Gamete Recognition Gene Divergence Yields a Robust Eutherian Phylogeny across Taxonomic Levels
Abstract
1. Introduction
2. Materials and Methods
2.1. Phylogenetic Analysis of Zan
2.2. Testing the Phylogenetic Utility of Zan
2.3. Tanglegram Comparisons
2.4. Topological Statistical Comparisons
2.5. Divergence/Selection Comparisons
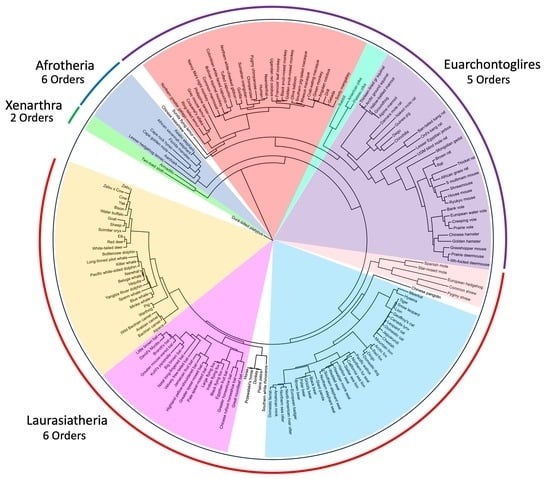
3. Results


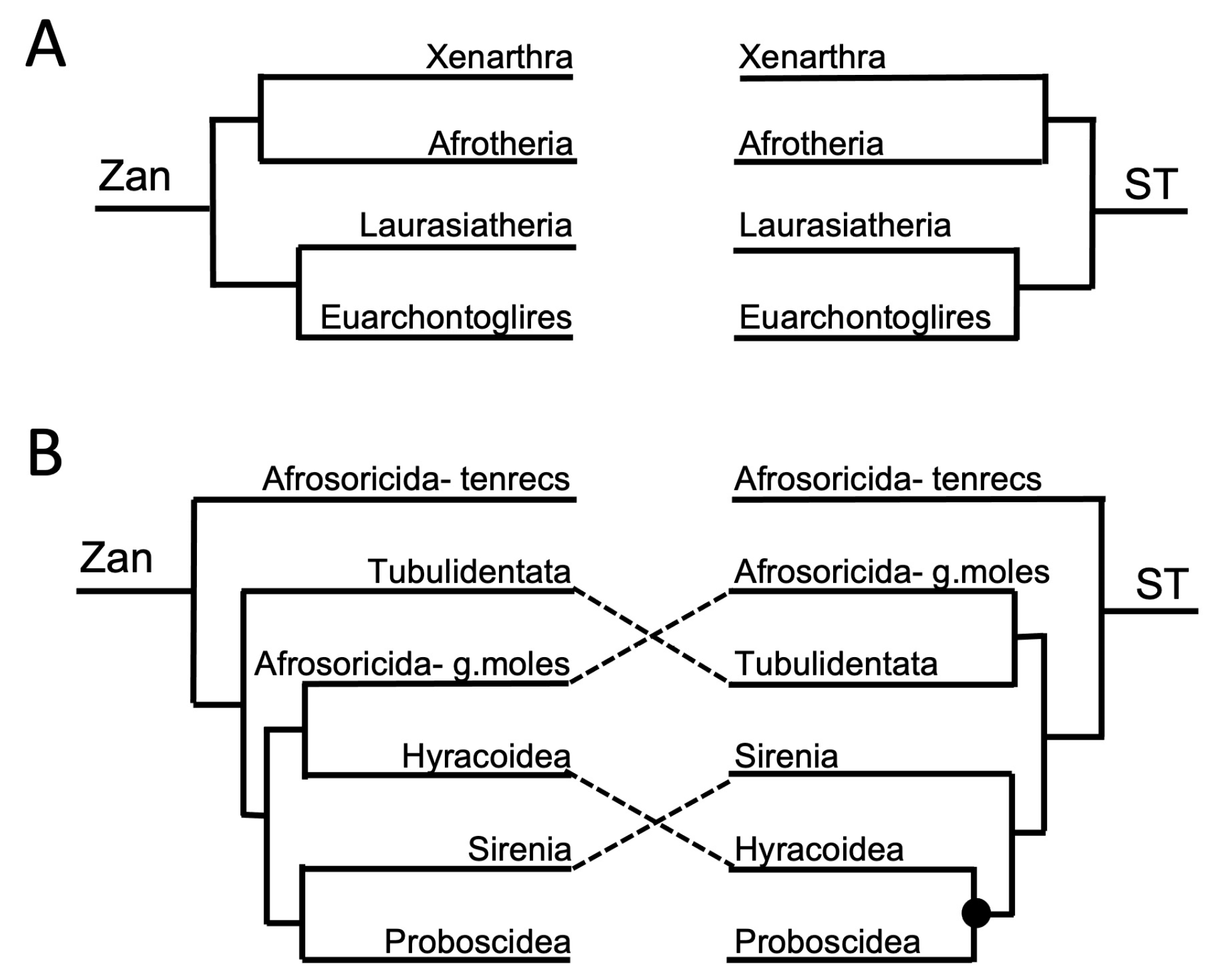
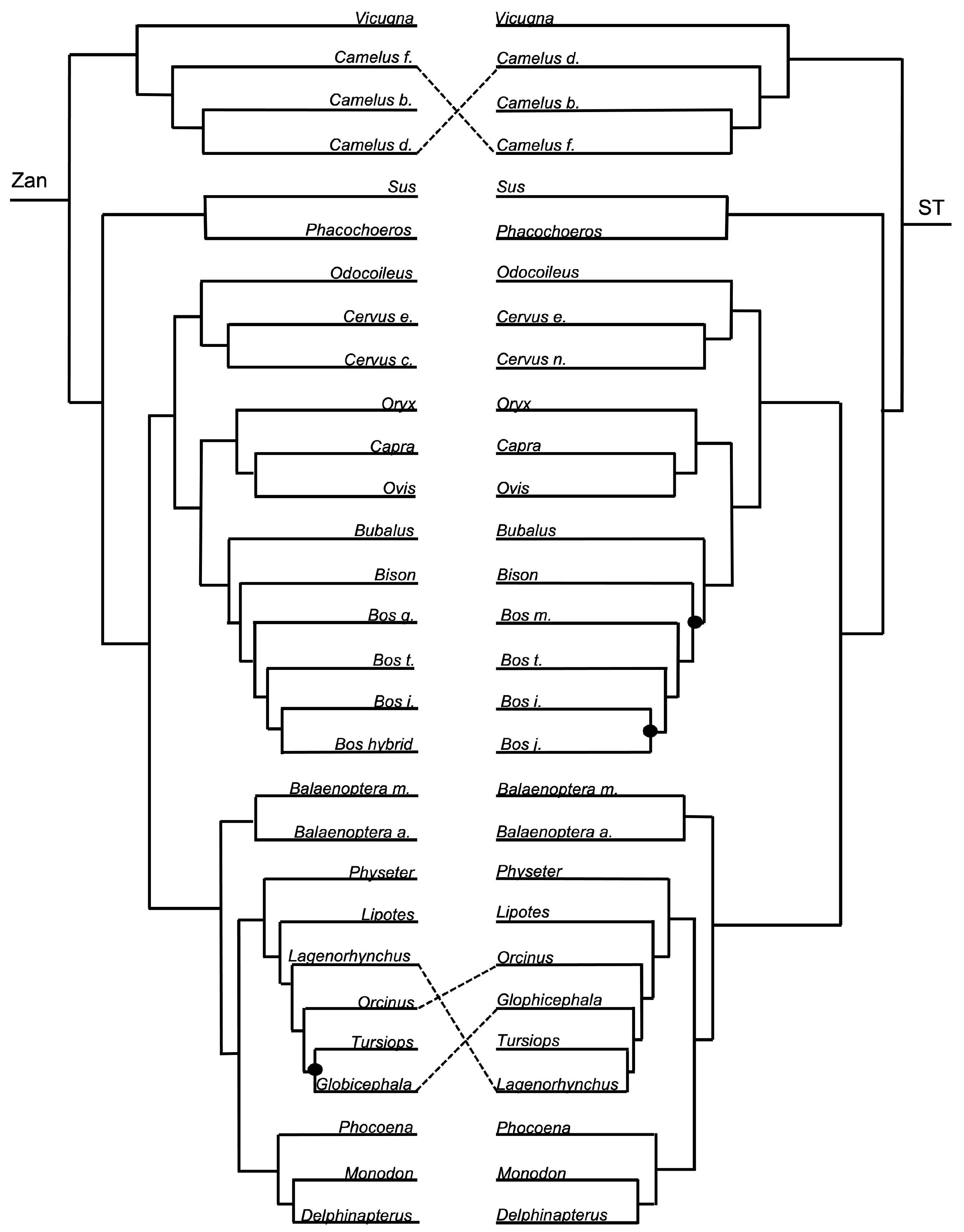
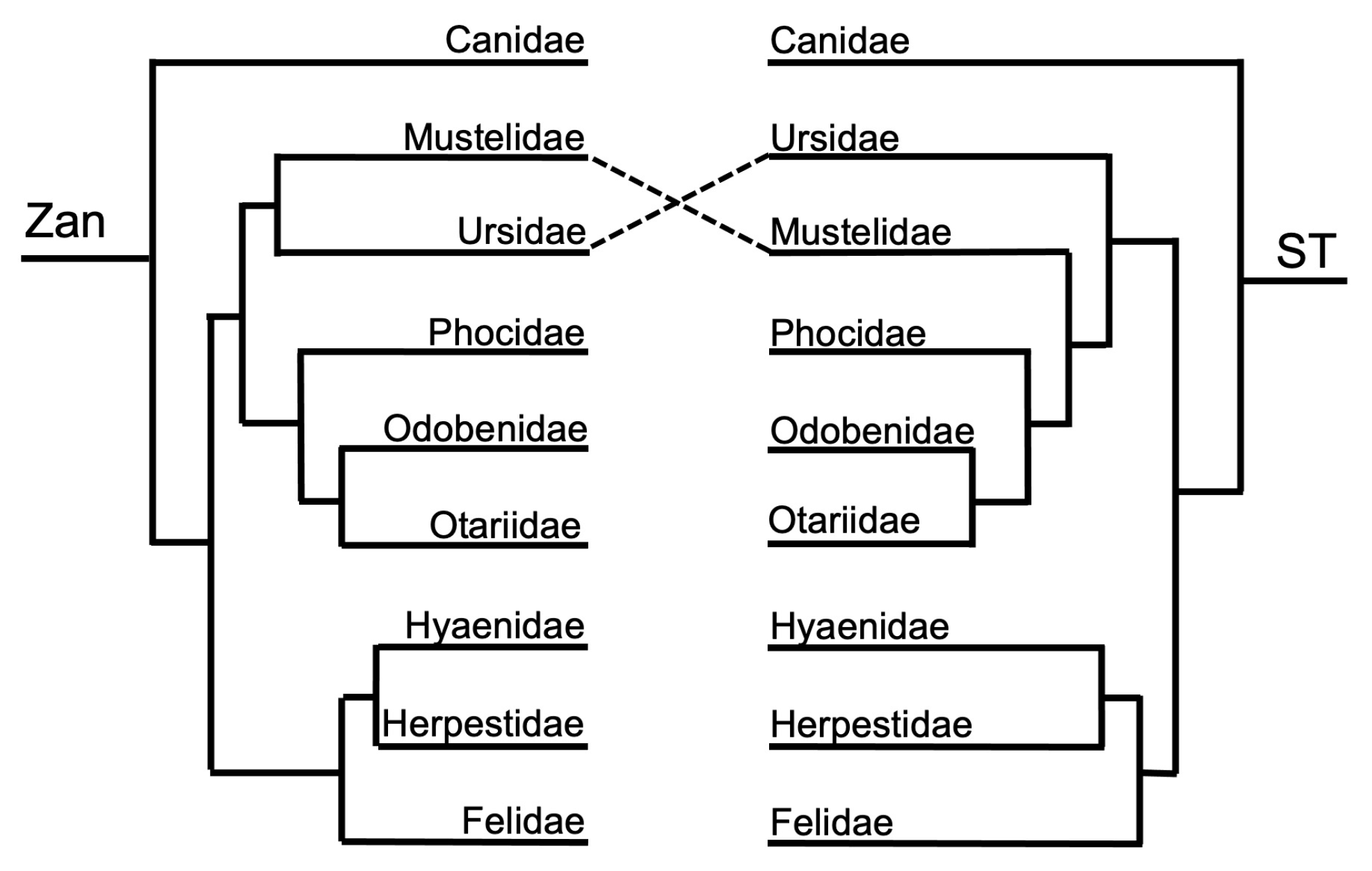
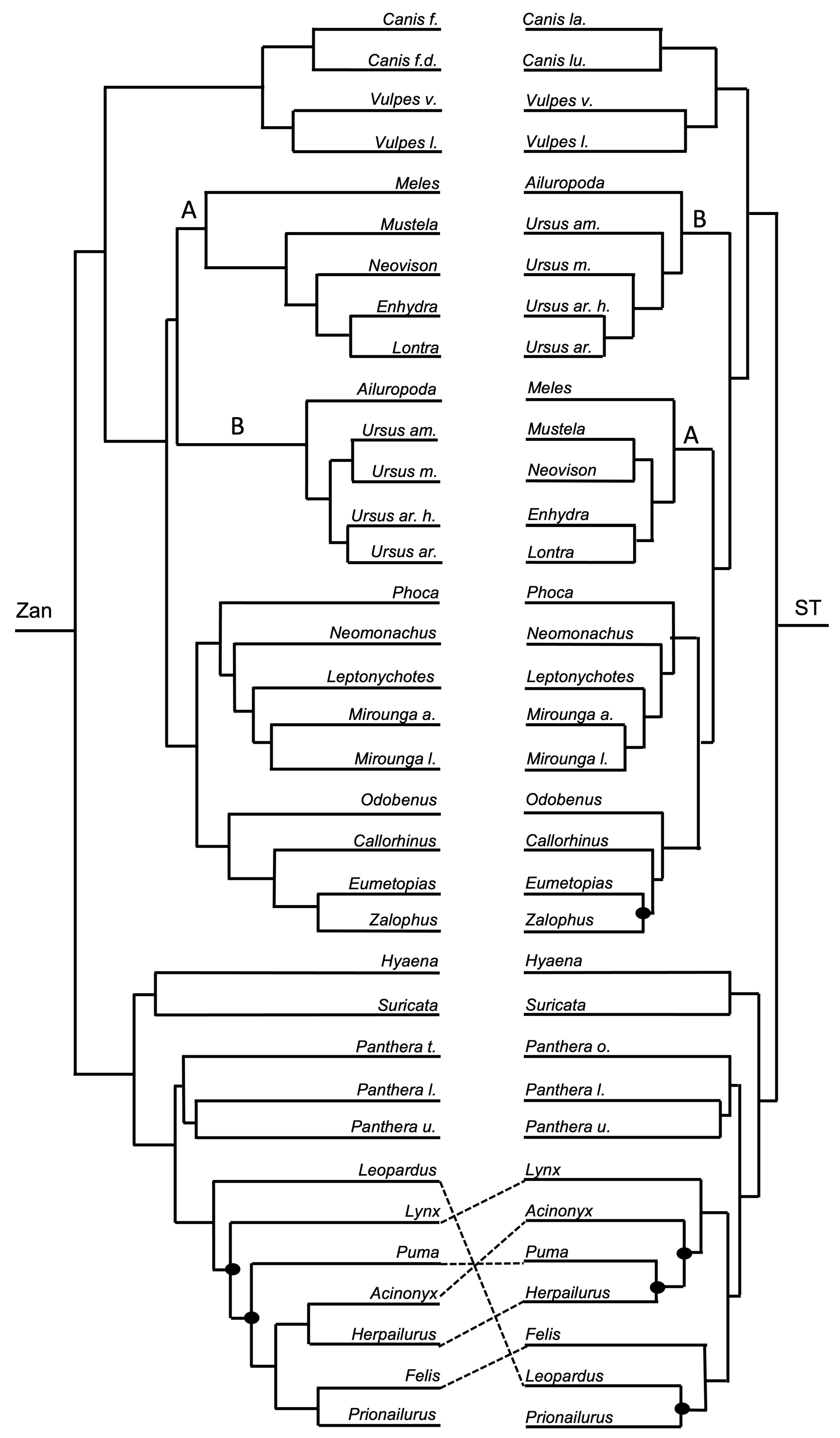
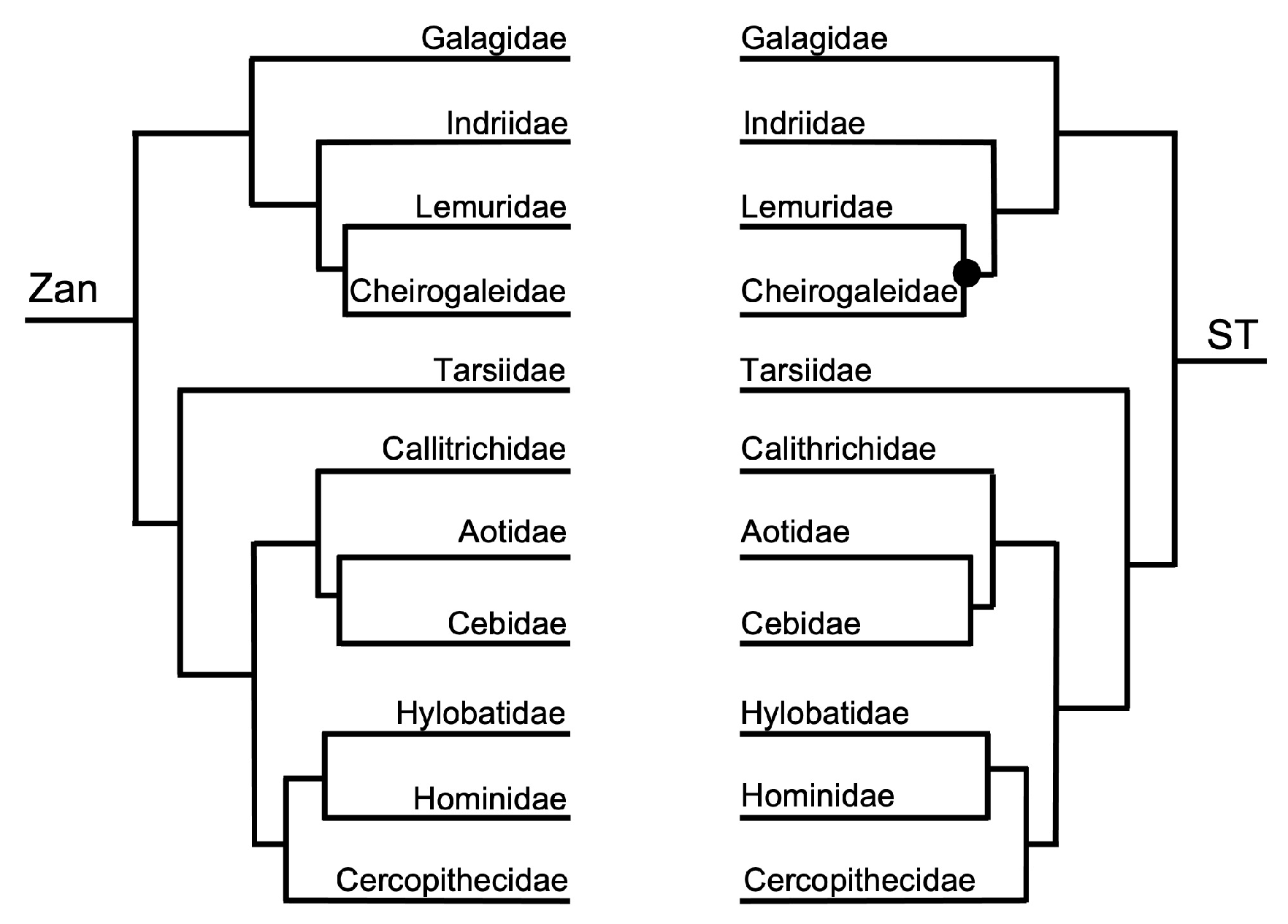
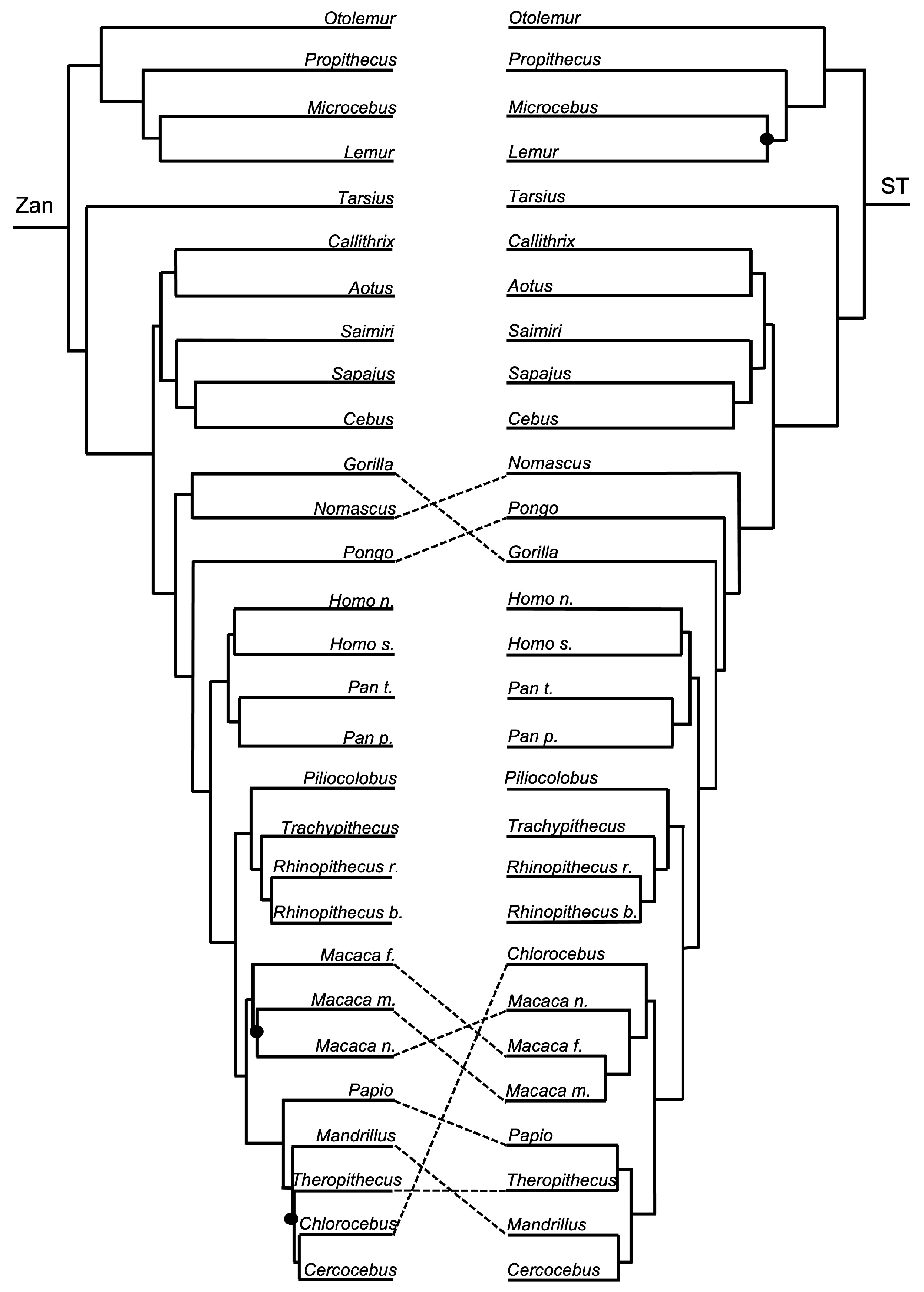
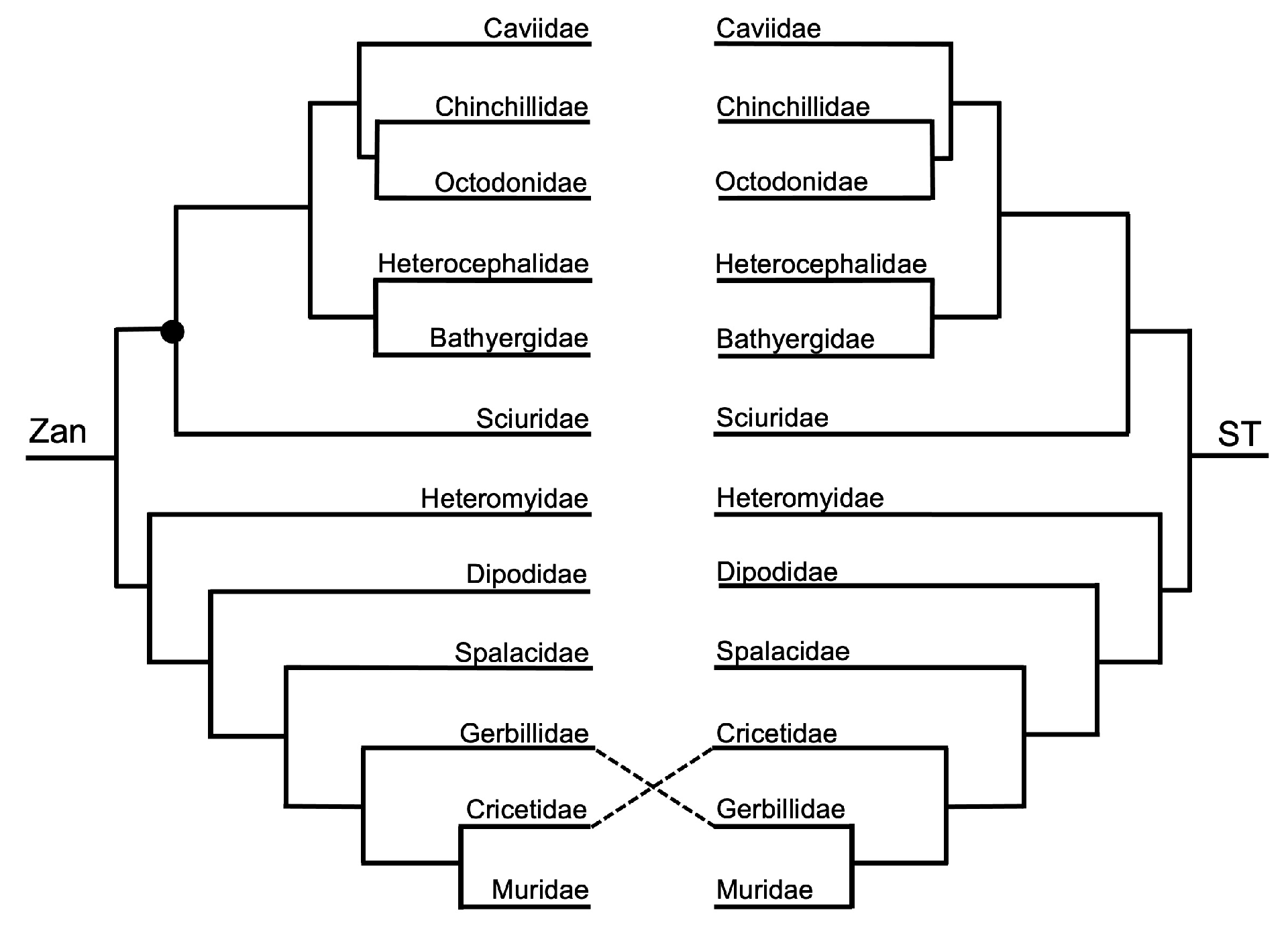
3.1. Tanglegram Comparisons
3.1.1. Magnordinal- and Superordinal-Levels
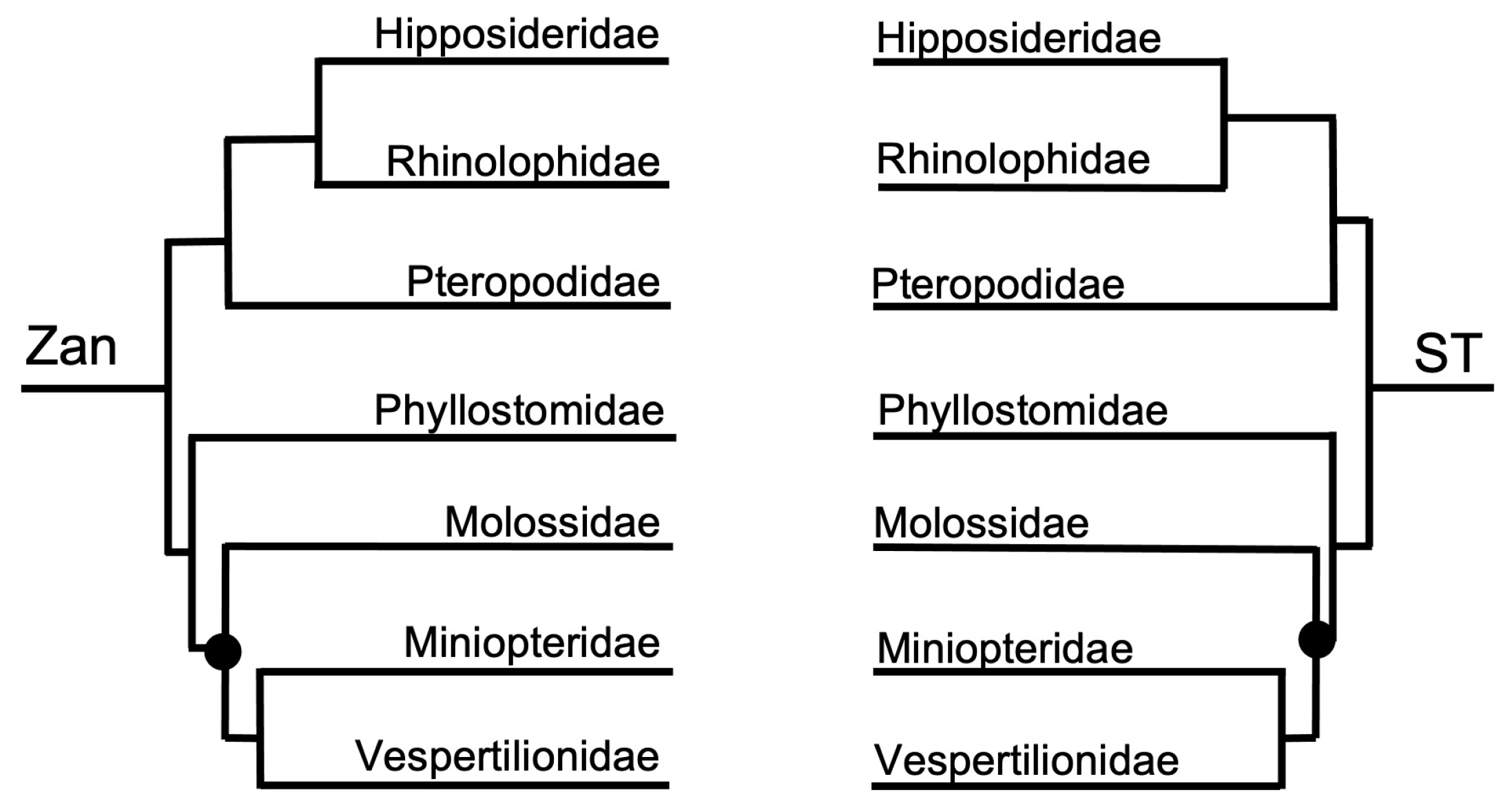
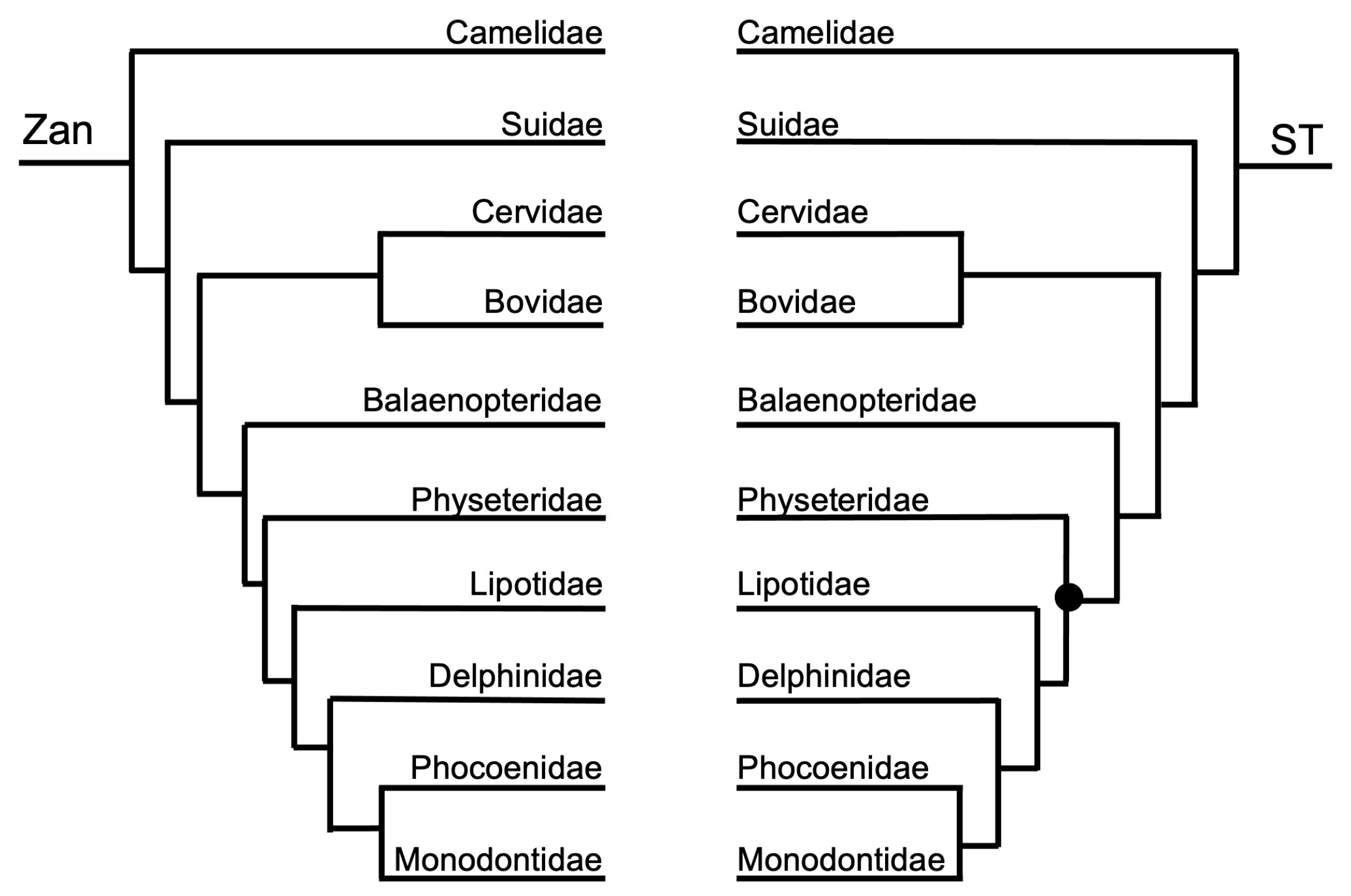
3.1.2. Ordinal-Level
3.1.3. Intra-Ordinal Level
3.2. Topology Statistical Comparisons
3.3. Divergence/Selection Comparisons
4. Discussion
4.1. Tanglegram Comparisons
4.1.1. Inter-Ordinal
4.1.2. Intra-Ordinal
4.1.3. Resolving Controversial Relationships
4.1.4. Zan as a Speciation Gene Phylogenetic Marker
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meredith, R.W.; Janecka, J.E.; Gatesy, J.; Ryder, O.A.; Fisher, C.A.; Teeling, E.C.; Goodbla, A.; Eizirik, E.; Simão, T.L.L.; Stadler, T.; et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science 2011, 334, 521–524. [Google Scholar] [CrossRef]
- dos Reis, M.; Inoue, J.; Hasegawa, M.; Asher, R.J.; Donoghue, P.C.; Yang, Z. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc. R. Soc. B 2012, 279, 3491–3500. [Google Scholar] [CrossRef]
- Song, S.; Liu, L.; Edwards, S.V.; Wu, S. Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc. Natl. Acad. Sci. USA 2012, 109, 14942–14947. [Google Scholar] [CrossRef]
- O’Leary, M.A.; Bloch, J.I.; Flynn, J.J.; Gaudin, T.J.; Giallombardo, A.; Giannini, N.P.; Goldberg, S.L.; Kraatz, B.P.; Luo, Z.; Meng, J.; et al. The placental mammal ancestor and the post-KPg radiation of placentals. Science 2013, 339, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Springer, M.S.; Meredith, R.W.; Teeling, E.C.; Murphy, W.J. Technical comment on ‘The placental mammal ancestor and the post-KPg radiation of placentals’. Science 2013, 341, 613. [Google Scholar] [CrossRef] [PubMed]
- Springer, M.S.; Gatesy, J. The gene tree delusion. Mol. Phy. Evol. 2016, 94, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Simpson, G.G. The Principles of Classification and a Classification of Mammals; American Museum of Natural History: New York, NY, USA, 1945; Volume 85. [Google Scholar]
- Shoshani, J.; McKenna, M.C. Higher Taxonomic Relationships among Extant Mammals Based on Morphology, with Selected Comparisons of Results from Molecular Data. Mol. Phy. Evol. 1998, 9, 572–584. [Google Scholar] [CrossRef]
- Novacek, M.J.; Wyss, A.R.; McKenna, M.C. The major groups of Eutherian mammals. In The Phylogeny and Classification of the Tetrapods; Benton, M.J., Ed.; Clarendon Press: Oxford, UK, 1988; Volume 2, pp. 31–71. [Google Scholar]
- Novacek, M.J. Higher mammal phylogeny: The morphological-molecular synthesis. In The Hierarchy of Life; Fernholm, B., Bremer, K., Jornvall, H., Eds.; Elsevier: Amsterdam, The Netherlands, 1989; pp. 421–435. [Google Scholar]
- Novacek, M.J. Mammalian phylogeny: Shaking the tree. Nature 1992, 356, 121–125. [Google Scholar] [CrossRef]
- MacPhee, R.D.E.; Novacek, M.J. Definition and relationships of Lipotyphla. In Mammal Phylogeny: Placentals; Szalay, F.S., Novacek, M.J., McKenna, M.C., Eds.; Springer: New York, NY, USA, 1993; pp. 13–31. [Google Scholar]
- Gaudin, T.J.; Wible, J.R.; Hopson, J.A.; Turnbull, W.D. Reexamination of the morphological evidence for the cohort Epitheria (Mammalia, Eutheria). J. Mamm. Evol. 1996, 3, 31–79. [Google Scholar] [CrossRef]
- Madsen, O.; Scally, M.; Douady, C.J.; Kao, D.J.; DeBry, R.W.; Adkins, R.; Amrine, H.M.; Stanhope, M.J.; de Jong, W.W.; Springer, M.S. Parallel adaptive radiations in two major clades of placental mammals. Nature 2001, 409, 610–614. [Google Scholar] [CrossRef]
- Murphy, W.J.; Eizirik, E.; Johnson, W.E.; Zhang, Y.P.; Ryder, O.A.; O’Brien, S.J. Molecular phylogenetics and the origins of placental mammals. Nature 2001, 409, 614–618. [Google Scholar] [CrossRef]
- Springer, M.S.; Cleven, G.C.; Madsen, O.; de Jong, W.W.; Waddell, V.G.; Amrine, H.M.; Stanhope, M.J. Endemic African mammals shake the phylogenetic tree. Nature 1997, 388, 61–64. [Google Scholar] [CrossRef] [PubMed]
- De Jong, W.W.; van Dijk, M.A.M.; Poux, C.; Kappé, G.; van Rheede, T.; Madsen, O. Indels in protein-coding sequences of Euarchontoglires constrain the rooting of the eutherian tree. Mol. Phy. Evol. 2003, 28, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Stanhope, M.J.; Waddell, V.G.; Madsen, O.; de Jong, W.W.; Hedges, S.B.; Cleven, G.C.; Kao, D.; Springer, M.S. Molecular evidence for multiple origins of Insectivora and for a new order of endemic African insectivore mammals. Proc. Natl. Acad. Sci. USA 1998, 95, 9967–9972. [Google Scholar] [CrossRef] [PubMed]
