761 (MO); (B): J.R. Swallen 4668 (US)]. Extracted from [70].
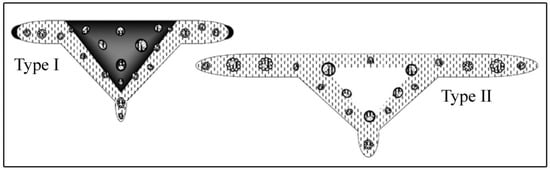
Figure 4.
Different anatomical types of rachises recognized in Paspalum: Basal type with a solid medulla (type I) and with spongy medulla (type II). Adapted from [69].
Paspalum acutifolium, here analyzed for the first time, was resolved in clade C and is closely related to P. juergensii (Figure 1). These species do not have a sympatric distribution, but they share the habitat (i.e., shady and humid places) and are similar in its general aspects (Figure 2C). Paspalum acutifolium is endemic to the Caribbean, characterized by the caespitose habit lacks conspicuous rhizomes, blades papillose-hirsute, ligules ovoid, and inflorescences with 2–4 racemes [31], while P. juerguensii is restricted to South America (i.e., Colombia to Ecuador, Bolivia to Brazil and northeast Argentina) [1,71] and has rhizomes and arched cataphylls, blades sparsely pilose, ligules laciniate, and inflorescences with 4–14 racemes [1]. In ref. [21]’s analyses, P. juergensii formed a moderately supported monophyletic group with Paspalum paniculatum L., Paspalum umbrosum Trin., and Paspalum squamulatum E. Fourn., other species of the Paniculata group, but our findings did not support these relationships. Ref. [31] also noted morphological similarities between P. acutifolium and P. galapageium; however, these two species are not phylogenetically related.
Species of clade D are morphologically quite distinct, although they share lanceolate to linear-lanceolate blades. Within this clade, the relationships remain unresolved or have low support (Figure 1). Paspalum saugetii and P. restingense have conflictive and, up to now, unresolved positions, as they were placed together (MLB 79/PB 97) but excluded from clade D by ML and MP approaches. All analyses indicate a close relationship between the two accessions of P. caespitosum and P. bakeri, but in the MP, they were grouped in a polytomy. Paspalum caespitosum 2 is a sequence added in this analysis to previously published sequences; it is nested with P. bakeri and differs from the published P. caespitosum sequence by five mutations in the ndhF (i.e., a T-to-C transition at position 82, a T-to-A transition at positions 84 and 493, and aT-to-G transition at positions 97 and 270). Whether the difference between the two sequences of P. caespitosum represents real biological variation, considering that it is a highly polymorphic taxon, or errors in sequencing is unclear, but in either case, a close relationship between P. caespitosum and P. bakeri is here suggested. On the other hand, a close relationship between P. caespitosum, P. mole, and P. corcovadense is not a surprising result since these three species share a distinctive combination of character states (e.g., general aspect, sheaths usually glabrous, blades lanceolate, lower glume absent, spikelets ellipsoid, delicately pubescent, with 3–5 nerves), and may grow sympatrically in the Caribbean ([1,31,66]; Figure 2D–F). The Bayesian analysis found weak support for a sister relationship between the two subclades within clade D, although neither ML nor MP retrieved this grouping. Paspalum molle and P. oligostachyum are distinct but morphologically similar, and they were unambiguously strongly supported as sisters in all combined analyses. Morphological similarities have also been noted between P. restingense and P. corcovadense, especially in terms of their general aspect [5]; however, the weak support in the BI suggests that additional analyses should be undertaken with more variable markers to test this rigorously.