- Waddell, P.J.; Okada, N.; Hasegawa, M. Towards resolving the interordinal relationships of placental mammals. Syst. Biol. 1999, 48, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Amrine-Madsen, H.; Koepfli, K.P.; Wayne, R.K.; Springer, M.S. A new phylogenetic marker, apoliprotein B, provides compelling evidence for eutherian relationships. Mol. Phylogenet. Evol. 2003, 28, 225–240. [Google Scholar] [CrossRef]
- Nishihara, H.; Hasegawa, M.; Okada, N. Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc. Natl. Acad. Sci. USA 2006, 103, 9929–9934. [Google Scholar] [CrossRef]
- Hallstrom, B.; Kullberg, M.; Nilsson, M.A.; Janke, A. Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sister groups. Mol. Biol. Evol. 2007, 24, 2059–2068. [Google Scholar] [CrossRef]
- Wildman, D.E.; Uddin, M.; Opazo, J.C.; Liu, G.; Lefort, V.; Guindon, S.; Gascuel, O.; Grossman, L.I.; Romero, R.; Goodman, M. Genomics, biogeography, and the diversification of placental mammals. Proc. Natl. Acad. Sci. USA 2007, 104, 14395–14400. [Google Scholar] [CrossRef]
- Asher, R.J.; Bennett, N.; Lehmann, T. The new framework for understanding placental mammal evolution. Bioessays 2009, 31, 853–864. [Google Scholar] [CrossRef]
- Liu, F.G.R.; Miyamoto, M.M.; Freire, N.P.; Ong, P.Q.; Tennant, M.R.; Young, T.S.; Gugel, K.F. Molecular and morphological supertrees for eutherian (placental) mammals. Science 2001, 291, 1786–1789. [Google Scholar] [CrossRef]
- Beck, R.M.D.; Bininda-Emonds, O.R.P.; Cardillo, M.; Liu, F.; Purvis, A. A higher-level MRP supertree of placental mammals. BMC Evol. Biol. 2006, 6, 93. [Google Scholar] [CrossRef][Green Version]
- Bininda-Emonds, O.R.P.; Cardillo, M.; Jones, K.E.; MacPhee, R.D.E.; Beck, R.M.D.; Grenyer, R.; Price, S.A.; Vos, R.A.; Gittleman, J.L.; Purvis, A. The delayed rise of present-day mammals. Nature 2007, 446, 507–512. [Google Scholar] [CrossRef]
- Upham, N.S.; Esselstyn, J.A.; Jetz, W. Inferring the mammal tree: Species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 2019, 17, e3000494. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.J.; Pevzner, P.A.; O’Brien, S.J. Mammalian phylogenomics comes of age. Tren. Genet. 2004, 20, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Fabre, P.; Hautier, L.; Dimitrov, D.; Douzery, E.J.P. A glimpse on the pattern of rodent diversification a phylogenetic approach. BMC Evol. Biol. 2012, 12, 88. [Google Scholar] [CrossRef] [PubMed]
- Halliday, T.J.D.; Upchurch, P.; Goswami, A. Resolving the relationships of Paleocene placental mammals. Biol. Rev. 2015, 92, 521–550. [Google Scholar] [CrossRef] [PubMed]
- Springer, M.S.; Stanhope, M.J.; Madsen, O.; de Jong, W.W. Molecules consolidate the placental mammal tree. Tren. Ecol. Evol. 2004, 19, 430–438. [Google Scholar] [CrossRef]
- Springer, M.S.; Murphy, W.J.; Eizirik, E.; Madsen, O.; Scally, M.; Douady, C.J.; Teeling, E.C.; Stanhope, M.J.; de Jong, W.W.; O’Brien, S.J. A molecular classification for the living orders of placental mammals and the phylogenetic placement of primates. In Primate Origins: Adaptations and Evolution; Springer: Boston, MA, USA, 2007; pp. 1–28. [Google Scholar]
- Huelsenbeck, J.P.; Bull, J.J.; Cunningham, C.W. Combining data in phylogenetic analysis. Tren. Ecol. Evol. 1996, 11, 152–158. [Google Scholar] [CrossRef]
- Paradis, E. Analysis of diversification: Combining phylogenetic and taxonomic data. Proc. R. Soc. Lond. B 2003, 270, 2499–2505. [Google Scholar] [CrossRef]
- Hahn, M.W.; Nakhleh, L. Irrational exuberance for resolved species trees. Evolution 2015, 70, 7–17. [Google Scholar] [CrossRef]
- Stadler, T.; Bokma, F. Estimating speciation and extinction rates for phylogenies of higher taxa. Syst. Biol. 2013, 62, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.J.; Eizirik, E.; O’Brian, S.J.; Madsen, O.; Scally, M.; Douady, C.J.; Teeling, E.; Ryder, O.A.; Stanhope, M.J.; de Jong, W.E.; et al. Resolution of the Early Placental Mammalian Radiation Using Bayesian Phylogenetics. Science 2001, 294, 2348–2351. [Google Scholar] [CrossRef] [PubMed]
- Scally, M.; Madsen, O.; Douady, C.J.; de Jong, W.W.; Stanhope, M.J.; Springer, M.S. Molecular evidence for the major clades of placental mammals. J. Mamm. Evol. 2001, 8, 239–277. [Google Scholar] [CrossRef]
- Delsuc, F.; Scally, M.; Madsen, O.; Stanhope, M.J.; de Jong, W.W.; Catzeflis, F.M.; Springer, M.S.; Douzery, E.J.P. Molecular phylogeny of living Xenarthrans and the impact of character and taxon sampling on the placental tree rooting. Mol. Biol. Evol. 2002, 19, 1656–1671. [Google Scholar] [CrossRef][Green Version]
- Springer, M.S.; Murphy, W.J.; Eizirik, E.; O’Brien, S.J. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. USA 2003, 100, 1056–1061. [Google Scholar] [CrossRef]
- Springer, M.S.; DeBry, R.W.; Amrine, H.M.; Madsen, O.; de Jong, W.W.; Stanhope, M.J. Mitochondrial versus nuclear gene sequences in deep-level mammalian phylogeny reconstruction. Mol. Biol. Evol. 2001, 18, 132–143. [Google Scholar] [CrossRef]
- Tarver, J.E.; dos Reis, M.; Mirarab, S.; Moran, R.J.; Parker, S.; O’Reilly, J.E.; King, B.L.; O’Connell, M.J.; Asher, R.J.; Warnow, T.; et al. The interrelationships of placental mammals and the limits of phylogenetic inference. Gen. Biol. Evol. 2016, 8, 330–344. [Google Scholar] [CrossRef]
- Foley, N.M.; Springer, M.S.; Teeling, E.C. Mammal madness: Is the mammal tree of life not yet resolved? Phil. Trans. R. Soc. B 2016, 371, 20150140. [Google Scholar] [CrossRef]