Paspalum laxum, here analyzed for the first time, is placed in clade E together with P. macrophyllum and P. commune (Figure 1), three species that are exclusively polyploid (Table S3; [72,73]). They share blades linear-lanceolate and panicles truncate with 3–15 racemes ([1,9,31]; Figure 2G), being P. laxum sometimes confused with P. commune [1]. The latter species differs from P. laxum mainly by the size and indumentum of the spikelets (i.e., spikelets 1.3–2 mm long, papillose-hirsute in P. laxum (Figure 2H) vs. spikelets 2.2–2.7 mm long, shortly pubescent in P. commune) and by the geographic distribution: Paspalum laxum is found from southern Florida, Belize, Malpelo Island, the Caribbean to French Guiana, and Colombia, while P. commune is restricted to Bolivia and northwest Argentina [1,31]. Previous molecular analyses recovered the group Macrophylla sensu ref. [9] in a strongly supported clade composed of P. macrophyllum, P. commune, and Paspalum regnellii Mez [21,45]. However, by including P. laxum in clade E, and P. regnellii related to Paspalum rufum Nees (groups Eriantha/Virgata; [1]) and Paspalum rupestre Trin. (group Rupestria), outside the clade E, our analyses showed the group Macrophylla polyphyletic, in disagreement with previous results. Morphological similarities between P. laxum and P. blodgettii were noted by ref. [31]; these species share the caespitose habit, axillary inflorescences, and spikelets papillose-scabrous [31] but they are not phylogenetically related. Paspalum blodgettii is placed in clade A, as sister to P. maritimum (Figure 1).
Within clade F, P. chacoense, P. indecorum, and P. ekmanianum were recovered as a robust group, as previously suggested [21]. These three species are commonly found in sandy, rocky, or humid soils of open areas, being characterized by the presence of basal thick and arched rhizomes ([1,31]; Figure 5A). Paspalum chacoense is easily separated from P. oligostachyum and P. indecorum by its typical 1-nerved upper glume (Figure 5B), while P. indecorum and P. oligostachyum have the upper glume 5-nerved and 5–7-nerved, respectively [31]. In P. indecorum, [67] it was observed that the abaxial epidermal cells have a papillose outer tangential wall (Figure 6A,B), which is characteristic of grasses from dry [74] or saline areas [75].
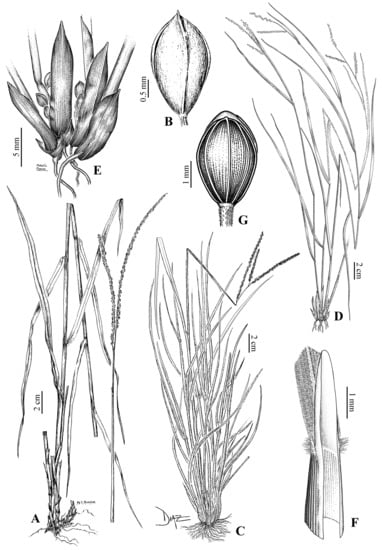
Figure 5.
Morphological variation in species of the Caespitosa group: (A,B) Paspalum chacoense: (A). Habit; (B) Spikelet, upper glume view; (C) Paspalum ligulare, habit; (D–F) Paspalum galapageium: (D) Habit; (E) Base of the culms and cleistogamous spikelets detail; (F) Ligule detail; (G) Paspalum redundans, spikelet, upper glume view [(A,B): A. Burkart 20258 (SI); (C): F.O. Zuloaga 4758 (SI); (D–F): J.T. Howell 10019 (US); (G): J.T. Howell 9902 (SI)].
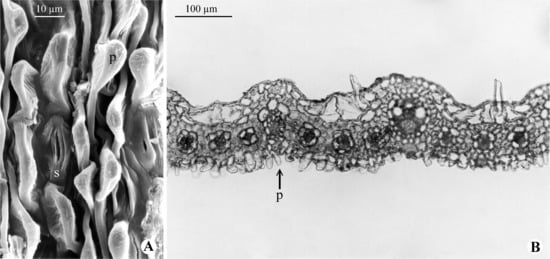
Figure 6.
Paspalum indecorum: (A) Scanning electron micrograph of the abaxial surface of leaf blade (s = stomata; p = papilla); (B) Light micrograph of leaf blade in cross section showing abaxial papillae (p) [(A): Montes 12739 (US); (B): F.O. Zuloaga 506 (SI)]. Extracted from [70].
Despite this, within Paspalum, the presence of papillae does not necessarily represent a direct adaptation to dry environments, as they are found in species of varied habitat [67]. For P. indecorum and P. chacoense, there is no register of polyploids [Table S3], and as might be expected, its two accessions placed together confirmed their monophyly.