- Novacek, M.J. Fossils, topologies, missing data, and the higher level phylogeny of Eutherian mammals. Syst. Biol. 1992, 41, 58–73. [Google Scholar] [CrossRef]
- Swanson, W.J.; Yang, Z.; Wolfner, M.F.; Aquadro, C.F. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc. Natl. Acad. Sci. USA 2001, 98, 2509–2514. [Google Scholar] [CrossRef]
- Torgerson, D.G.; Kulathinal, R.J.; Singh, R.S. Mammalian Sperm Proteins Are Rapidly Evolving: Evidence of Positive Selection in Functionally Diverse Genes. Mol. Biol. Evol. 2002, 19, 1973–1980. [Google Scholar] [CrossRef] [PubMed]
- Sabeti, P.C.; Schaffner, S.F.; Fry, B.; Lohmueller, J.; Varilly, P.; Shamovsky, O.; Palma, A.; Mikkelsen, T.S.; Altshuler, D.; Lander, E.S. Positive natural selection in the human lineage. Science 2006, 312, 1614–1620. [Google Scholar] [CrossRef]
- Resch, A.M.; Carmel, L.; Mariño-Ramírez, L.; Ogurtsoc, A.Y.; Shabalina, S.A.; Rogozin, I.B. Widespread Positive Selection in Synonymous Sites of Mammalian Genes. Mol. Biol. Evol. 2007, 24, 1821–1831. [Google Scholar] [CrossRef] [PubMed]
- Massey, S.E.; Churbanov, A.; Tastogi, S.; Liberles, D.A. Characterizing positive and negative selection and their phylogenetic effects. Gene 2008, 418, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Koisol, C.; Vinař, T.; da Fonseca, R.R.; Hubisz, M.J.; Bustamante, C.D.; Nielsen, R.; Siepel, A. Patterns of Positive Selection in Six Mammalian Genomes. PLoS Genet. 2008, 4, e1000144. [Google Scholar] [CrossRef]
- Pollard, K.S.; Hubisz, M.J.; Rosembloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Gen. Res. 2010, 20, 110–121. [Google Scholar] [CrossRef]
- McCandlish, D.M.; Stoltzfus, A.; Dykhuizen, D.E. Modeling Evolution Using the Probability of Fixation: History and Implications. Quart. Rev. Biol. 2014, 89, 225–252. [Google Scholar] [CrossRef]
- Young, A.D.; Gillung, J.P. Phylogenomics—Principles, opportunities, and pitfalls of big-data phylogenetics. Syst. Entom. 2019, 45, 225–247. [Google Scholar] [CrossRef]
- Molloy, E.K.; Warnow, T. To Include or Not to Include: The Impact of Gene Filtering on Species Tree Estimation Methods. Syst. Biol. 2017, 67, 285–303. [Google Scholar] [CrossRef]
- Latrille, T.; Rodrigue, N.; Lartillot, N. Genes and sites under adaptation at the phylogenetic scale also exhibit adaptation at the population-genetic scale. Proc. Natl. Acad. Sci. USA 2023, 120, e2214977120. [Google Scholar] [CrossRef] [PubMed]
- Yi, S. Neutrality and Molecular Clocks. Nat. Educ. Knowl. 2013, 4, 3. [Google Scholar]
- Ting, C.T.; Takahashi, A.; Wu, C.I. Incipient speciation by sexual isolation in Drosophila: Concurrent evolution at multiple loci. Proc. Natl. Acad. Sci. USA 2001, 98, 6709–6713. [Google Scholar] [CrossRef]
- Wolf, J.B.W.; Lindell, J.; Backstrom, N. Speciation genetics: Current status and evolving approaches. Phil. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 1717–1733. [Google Scholar] [CrossRef] [PubMed]
- Phifer-Rixley, M. Searching for the genes that separate species. eLife 2014, 3, e05377. [Google Scholar] [CrossRef] [PubMed]
- Roberts, E.K.; Tardif, S.; Wright, E.A.; Platt II, R.N.; Bradley, R.D.; Hardy, D.M. Rapid divergence of a gamete recognition gene promoted macroevolution of Eutheria. Gen. Biol. 2022, 23, 155. [Google Scholar] [CrossRef]
- Hardy, D.M.; Garbers, D.L. Species-specific binding of sperm proteins to the extracellular matrix (zona pellucida) of the egg. J. Biol. Chem. 1994, 269, 19000–19004. [Google Scholar] [CrossRef]
- Hardy, D.M.; Garbers, D.L. A sperm membrane protein that binds in a species-specific manner to the egg extracellular matrix is homologous to von Willebrand factor. J. Biol. Chem. 1995, 270, 26025–26028. [Google Scholar] [CrossRef]
- Gao, Z.; Garbers, D.L. Species diversity in the structure of zonadhesin, a sperm-specific membrane protein containing multiple cell adhesion molecule-like domains. J. Biol. Chem. 1998, 273, 3415–3421. [Google Scholar] [CrossRef]
- Herlyn, H.; Zischler, H. Tandem repetitive D domains of the sperm ligand zonadhesin evolve faster in the paralogue than in the orthologue comparison. J. Mol. Evol. 2006, 63, 602–611. [Google Scholar] [CrossRef][Green Version]
- Tardif, S.; Wilson, M.D.; Wagner, R.; Hunt, P.; Gertsenstein, M.; Nagy, A.; Lobe, C.; Koop, B.F.; Hardy, D.M. Zonadhesin is essential for species specificity of sperm adhesion to the egg zona pellucida. J. Biol. Chem. 2010, 285, 24863–24870. [Google Scholar] [CrossRef]
- Legan, P.K.; Rau, A.; Kee, J.N.; Richardson, G.P. The mouse tectorins modular matrix proteins of the inner ear homologous to components of the sperm-egg adhesion system. J. Biol. Chem. 1997, 272, 8791–8801. [Google Scholar] [CrossRef] [PubMed]
- Wyckoff, G.J.; Wang, W.; Wu, C.-I. Rapid evolution of male reproductive genes in the descent of man. Nature 2000, 403, 304–309. [Google Scholar] [CrossRef]
- Swanson, W.J.; Vacquier, V.D. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 2002, 3, 137–144. [Google Scholar] [CrossRef]
- Swanson, W.J.; Nielsen, R.; Yang, Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 2003, 20, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Dufourny, L.; Levasseur, A.; Martine, M.; Callebaut, I.; Pontarotti, P.; Malpaux, B.; Monget, P. GPR50 is the mammalian ortholog of Mel1c: Evidence of rapid evolution in mammals. BMC Evol. Biol. 2008, 8, 105. [Google Scholar] [CrossRef]
- Raterman, D.; Springer, M.S. The molecular evolution of acrosin in placental mammals. Mol. Reprod. Dev. 2008, 75, 1196–1207. [Google Scholar] [CrossRef]
- Turner, L.M.; Chuong, E.B.; Hoekstra, H.E. Comparative analysis of testis protein evolution in rodents. Genetics 2008, 179, 2075–2089. [Google Scholar] [CrossRef]
- Waddell, L.A.; Lefevre, L.; Bush, S.J.; Raper, A.; Young, R.; Lisowski, Z.M.; McCulloch, M.E.B.; Muriuki, C.; Sauter, K.A.; Clark, E.L.; et al. ADGRE1 (EMR1, F4/80) is a rapidly-evolving gene expressed in mammalian monocyte-macrophages. Front. Immunol. 2008, 9, 2246. [Google Scholar] [CrossRef]
- Oliver, P.L.; Goodstadt, L.; Bayes, J.J.; Birtle, Z.; Roach, K.C.; Phadnis, N.; Beatson, S.A.; Lunter, G.; Malik, H.S.; Ponting, C.P. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009, 5, e1000753. [Google Scholar] [CrossRef] [PubMed]
- Finn, S.; Civetta, A. Sexual selection and the molecular evolution of ADAM proteins. J. Mol. Evol. 2010, 71, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Grayson, P.; Civetta, A. Positive selection in the adhesion domain of Mus sperm Adam genes through gene duplications and function-driven gene complex formations. BMC Evol. Biol. 2013, 13, 217. [Google Scholar] [CrossRef] [PubMed]
- Grayson, P. Izumo1 and Juno: The evolutionary origins and coevolution of essential sperm-egg binding partners. R. Soc. Open Sci. 2015, 2, 150296. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, H.Q.; Yao, Y.F.; Liu, W.; Ni, Q.Y.; Zhang, M.W.; Xu, H.L. Uneven evolutionary rate of the melatonin-related receptor gene (GPR50) in primates. Genet. Mol. Res. 2015, 14, 680–690. [Google Scholar] [CrossRef]
- Goodwin, Z.A.; de Guzman, S.C. Recent positive selection in genes of the mammalian epidermal differentiation complex locus. Front. Genet. 2017, 7, 227. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Campbell, M.C.; Li, N.; Lee, D.S.W.; Zhang, Z.; Townsend, J.P. Detection of regional variation in selection intensity within protein-coding genes using DNA sequence polymorphism and divergence. Mol. Biol. Evol. 2017, 34, 3006–3022. [Google Scholar] [CrossRef] [PubMed]
- Cantsilieris, S.; Nelson, B.J.; Huddleston, J.; Baker, C.; Harshman, L.; Penewit, K.; Munson, K.M.; Sorenson, M.; Welch, A.E.; Dang, V.; et al. Recurrent structural variation, clustered sites of selection, and disease risk for the complement factor H (CFH) gene family. Proc. Natl. Acad. Sci. USA 2018, 115, E4433–E4442. [Google Scholar] [CrossRef]
- Moros-Nicolás, C.; Fouchécourt, S.; Goudet, G.; Monget, P. Genes encoding mammalian oviductal proteins involved in fertilization are subjected to gene death and positive selection. J. Mol. Evol. 2018, 86, 655–667. [Google Scholar] [CrossRef]
- Campbell, V.; Lapointe, F. An application of supertree methods to Mammalian mitogenomic sequences. Evol. Bioinform. Online 2010, 6, 57–71. [Google Scholar] [CrossRef]
- Scornavacca, C.; Zickmann, F.; Huson, D.H. Tanglegrams for rooted phylogenetic trees and networks. Bioinformatics 2011, 27, i248–i256. [Google Scholar] [CrossRef]
- Venkatachalam, B.; Gusfield, D. Generalizing Tanglegrams. 2018. Available online: https://scholarship.org/uc/item/0bg5p8ch (accessed on 3 October 2023).
- Shimodaira, H. An Approximately Unbiased Test of Phylogenetic Tree Selection. Syst. Biol. 2002, 51, 492–508. [Google Scholar] [CrossRef]
- Webb, C.O.; Donoghue, M.J. Phylomatic: Tree assembly for applied phylogenetics. Mol. Ecol. Notes 2005, 5, 181–183. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony, Version 4.0 b10. 2003. Available online: http://phylosolutions.com/paup-test/ (accessed on 11 October 2023).
- Xu, B.; Yang, Z. PAMLX: A graphical user interface for PAML. Mol. Biol. Evol. 2013, 30, 2723–2724. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Rose, K.D.; Archibald, J.D. The Rise of Placental Mammals: Origins and Relationships of the Major Extant Clades; JHU Press: Baltimore, MD, USA, 2005; p. 65. [Google Scholar]
- Seiffert, E.; Guillon, J.M. A new estimate of afrotherian phylogeny based on simultaneous analysis of genomic, morphological, and fossil evidence. BMC Evol. Biol. 2007, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Rose, K.D. The Beginning of the Age of Mammals; JHU Press: Baltimore, MD, USA, 2006; pp. 242–243. [Google Scholar]
- Cooper, L.N.; Seiffert, E.R.; Clementz, M.; Madar, S.I.; Bajpai, S.; Hussain, S.T.; Thewissen, J.G.M. Anthracobunids from the Middle Eocene of India and Pakistan Are Stem Perissodactyls. PLoS ONE 2014, 9, e109232. [Google Scholar] [CrossRef]
- Zhou, X.; Xu, S.; Xu, J.; Chen, B.; Zhou, K.; Yang, G. Phylogenomic analysis resolves the interordinal relationships and rapid diversification of the Laurasiatherian mammals. Syst. Biol. 2012, 61, 150–164. [Google Scholar] [CrossRef]
- Ursing, B.M.; Arnason, U. Analyses of mitochondrial genomes strongly support a hippopotamus-whale clade. Proc. R. Soc. B 1998, 265, 2251–2255. [Google Scholar] [CrossRef] [PubMed]
- Asher, R.J.; Helgen, K.M. Nomenclature and placental mammal phylogeny. BMC Evol. Biol. 2010, 6, 93. [Google Scholar] [CrossRef] [PubMed]