The second robust group within clade F (i.e., Paspalum ligulare and P. pleostachyum), newly identified in this study, is quite morphologically distinct from the first one. It includes highly plastic species in relation to their habitats, characterized by having a caespitose habit without conspicuous rhizomes and forming dense populations ([28,31]; Figure 5C).
Paspalum ligulare and P. pleostachyum are in a morphologically intricate species complex, with chromosome counts ranging from 2n = 20 to 2n = 40 [Table S3]. Due to the minor morphological differences in size of ligules and spikelets, as well as the overlap in geographic distribution, they have been a cause of controversy in different taxonomic treatments. Paspalum pleostachyum was treated either as a related but distinct species of P. ligulare [9,30,66] or, more recently, as its synonym [31]. Although additional analyses should be undertaken including multiple accessions of both taxa, our findings support a very close relationship between P. ligulare and P. pleostachyum and thus, ref. [31]’s proposal based on morphological data.
Clade G, newly identified in this study, includes P. galapageium and P. redundans, two species endemic to the Galapagos Archipelago, characterized by the linear-lanceolate blades, ligules long ovate, and lower glume and cleistogamous spikelets usually present ([9,31]; Figure 5D–G). While recognizing the close morphological similarity between these taxa, A. Chase in ref. [76] treated both as valid species and proposed two varieties for P. galapageium (i.e., Paspalum galapageium var. galapageium, P. galapageium var. minoratum Chase) based on the size and number of spikelets nerves. Because the two species have only minor morphological differences and overlap in geographic distribution, ref. [31] recognized P. redundans as a new variety for P. galapageium. This is the most supported result and corroborates ref. [31]’s proposal to consider them as varieties of the same species; their close relationship also suggests that speciation in clade G is associated with geographic isolation.
4.3. Needs for Future Molecular Studies in Paspalum
This article added missing information on the relationships of the Caespitosa group, hitherto unknown. Although we have confirmed that the Caespitosa is not a natural group, as are most informal groups within subgenus Paspalum, we were able to retrieve new clades, provide stronger support for the relationships previously proposed (e.g., clade B), and characterize them based on morphological, anatomical, and ecological evidence. By adding new accessions to the analyses, our findings also showed the group Macrophylla to be polyphyletic, which had been recovered as a strongly supported clade in previous analyses (i.e., [21,45]). A stable and workable infrageneric treatment for Paspalum is needed to facilitate its species identification and to enable the new ones to be classified, as well as for further revisionary studies to have long-term relevance and utility. The nuclear ribosomal ETS and the plastid markers ndhF, rpl16, and trnH-psbA used in our analyzes, as well as the rbcL, rpoA, trnG, and trnL-F [3,21,45], seem to be inefficient in recovering the complex relationships within the genus. Therefore, future molecular phylogenies will need to use markers that allow capturing more mutations, such as other chloroplast protein-coding genes or whole plastomes [77,78,79] or low-copy nuclear genes (LCNGs) [35,80], or from Hyb Seq-type massive sequencing analysis. As it is well known that the chloroplast genome cannot recover reticulations caused by allopolyploids and that plastome phylogenies give an incomplete picture of the history of any group with hybridization [81,82], the results should be confirmed by studies that include LCNGs, which hold great potential to improve the robustness of phylogenetic trees [83] and, therefore, may be a key to providing information on events of reticulate evolution, species boundaries, as well as helping to develop a more satisfactory infrageneric resolution for Paspalum.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15020134/s1, Table S1: Taxa, voucher information, infrageneric category, and GenBank accession numbers for ETS, ndhF, rpl16, and trnH-psbA, respectively. Sequences obtained for this study are indicated with an asterisk (*) and regions with no sequence data are indicated with a dash (–); herbarium acronyms follow [84]; Table S2: Descriptive statistics for each separate data partition and combined matrices used in parsimony analyses; Table S3: Chromosome numbers known in taxa of the group Caespitosa of Paspalum L. [85,86,87,88,89].