- Kriegs, J.O.; Churakov, G.; Jurka, J.; Brosius, J.; Schmitz, J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Tren. Genet. 2007, 23, 158–161. [Google Scholar] [CrossRef]
- Meng, J. The osteology of Rhombomylus (Mammalia, Glires): Implications for phylogeny and evolution of Glires. Bull. Am. Mus. Nat. Hist. 2003, 275, 1–247. [Google Scholar] [CrossRef]
- Meng, J.; Wyss, A.R. The morphology of Tribosphenomys (Rodentiaformes, Mammalia): Phylogenetic Implications for Basal Glires. J. Mam. Evol. 2001, 8, 1–71. [Google Scholar] [CrossRef]
- McKenna, M.C.; Bell, S.K. Classification of Mammals: Above the Species Level; Columbia University Press: New York, NY, USA, 1997. [Google Scholar]
- Burgin, C.J.; Colella, J.P.; Kahn, P.L.; Upham, N.S. How many mammal species are there? J. Mamm. 2019, 99, 1–14. [Google Scholar] [CrossRef]
- Teeling, E.C.; Hedges, S.B. Making the impossible possible: Rooting the tree of placental mammals. Mol. Biol. Evol. 2013, 30, 1999–2000. [Google Scholar] [CrossRef]
- Van Dijk, M.A.M.; Paradis, E.; Catzeflis, F.; de Jong, W.W. The Virtues of Gaps: Xenarthran (Edentate) Monophyly Supported by a Unique Deletion in αA-Crystallin. Syst. Biol. 1999, 48, 94–106. [Google Scholar] [CrossRef]
- Waddell, P.J.; Cao, Y.; Hauf, J.; Hasegawa, M. Using novel phylogenetic methods to evaluate mammalian mtDNA, including amino acid-invariant sites-LogDet plus site stripping, to detect internal conflicts in the data, with special reference to the positions of hedgehog, armadillo, and elephant. Syst. Biol. 1999, 48, 31–53. [Google Scholar] [CrossRef]
- Madsen, O.; Deen, P.M.; Pesole, G.; Saccone, C.; de Jong, W.W. Molecular evolution of mammalian aquaporin-2: Further evidence that elephant shrew and aardvark join the paenungulate clade. Mol. Biol. Evol. 1997, 14, 363–371. [Google Scholar] [CrossRef]
- Nikaido, M.; Nishihara, H.; Hukumoto, Y.; Okada, N. Ancient SINEs from African Endemic Mammals. Mol. Biol. Evol. 2003, 20, 522–527. Erratum in Mol. Biol. Evol. 2003, 20, 1181. https://doi.org/10.1093/molbev/msg161. [CrossRef]
- Murphy, W.J.; O’Brien, S. Designing and optimizing comparative anchor primers for comparative gene mapping and phylogenetic inference. Nat. Protoc. 2007, 2, 3022–3030. [Google Scholar] [CrossRef]
- Yang, F.; Alkalaeva, E.Z.; Perelman, P.L.; Pardini, A.T.; Harrison, W.R.; O’Brien, P.M.C.; Fu, B.; Graphodatsky, A.S.; Ferguson-Smith, M.A.; Robinson, T.J. Reciprocal chromosome painting among human, aardvark, and elephant (superorder Afrotheria) reveals the likely eutherian ancestral karyotype. Proc. Natl. Acad. Sci. USA 2003, 100, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- Fröenicke, L.; Wienberg, J.; Stone, G.; Adams, L.; Stanyon, R. Towards the delineation of the ancestral eutherian genome organization: Comparative genome maps of human and the African elephant (Loxodonta africana) generated by chromosome painting. Proc. R. Soc. Lond. B 2003, 207, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Douady, C.J.; Chatelier, P.I.; Madsen, O.; de Jong, W.W.; Catzeflis, F.; Springer, M.S.; Stanhope, M.J. Molecular phylogenetic evidence confirming the Eulipotyphla concept and in support of hedgehogs as the sister group to shrews. Mol. Phylogenet. Evol. 2002, 25, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Puttick, M.N.; Thomas, G.H. Fossils and living taxa agree on patterns of body mass evolution: A case study with Afrotheria. Proc. R. Soc. Lond. B 2015, 282, 20152023. [Google Scholar] [CrossRef]
- Eizirik, E.; Murphy, W.J.; Koepfli, K.; Johnson, W.E.; Dragoo, J.W.; Wayne, R.K.; O’Brien, S.J. Pattern and timing of diversification of the mammalian order Carnivora inferred from multiple nuclear gene sequences. Mol. Phylogenet. Evol. 2010, 56, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Nash, W.G.; Menninger, J.C.; Wienberg, J.; Padilla-Nash, H.M.; O’Brien, S.J. The pattern of phylogenomic evolution of the Canidae. Cytogenet. Cell Genet. 2001, 95, 210–224. [Google Scholar] [CrossRef]
- Van Rheede, T.; Bastiaans, T.; Boone, D.N.; Hedges, S.B.; de Jong, W.W.; Madsen, O. The Platypus Is in Its Place: Nuclear Genes and Indels Confirm the Sister Group Relation of Monotremes and Therians. Mol. Biol. Evol. 2006, 23, 587–597. [Google Scholar] [CrossRef]
- Murphy, W.J.; Pringle, T.H.; Crider, T.A.; Springer, M.S.; Miller, W. Using genomic data to unravel the root of the placental mammal phylogeny. Genom. Res. 2007, 17, 413–421. [Google Scholar] [CrossRef]
- Waters, P.D.; Dobigny, G.; Waddell, P.J.; Robinson, T.J. Evolutionary History of LINE-1 in the Major Clades of Placental Mammals. PLoS ONE. 2007, 2, e158. [Google Scholar] [CrossRef]
- Cao, Y.; Fujiwara, M.; Nikaido, M.; Okada, N.; Hasegawa, M. Interordinal relationships and timescale of Eutherian evolution as inferred from mitochondrial genome data. Gene 2000, 259, 149–158. [Google Scholar] [CrossRef]
- Easteal, S. The pattern of mammalian evolution and the relative rate of molecular evolution. Genetics 1990, 124, 165–173. [Google Scholar] [CrossRef]
- Kullberg, M.; Nilsson, M.A.; Arnason, U.; Harley, E.H.; Janke, A. Housekeeping Genes for Phylogenetic Analysis of Eutherian Relationships. Mol. Biol. Evol. 2006, 23, 1493–1503. [Google Scholar] [CrossRef] [PubMed]
- Huttley, G.A.; Wakefield, M.J.; Easteal, S. Rates of Genome Evolution and Branching Order from Whole Genome Analysis. Mol. Biol. Evol. 2007, 24, 1722–1730. [Google Scholar] [CrossRef]
- Poux, C.; van Rheede, T.; Madsen, O.; de Jong, W.W. Sequence Gaps Join Mice and Men: Phylogenetic Evidence from Deletions in Two Proteins. Mol. Biol. Evol. 2002, 19, 2035–2037. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thomas, J.W.; Touchman, J.W.; Blakesley, R.W.; Bouffard, G.G.; Beckstrom-Sternberg, S.M.; Margulies, E.H.; Blanchette, M.; Siepel, A.C.; Thomas, P.J.; McDowell, J.C.; et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 2003, 424, 788–793. [Google Scholar] [CrossRef] [PubMed]
- Kirkness, E.F.; Bafna, V.; Halpern, A.L.; Levy, S.; Remington, K.; Rusch, D.B. The Dog Genome: Survey Sequencing and Comparative Analysis. Science 2003, 301, 1898–1903. [Google Scholar] [CrossRef]
- Esselstyn, J.A.; Oliveros, C.H.; Swanson, M.T.; Faircloth, B.C. Investigating Difficult Nodes in the Placental Mammal Tree with Expanded Taxon Sampling and Thousands of Ultraconserved Elements. Gen. Biol. Evol. 2017, 9, 2308–2321. [Google Scholar] [CrossRef]
- Zhang, L.; Dong, X.; Tian, X.; Lee, M.; Ablaeva, J.; Firsanov, D.; Lee, S.; Maslov, A.Y.; Gladyshev, V.N.; Seluanov, A.; et al. Maintenance of genome sequence integrity in long- and short-lived rodent species. Sci. Adv. 2021, 7, eabj3284. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, F.; Xu, S.; Yang, G.; Li, M. The position of tree shrews in the mammalian tree: Comparing multi-tree analyses with phylogenomic results leaves monophyly of Euarchonta doubtful. Integr. Zool. 2015, 10, 186–198. [Google Scholar] [CrossRef]
- Kumar, V.; Hallstrom, B.M.; Janke, A. Coalescent-based genome analyses resolve the early branches of the Euarchontoglires. PLoS ONE 2013, 8, e60019. [Google Scholar] [CrossRef]
- Misawa, K.; Nei, M. Reanalysis of Murphy et al.’s Data Gives Various Mammalian Phylogenies and Suggests Overcredibility of Bayesian Trees. J. Mol. Evol. 2003, 57, S290–S296. [Google Scholar] [CrossRef]
- Misawa, K.; Nei, A. Revisiting the Glires concept—Phylogenetic analysis of nuclear sequences. Mol. Phy. Evol. 2003, 28, 320–327. [Google Scholar] [CrossRef]
- Roca, A.L.; Bar-Gal, G.K.; Eizirik, E.; Helgen, K.M.; Maria, R.; Springer, M.S.; O’Brien, S.J.; Murphy, W.J. Mesozoic origin for West Indian insectivores. Nature 2004, 429, 649–651. [Google Scholar] [CrossRef]
- Brace, S.; Thomas, J.A.; Dalén, L.; Burger, J.; MacPhee, R.D.E.; Barnes, I.; Turvey, S.T. Evolutionary History of the Nesophontidae, the Last Unplaced Recent Mammal Family. Mol. Biol. Evol. 2016, 33, 3095–3103. [Google Scholar] [CrossRef]
- Springer, M.S.; Teeling, E.C.; Madsen, O.; de Jong, W.W. Integrated fossil and molecular data reconstruct bat echolocation. Proc. Natl. Acad. Sci. USA 2001, 98, 6241–6246. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Purvis, A.; MacLarnon, A.; Bininda-Emonds, O.R.P.; Simmons, N.B. A phylogenetic supertree of the bats (Mammalia: Chiroptera). Biol. Rev. 2002, 77, 223–259. [Google Scholar] [CrossRef] [PubMed]
- Teeling, E.C.; Springer, M.S.; Madsen, O.; Bates, P.; O’Brien, S.J.; Murphy, W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 2005, 307, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Tsagkogeorga, G.; Parker, J.; Stupka, E.; Cotton, J.A.; Rossiter, S.J. Phylogenomic analyses elucidate the evolutionary relationships of bats. Curr. Biol. 2013, 23, 2262–2267. [Google Scholar] [CrossRef]
- Simmons, N.B.; Seymour, K.L.; Habersetzer, J.; Gunnell, G.F. Primitive Early Eocene bat from Wyoming and the evolution of flight and echolocation. Nature 2008, 451, 818–821. [Google Scholar] [CrossRef]
- Pumo, D.E.; Finamore, P.S.; Franek, W.R.; Phillips, C.J.; Tarzami, S.; Balzarano, D. Complete mitochondrial genome of a neotropical fruit bat, Artibeus jamaicensis, and a new hypothesis of the relationships of bats to other eutherian mammals. J. Mol. Evol. 1998, 47, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Mouchaty, S.K.; Gullberg, A.; Janke, A.; Arnason, U. The phylogenetic position of the Talpidae within Eutheria based on analysis of complete mitochondrial sequences. Mol. Biol. Evol. 2000, 17, 60–67. [Google Scholar] [CrossRef]
- Nikaido, M.; Kawai, K.; Cao, Y.; Harado, M.; Tomita, S.; Tomita, S.; Okada, N.; Hasegawa, M. Maximum Likelihood Analysis of the Complete Genomes of Eutherians and a Reevaluation of the Phylogeny of Bats and Insectivores. J. Mol. Evol. 2001, 53, 508–516. [Google Scholar] [CrossRef]
- Lapointe, F.J.; Kirsch, J.A.W.; Hutcheon, J.M. Total evidence, consensus, and bat phylogeny: A distance-based approach. Mol. Phylogenet. Evol. 1999, 11, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Van den Bussche, R.A.; Hoofer, S.R. Phylogenetic relationships among recent chiropteran families and the importance of choosing appropriate out-group taxa. J. Mamm. 2004, 85, 321–330. [Google Scholar] [CrossRef]
- Agnarsson, I.; Zanrana-Torrelio, C.M.; Flores-Saldana, N.P.; May-Collado, L.J. A time-calibrated species-level phylogeny of bats (Chiroptera, Mammalia). PLoS Curr. 2011, 3, RRN1212. [Google Scholar] [CrossRef] [PubMed]