Author Contributions
Conceptualization, C.D. and F.O.Z.; Data curation, C.D.; Formal analysis, C.D.; Funding acquisition, F.O.Z.; Investigation, C.D., S.S.A. and F.O.Z.; Methodology, C.D., J.M.A. and S.S.A.; Resources, F.O.Z.; Supervision, V.C.S. and F.O.Z.; Visualization, S.S.A.; Writing—original draft, C.D. and F.O.Z.; Writing—review & editing, C.D., J.M.A., S.S.A., V.C.S. and F.O.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Agencia Nacional de Promoción Científica y Tecnológica, Argentina (grants PICT2016-2418 and PICT-2020-SERIEA-03097) and by the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET grant PID0782).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
All aligned data matrices and trees of separate and combined datasets from the three methods of analysis are available at the Repositorio Institucional CONICET Digital under the following link: http://hdl.handle.net/11336/179173, accessed on 23 December 2022.
Acknowledgments
The authors thank to Francisco Rojas, Marcelo A. Díaz, Marcelo A. Moreno, and María A. Marino, for preparing illustrations, and the editors and anonymous reviewers for their suggestions and comments on drafts of this article.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Zuloaga, F.O.; Morrone, O. Revisión de las especies de Paspalum para América del Sur Austral (Argentina, Bolivia, sur de Brasil, Chile, Paraguay y Uruguay). Monogr. Syst. Bot. Mo. Bot. Gard. 2005, 102, 304. [Google Scholar] [CrossRef]
- Bennett, H.W. Dallisgrass, bahiagrass, and vaseygrass. In Forages: The Science of Grassland Agriculture; Hudges, H.D., Heath, M.E., Metcalfe, D.S., Eds.; The Iowa State University Press: Ames, IA, USA, 1962; pp. 281–285. [Google Scholar]
- Filguerias, T.S. Gramíneas forrageiras nativas do Distrito Federal, Brasil. Pesqui. Agropecu. Bras. 1992, 27, 1103–1111. [Google Scholar]
- Rua, G.H.; Arakaki, P.R.; Speranza, M.; Vaio, M. A phylogenetic analysis of the genus Paspalum (Poaceae) based on cpDNA and morphology. Plant Syst. Evol. 2010, 288, 227–243. [Google Scholar] [CrossRef]
- BFG (The Brazil Flora Group). Brazilian Flora 2020: Leveraging the power of a collaborative scientific network. Taxon 2021, 71, 178–198. [Google Scholar] [CrossRef]
- Nees von Esenbeck, C.G.D. III. Paspalus. In Flora Brasiliensis Seu Enumeratio Plantarum; Martius, C.F.P., Ed.; Sumptibus JG Cottage: Tuebingen, Germany, 1829; Volume 2, pp. 18–83. [Google Scholar]
- Döll, J.C. Gramineae. In Flora Brasiliensis; Martius, C.F.P., Eichler, A.W., Eds.; Frid. Fleischer: Germany, 1877; Volume 2, pp. 1–358. [Google Scholar]
- Chase, A. The North American species of Paspalum. Contr. U.S. Natl. Herb. 1929, 28, 1–310. [Google Scholar]
- Chase, A. Paspalum of South America; Smithsonian Institution, Hitchcock and Chase Library: Washington, DC, USA, 1939; unpublsihed manuscript. [Google Scholar]
- Pilger, R.K.F. Bemerkungen zur Systematik der Gattung Paspalum L. Repert. Spec. Nov. Regni Veg. 1929, 26, 228–231. [Google Scholar]
- Pilger, R.K.F. Gramineae III. Unterfamilie Panicoideae. In Die Natürlichen Pflanzenfamilien; Engler, A., Prantl, K., Eds.; W Engelmann: Leipzig, Germany, 1940; Volume 14e, pp. 01–208. [Google Scholar]
- Clayton, W.D.; Renvoize, S.A. Genera graminum. Grasses of the World. Kew Bull. Addit. Ser. 1986, 13, 1–389. [Google Scholar]