- Price, S.A.; Bininda-Emonds, O.R.P.; Gittleman, J.L. A complete phylogeny of the whales, dolphins and even-toed hoofed mammals (Cetartiodactyla). Biol. Rev. 2005, 80, 445–473. [Google Scholar] [CrossRef]
- Bininda-Emonds, O.R.P.; Gittleman, J.L.; Purvis, A. Building large trees by combining phylogenetic information: A complete phylogeny of the extant Carnivora (Mammalia). Biol. Rev. Camb. Philos. Soc. 1999, 74, 143–175. [Google Scholar] [CrossRef]
- Finarelli, J.A.; Goswami, A. The evolution of orbit orientation and encephalization in the Carnivora (Mammalia). J. Anat. 2009, 214, 671–678. [Google Scholar] [CrossRef]
- Flynn, J.J.; Finarelli, J.A.; Zehr, S.; Hsu, J.; Nedbal, M.A. Molecular phylogeny of the Carnivora (Mammalia): Assessing the impact of increased sampling on resolving enigmatic relationships. Syst. Biol. 2005, 54, 317–337. [Google Scholar] [CrossRef]
- Arnason, U.; Gullberg, A.; Janke, A.; Kullberg, M. Mitogenomic analyses of caniform relationships. Mol. Phylogenet. Evol. 2007, 45, 863–874. [Google Scholar] [CrossRef]
- Barycka, E. Evolution and systematics of the feliform Carnivora. Mamm. Biol. 2007, 72, 257–282. [Google Scholar] [CrossRef]
- Werdelin, L.; Yamaguchi, N.; Johnson, W.E.; O’Brien, S.J. Phylogeny and evolution of cats (Felidae). In Biology and Conservation of Wild Felids; MacDonald, D.W., Loveridge, A.J., Eds.; Oxford University Press: Oxford, UK, 2010; pp. 59–82. [Google Scholar]
- Wozencraft, W.C. Order Carnivora. In Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed.; Wilson, D.E., Reeder, D.M., Eds.; Johns Hopkins University Press: Baltimore, MD, USA, 2005; pp. 548–559. [Google Scholar]
- Mattern, M.Y.; McLennan, D.A. Phylogeny and Speciation of Felids. Cladistics 2000, 16, 232–253. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Davis, B.W.; Eizirik, E.; Murphy, W.J. Phylogenomic evidence for ancient hybridization in the genomics of living cats (Felidae). Genome Res. 2016, 26, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nyakatura, K.; Bininda-Emonds, O.R.P. Updating the evolutionary history of Carnivora (Mammalia): A new species-level supertree complete with divergence time estimates. BMC Biol. 2012, 10, 12. [Google Scholar] [CrossRef]
- Zinner, D.; Arnold, M.L.; Roos, C. The strange blood: Natural hybridization in primates. Evol. Anthropol. 2011, 20, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Finstermeier, K.; Zinner, D.; Brameier, M.; Meyer, M.; Kreuz, E.; Hofreiter, M. A Mitogenomic Phylogeny of Living Primates. PLoS ONE 2013, 8, e69504. [Google Scholar] [CrossRef]
- Pozzi, L.; Hodgson, J.A.; Burrell, A.S.; Sterner, K.N.; Raaum, R.L.; Disotell, T.R. Primate phylogenetic relationships and divergence dates inferred from complete mitochondrial genomes. Mol. Phylogenet. Evol. 2014, 75, 165–183. [Google Scholar] [CrossRef]
- Cao, Y.; Adachi, J.; Yano, T.; Hasegawa, M. Phylogenetic place of guinea pigs: No support of the rodent-polyphyly hypothesis from maximum-likelihood analyses of multiple protein sequences. Mol. Biol. Evol. 1994, 11, 593–604. [Google Scholar] [CrossRef]
- D’Erchia, A.; Gissi, C.; Pesole, G.; Saccone, C.; Arnason, U. The guinea-pig is not a rodent. Nature 1996, 381, 597–600. [Google Scholar] [CrossRef]
- Graur, D.; Hide, W.; Li, W. Is the guinea-pig a rodent? Nature 1991, 351, 649–652. [Google Scholar] [CrossRef]
- Huchon, D.; Catzeflis, F.M.; Douzery, E.J.P. Variance of molecular datings, evolution of rodents and the phylogenetic affinities between Ctenodactylidae and Hystricognathi. Proc. R. Soc. Lond. B 2000, 267, 393–402. [Google Scholar] [CrossRef]
- Montgelard, C.; Forty, E.; Arnal, V.; Matthee, C.A. Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast-evolving nucleotides in mitochondrial, exon and intron fragments. BMC Evol. Biol. 2008, 8, 321. [Google Scholar] [CrossRef] [PubMed]
- Honeycutt, R.L.; Frabotta, L.J.; Rowe, D.L. Rodent evolution, phylogenetics, and biogeography. In Rodent Societies: An Ecological and Evolutionary Perspective; University of Chicago Press: Chicago, IL, USA, 2007; pp. 8–23. [Google Scholar]
- Honeycutt, R.L. Rodents (Rodentia). In The Timetree of Life; OUP: Oxford, UK, 2009; pp. 490–494. [Google Scholar]
- Alhajeri, B.H.; Hunt, P.J.; Steppan, S.J. Molecular systematics of gerbils and deomyines (Rodentia: Gerbillinae, Deomyinae) and a test of desert adaptation in the tympanic bulla. J. Zoolog. Syst. Evol. Res. 2015, 53, 312–330. [Google Scholar] [CrossRef]
- Li, C.L.; Du, X.Y.; Gao, J.; Wang, C.; Guo, H.G.; Dai, F.W.; Sa, X.Y.; An, W.; Chen, Z.W. Phylogenetic analysis of the Mongolian gerbil (Meriones unguiculatus) from China based on mitochondrial genome. Genet. Mol. Res. 2016, 15, gmr.15037703. [Google Scholar] [CrossRef] [PubMed]
- Ruedi, M.; Mayer, F. Molecular Systematics of Bats of the Genus Myotis (Vespertilionidae) Suggests Deterministic Ecomorphological Convergences. Mol. Phylogenet. Evol. 2001, 21, 436–448. [Google Scholar] [CrossRef]
- Arnold, M.L.; Meyer, A. Natural hybridization in primates: One evolutionary mechanism. Zoology 2006, 109, 261–276. [Google Scholar] [CrossRef]
- Ji, R.; Cui, P.; Ding, F.; Geng, J.; Gao, H.; Zhang, H.; Yu, J.; Hu, S.; Meng, H. Monophyletic origin of domestic Bactrian camel (Camelus bactrianus) and its evolutionary relationship with the extant wild camel (Camelus bactrianus ferus). Anim. Genet. 2009, 40, 377–382. [Google Scholar] [CrossRef]