- Rodríguez, H. Pectinata chase ex Rodríguez, nueva sección en el género Paspalum L. (Gramineae). Ernstia 1992, 2, 21–23. [Google Scholar]
- Rodríguez, H. El subgénero Ceresia (Pers.) Reichenb. del género Paspalum L. (Gramineae) en Venezuela. Ernstia 1998, 8, 7–50. [Google Scholar]
- Morrone, O.; Denham, S.S.; Aliscioni, S.S.; Zuloaga, F.O. Revisión de las especies de Paspalum (Panicoideae: Paniceae), subgénero Anachyris. Candollea 2000, 55, 105–155. [Google Scholar]
- Denham, S.S.; Zuloaga, F.O.; Morrone, O. Systematic revision and phylogeny of Paspalum subgenus Ceresia (Poaceae: Panicoideae: Paniceae). Ann. Mo. Bot. Gard. 2002, 89, 337–399. [Google Scholar] [CrossRef]
- Rua, G.H.; Aliscioni, S.S. A morphology-based cladistic analysis of Paspalum sect. Pectinata (Poaceae). Syst. Bot. 2002, 27, 489–501. [Google Scholar] [CrossRef]
- Zuloaga, F.O.; Pensiero, J.; Morrone, O. Systematics of Paspalum grupo Notata (Poaceae, Panicoideae, Paniceae). Syst. Bot. Monogr. 2004, 71, 1–75. [Google Scholar] [CrossRef]
- Denham, S.S. Revisión sistemática del subgénero Harpostachys de Paspalum (Poaceae: Panicoideae: Paniceae). Ann. Mo. Bot. Gard. 2005, 92, 463–532. [Google Scholar]
- Delfini, C.; Souza, V.C.; Zuloaga, F.O. Taxonomic revision and nomenclatural update of Paspalum sect. Pectinata (Poaceae, Panicoideae, Paspaleae). Phytotaxa 2017, 323, 1–26. [Google Scholar] [CrossRef]
- Scataglini, M.A.; Zuloaga, F.O.; Giussani, L.M.; Denham, S.S.; Morrone, O. Phylogeny of new world Paspalum (Poaceae, Panicoideae, Paspaleae) based on plastid and nuclear markers. Plant Syst. Evol. 2014, 300, 1051–1070. [Google Scholar] [CrossRef]
- Cialdella, A.M.; Morrone, O.; Zuloaga, F.O. Revisión de las especies del género Paspalum (Poaceae: Panicoideae: Paniceae) grupo Bonplandiana. Darwiniana 1995, 33, 67–95. [Google Scholar]
- Morrone, O.; Zuloaga, F.O.; Carbonó, E. Revisión del grupo Racemosa del género Paspalum (Poaceae: Panicoideae: Paspaleae). Ann. Mo. Bot. Gard. 1995, 82, 82–116. [Google Scholar] [CrossRef]
- Morrone, O.; Vega, A.S.; Zuloaga, F.O. Revisión de las especies del género Paspalum L. (Poaceae: Panicoideae: Paniceae), grupo Dissecta (s. str.). Candollea 1996, 51, 2–34. [Google Scholar]
- Morrone, O.; Denham, S.S.; Zuloaga, F.O. Revisión taxonómica del género Paspalum grupo Eriantha (Poaceae, Panicoideae, Paniceae). Ann. Mo. Bot. Gard. 2004, 91, 225–246. [Google Scholar]
- Oliveira, R.C.; Valls, J.F.M. Taxonomia de Paspalum L., grupo Linearia (Gramineae–Paniceae) do Brasil. Rev. Bras. Bot. 2002, 25, 371–389. [Google Scholar] [CrossRef]
- Denham, S.S.; Morrone, O.; Zuloaga, F.O. Estudios en el género Paspalum (Poaceae, Panicoideae, Paniceae). Paspalum denticulatum y especies afines. Ann. Mo. Bot. Gard. 2010, 97, 11–33. [Google Scholar] [CrossRef]
- Pozzobon, M.T.; Paganella, M.B.; Santos, S.; Valls, J.F.M. Aspectos citológicos e reprodutivos no grupo Caespitosa do gênero Paspalum. Cienc. Rural 2013, 43, 2004–2010. [Google Scholar] [CrossRef]
- Delfini, C.; Souza, V.C.; Zuloaga, F.O. Notas Nomenclaturales en Paspalum grupo Caespitosa (Poaceae, Panicoideae, Paspaleae). Novon 2019, 27, 156–161. [Google Scholar] [CrossRef]
- Zuloaga, F.O.; Morrone, O.; Davidse, G.; Filgueiras, T.S.; Peterson, P.M.; Soreng, R.J.; Judziewicz, E.J. Catalogue of New World Grasses (Poaceae): III. Subfamilies Panicoideae, Aristidoideae, Arundinoideae, and Danthonioideae. Contr. U.S. Natl. Herb. 2003, 46, 1–662. [Google Scholar]
- Delfini, C.; Souza, V.C.; Zuloaga, F.O. Taxonomic Revision of Paspalum group Caespitosa (Poaceae, Panicoideae, Paspaleae). Ann. Mo. Bot. Gard. 2023, 108, 1–50. [Google Scholar] [CrossRef]
- Nash, G.V. Poaceae. In North American Flora; Britton, N.L., Murrill, W.A., Barnhart, J.H., Eds.; The New York Botanical Garden: New York, NY, USA, 1912; Volume 17, pp. 99–196. [Google Scholar]
- Hiratsuka, J.; Shimada, H.; Whittier, R.; Ishibashi, T.; Sakamoto, M.; Mori, M.; Kondo, C.; Honji, Y.; Sun, C.-R.; Meng, B.-Y.; et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: Intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Genet. Genom. 1989, 217, 185–194. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, H.-C.; Lin, I.-P.; Chow, T.-Y.; Chen, H.-H.; Chen, W.-H.; Cheng, C.-H.; Lin, C.-Y.; Liu, S.-M.; Chang, C.-C.; et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): Comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 2006, 23, 279–291. [Google Scholar] [CrossRef]
- Acosta, J.M.; Zuloaga, F.O.; Reinheimer, R. Nuclear phylogeny and hypothesized allopolyploidization events in the subtribe Otachyriinae (Paspaleae, Poaceae). Syst. Biodivers. 2019, 17, 277–294. [Google Scholar] [CrossRef]
- Acosta, J.M.; Scataglini, M.A.; Reinheimer, R.; Zuloaga, F.O. A phylogenetic study of subtribe Otachyriinae (Poaceae, Panicoideae, Paspaleae). Plant Syst. Evol. 2014, 300, 2155–2166. [Google Scholar] [CrossRef]
- Delfini, C.; Acosta, J.M.; Souza, V.C.; Zuloaga, F.O. Molecular phylogeny of Axonopus (Poaceae, Panicoideae, Paspaleae): Monophyly, synapomorphies and taxonomic implications for infrageneric classification and species complexes. Ann. Mo. Bot. Gard. 2020, 105, 459–480. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. Bot. Soc. Am. 1987, 19, 11–15. [Google Scholar]
- Baldwin, B.G.; Markos, S. Phylogenetic utility of the external transcribed spacers (ETS) of 18S-26S rDNA: Congruence of ETS and ITS trees of Calycadenia (Compositae). Mol. Phylogenet. Evol. 1998, 10, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.; Beck, J.T.; Farmer, S.B.; Liu, W.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef] [PubMed]
- Kelchner, S.; Clark, L.G. Molecular evolution and phylogenetic utility of the chloroplast rpl16 intron in Chusquea and the Bambusoideae (Poaceae). Mol. Phylogenet. Evol. 1997, 8, 385–397. [Google Scholar] [CrossRef]
- Zhang, W. Phylogeny of the grass family (Poaceae) from rpl16 intron sequence data. Mol. Phylogenet. Evol. 2000, 15, 385–397. [Google Scholar] [CrossRef]
- Cialdella, A.M.; Giussani, L.M.; Aagesen, L.; Zuloaga, F.O.; Morrone, O. A phylogeny of Piptochaethium based on a combined analysis including trnL-F, rpl16 and morphology. Syst. Bot. 2007, 32, 545–559. [Google Scholar] [CrossRef]
- Jordan, W.C.; Courtney, W.M.; Neigel, E.J. Low levels of intraspecific genetic variation at a rapidly evolving chloroplast DNA locus in North American duckwoods (Lemnaceae). Am. J. Bot. 1996, 83, 430–439. [Google Scholar] [CrossRef]
- Giussani, L.M.; Zuloaga, F.O.; Quarín, C.L.; Cota-Sánchez, J.H.; Ubayasena, K.; Morrone, O. Phylogenetic relationships in the genus Paspalum (Poaceae: Panicoideae: Paniceae): An assessment of the Quadrifaria and Virgata informal groups. Syst. Bot. 2009, 34, 32–43. [Google Scholar] [CrossRef]
- Olmstead, R.G.; Sweere, J.A. Combining data in phylogenetic systematics: An empirical approach using three molecular data sets in the Solanaceae. Syst. Biol. 1994, 43, 467–481. [Google Scholar] [CrossRef]
- Aliscioni, S.S.; Giussani, L.M.; Zuloaga, F.O.; Kellogg, E.A. A molecular phylogeny of Panicum (Poaceae: Paniceae). Test of monophyly and phylogenetic placement within the Panicoideae. Am. J. Bot. 2003, 90, 796–821. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Fitch, W.M. Toward defining the course of evolution: Minimal change for a specific tree topology. Syst. Zool. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Crandall, K.A. Phylogeny estimation and hypothesis testing using maximum likelihood. Annu. Rev. Ecol. Evol. Syst. 1997, 28, 437–466. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Larget, B.; Miller, R.E.; Ronquist, F. Potential applications and pitfalls of Bayesian inference of phylogeny. Syst. Biol. 2002, 51, 673–688. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Nixon, K.C.; Carpenter, J.M. On simultaneous analysis. Cladistics 1996, 12, 221–241. [Google Scholar] [CrossRef]
- Farris, J.S.; KAllersjö, M.; Kluge, A.G.; Bult, C. Constructing a significance test for incongruence. Syst. Bot. Monogr. 1995, 44, 570–572. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic analysis using parsimony (and other methods), version 4.0 b10. Evolution 2002, 56, 1776–1788. [Google Scholar] [CrossRef]
- Goloboff, P.A.; Farris, J.S.; Nixon, K.C. TNT, a free program for phylogenetic analysis. Cladistics 2008, 24, 774–786. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihoodbased phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Adamowski, E.V.; Pagliarini, M.S.; Batista, L.A.R. Chromosome number and microsporogenesis in Paspalum maritimum (Caespitosa group; Gramineae). Braz. Arch. Biol. Technol. 2000, 43, 301–305. [Google Scholar] [CrossRef]
- Maciel, J.R.; Oliveira, R.C.; Alves, M. Paspalum L. (Poaceae: Panicoideae: Paniceae) no estado de Pernambuco, Brasil. Acta Bot. Bras. 2009, 23, 1145–1161. [Google Scholar] [CrossRef]
- Aliscioni, S.S. Anatomía ecológica de algunas especies del género Paspalum (Poaceae, Panicoideae, Paniceae). Darwiniana 2000, 38, 187–207. [Google Scholar] [CrossRef]
- Liphschitz, Ν.; Waisel, Y. Adaptation of plants to saline environments: Salt excretion and glandular structure. In Contributions of the Ecology of Halophytcs. Tasks for Vegetation Science; Sen, D.N., Rajpurohit, K.S., Eds.; Springer: Dordrecht, Netherlands, 1982; Volume 2, pp. 187–214. [Google Scholar]
- Aliscioni, S.S.; Denham, S.S. Rachis of the genus Paspalum L. (Poaceae: Panicoideae: Paniceae): Anatomy and taxonomic significance of the primary branches of the inflorescences. Flora 2008, 203, 60–76. [Google Scholar] [CrossRef]
- Aliscioni, S.S. Estudio Histofoliar Comparado de Especies Americanas del Género Paspalum L. (Poaceae: Panicoideae: Paniceae). PhD Thesis, Facultad de Ciencias Naturales y Museo, Universidad Nacional de La Plata,, La Plata, Argentina, 1999. [Google Scholar]
- POWO (Plants of the World Online). Facilitated by the Royal Botanic Gardens, Kew. Available online: http://www.plantsoftheworldonline.org/ (accessed on 8 July 2022).
- Hunziker, J.H.; Zuloaga, F.O.; Morrone, O.; Escobar, A. Estudios cromosómicos en Paniceae sudamericanas (Poaceae: Panicoideae). Darwiniana 1998, 35, 29–36. [Google Scholar]
- Morrone, O.; Escobar, A.; Zuloaga, F.O. Chromosome studies in American Panicoideae (Poaceae). Ann. Mo. Bot. Gard. 2006, 93, 647–657. [Google Scholar] [CrossRef]
- Metcalfe, C.R. Anatomy of the Monocotyledons, I: Gramineae, 1st ed.; Clarendon Press: Oxford, MS, USA, 1960; 794p. [Google Scholar]
- Ellis, R.P. A procedure for standardizing comparative leaf anatomy in the Poaceae. II. The epidermis as seen in surface view. Bothalia 1979, 12, 641–671. [Google Scholar] [CrossRef]
- Hitchcock, A.S. New species of grasses from the Galapagos and the Revillagigedo Islands. Proc. Calif. Acad. Sci. 1935, 21, 295–300. [Google Scholar]
- Cotton, J.L.; Wysocki, W.P.; Clark, L.G.; Kelchner, S.A.; Pires, J.C.; Edger, P.P.; Mayfield-Jones, D.; Duvall, M.R. Resolving deep relationships of PACMAD grasses: A phylogenomic approach. BMC Plant Biol. 2015, 15, 178. [Google Scholar] [CrossRef]
- Burke, S.V.; Wysocki, W.P.; Zuloaga, F.O.; Craine, J.M.; Pires, J.C.; Edger, P.P.; Mayfield-Jones, D.; Clark, L.G.; Kelchner, S.A.; Duvall, M.R. Evolutionary relationships in Panicoid grasses based on plastome phylogenomics (Panicoideae; Poaceae). BMC Plant Biol. 2016, 16, 140. [Google Scholar] [CrossRef]
- Saarela, J.M.; Burke, S.V.; Wysocki, W.P.; Barrett, M.D.; Clark, L.G.; Craine, J.M.; Peterson, P.M.; Soreng, R.J.; Vorontsova, M.S.; Duvall, M.R. A 250 plastome phylogeny of the grass family (Poaceae): Topological support under different data partitions. PeerJ 2018, 6, e4299. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, L.; Columbus, J.T.; Hu, Y.; Zhao, Y.; Tang, L.; Guo, Z.; Chen, W.; McKain, M.; Bartlett, M.; et al. A well-supported nuclear phylogeny of Poaceae and implications for the evolution of C4 photosynthesis. Mol. Plant 2022, 15, 755–777. [Google Scholar] [CrossRef]
- Estep, M.C.; McKain, M.R.; Diaz, D.V.; Zhong, J.; Hodge, J.G.; Hodkinson, T.R.; Layton, D.J.; Malcomber, S.T.; Pasquet, R.; Kellogg, E.A. Allopolyploidy, diversification, and the Miocene grassland expansion. Proc. Natl. Acad. Sci. USA 2014, 111, 15149–15154. [Google Scholar] [CrossRef]
- Kellogg, E.A. Has the connection between polyploidy and diversification actually been tested? Curr. Opin. Plant Biol. 2016, 30, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Sang, T. Utility of low-copy nuclear gene sequences in plant phylogenetics. Crit. Rev. Biochem. Mol. Biol. 2002, 37, 121–147. [Google Scholar] [CrossRef] [PubMed]
- Thiers, B. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff, New York Botanical Garden’s Virtual Herbarium, continuously updated. Available online: http://sweetgum.nybg.org/science/ih/ (accessed on 8 March 2022).
- Morrone, O.; Hunziker, J.H.; Zuloaga, F.O.; Escobar, A. Números cromosómicos en Paniceae sudamericanas (Poaceae: Panicoideae). Darwiniana 1995, 3, 53–60. [Google Scholar]
- Burson, B.L. Cytology of Paspalum chacoense and P. durifolium and their relationship to P. dilatatum. Bot. Gaz. 1985, 146, 124–129. [Google Scholar] [CrossRef]
- Quarín, C.L. Recuentos cromosómicos en gramíneas de Argentina subtropical. Hickenia 1977, 1, 73–78. [Google Scholar]
- Quarín, C.L.; Burson, B.L. Cytogenetic relations among Paspalum notatum var. saurae, P. pumilum, P. indecorum, and P. vaginatum. Bot. Gaz. 1983, 144, 433–438. [Google Scholar]
- Quarín, C.L.; Hanna, W.W.; Fernandez, A. Genetic studies in diploid and tetraploid Paspalum species. Embryo sac development, chromosome behavior, and fertility in P. cromyorhizon, P. laxum and P. proliferum. J. Hered. 1982, 73, 254–256. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).