- Mohandesan, E.; Fitak, R.R.; Corander, J.; Yadamsuren, A.; Chuluunbat, B.; Abdelhadi, O.; Raziq, A.; Nagy, P.; Stalder, G.; Walzer, C.; et al. Mitogenome Sequencing in the Genus Camelus Reveals Evidence for Purifying Selection and Long-term Divergence between Wild and Domestic Bactrian Camels. Sci. Rep. 2017, 7, 9970. [Google Scholar] [CrossRef]
- Legesse, Y.W.; Dunn, C.D.; Mauldin, M.R.; Ordoñez-Garza, N.; Rowden, G.R.; Mekasha Gebre, Y.; Kurtu, M.Y.; Ali, S.M.; Whibesilassie, W.D.; Ballou, M.; et al. Morphometric and genetic variation in 8 breeds of Ethiopian camels (Camelus dromedarius). J. Anim. Sci. 2018, 96, 4925–4934. [Google Scholar] [CrossRef]
- Burger, P.A.; Ciani, E.; Faye, B. Old World camels in a modern world—A balancing act between conservation and genetic improvement. Anim. Genet. 2019, 50, 598–612. [Google Scholar] [CrossRef]
- Zeller, U.; Göttert, T. The relations between evolution and domestication reconsidered—Implications for systematics, ecology, and nature conservation. Glob. Ecol. Cons. 2019, 20, e00756. [Google Scholar] [CrossRef]
- Groves, C.P. Systematic relationships in the Bovini (Artiodactyla, Bovidae). J. Zool. Syst. Evol. Res. 1981, 19, 264–278. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.; Wang, X.; Jiang, Q.; Zhao, H.; Wang, J.; Ju, Z.; Yang, L.; Gao, Y.; Wei, X.; et al. Population Structure and Selection Signatures Underlying High-Altitude Adaptation Inferred From Genome-Wide Copy Number Variations in Chinese Indigenous Cattle. Front. Genet. 2019, 10, 1404. [Google Scholar] [CrossRef] [PubMed]
- D’Elia, G.; Fabre, P.; Lessa, E.P. Rodent systematics in an age of discovery: Recent advances and prospects. J. Mamm. 2019, 100, 852–871. [Google Scholar] [CrossRef]
- Shen, Y.; Liang, L.; Zhu, Z.; Zhou, E.; Irwin, D.M.; Zhang, Y. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. USA 2010, 11, 8666–8671. [Google Scholar] [CrossRef] [PubMed]
- Hecker, N.; Sharma, V.; Hiller, M. Convergent gene losses illuminate metabolic and physiological changes in herbivores and carnivores. Proc. Natl. Acad. Sci. USA 2019, 116, 3036–3041. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. Diet evolution of carnivorous and herbivorous mammals in Laurasiatheria. BMC Ecol. Evol. 2022, 22, 82. [Google Scholar] [CrossRef]
- Fu, Y. Estimating Mutation Rate and Generation Time from Longitudinal Samples of DNA Sequences. Mol. Biol. Evol. 2001, 18, 620–626. [Google Scholar] [CrossRef][Green Version]
- Okazaki, A.; Yamazaki, S.; Inoue, I.; Ott, J. Population genetics: Past, present, and future. Hum. Genet. 2021, 140, 231–240. [Google Scholar] [CrossRef]
- Springer, M.S.; Burk, A.; Kavanagh, J.R.; Waddell, V.G.; Stanhope, M.J. The interphotoreceptor retinoid binding protein gene in therian mammals: Implications for higher level relationships and evidence for loss of function in the marsupial mole. Proc. Natl. Acad. Sci. USA 1997, 9, 13754–13759. [Google Scholar] [CrossRef]
- Bradley, R.D.; Baker, R.J. A Test of the Genetic Species Concept: Cytochrome-b Sequences and Mammals. J. Mamm. 2001, 82, 960–973. [Google Scholar] [CrossRef]
- Chan, A.H.; Chaisiri, K.; Saralamba, S.; Morand, S.; Thaenkham, U. Assessing the suitability of mitochondrial and nuclear DNA genetic markers for molecular systematics and species identification. Parasites Vectors 2021, 14, 233. [Google Scholar] [CrossRef] [PubMed]



| Gene | Species | Families | Length (nt) | Gene | Species | Families | Length (nt) |
|---|---|---|---|---|---|---|---|
| Acr | 145 | 60 | 2133 | Spaca6 | 133 | 51 | 2787 |
| Adam2 | 105 | 51 | 2413 | Spam1 | 120 | 50 | 2288 |
| Adam18 | 82 | 41 | 2939 | Spink2 | 147 | 55 | 646 |
| Adam32 | 106 | 47 | 3378 | Sprr4 | 119 | 50 | 414 |
| Adgre1 | 147 | 57 | 6871 | Tas1r2 | 85 | 42 | 2370 |
| C5ar1 | 146 | 59 | 1812 | Tchhl1 | 137 | 54 | 3294 |
| Ccdc54 | 155 | 62 | 1749 | Tcn1 | 132 | 60 | 1833 |
| Ccl1 | 140 | 55 | 465 | Tcte1 | 162 | 65 | 1972 |
| Cfh | 95 | 45 | 4884 | Tecta | 110 | 61 | 4317 |
| Cr2 | 118 | 46 | 8016 | Tectb | 166 | 63 | 1102 |
| Cytb | 114 | 62 | 1095 | Tpx1 | 87 | 38 | 846 |
| Gpr50 | 155 | 62 | 2798 | Tex11 | 130 | 53 | 4760 |
| Izumo1 | 144 | 60 | 2150 | Tex14 | 166 | 67 | 4518 |
| Izumo1R | 159 | 59 | 1605 | Tnp2 | 124 | 49 | 1213 |
| Lelp1 | 131 | 55 | 549 | Usp26 | 139 | 58 | 3389 |
| Man2b1 | 103 | 48 | 4368 | Wbp2nl | 155 | 60 | 1916 |
| Prdm9 | 57 | 27 | 2872 | Zan | 170 | 66 | 6873 |
| Prm1 | 66 | 34 | 890 | Zp2 | 125 | 59 | 3639 |
| Prm2 | 77 | 28 | 609 | Zp3 | 159 | 64 | 1667 |
| S100a2 | 137 | 59 | 394 | ||||
| Slc6a5 | 103 | 57 | 3128 | Total: | 104,962 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roberts, E.K.; Wright, E.A.; Worsham, A.E.; Hardy, D.M.; Bradley, R.D. Gamete Recognition Gene Divergence Yields a Robust Eutherian Phylogeny across Taxonomic Levels. Diversity 2023, 15, 1145. https://doi.org/10.3390/d15111145
Roberts EK, Wright EA, Worsham AE, Hardy DM, Bradley RD. Gamete Recognition Gene Divergence Yields a Robust Eutherian Phylogeny across Taxonomic Levels. Diversity. 2023; 15(11):1145. https://doi.org/10.3390/d15111145
Chicago/Turabian StyleRoberts, Emma K., Emily A. Wright, Asha E. Worsham, Daniel M. Hardy, and Robert D. Bradley. 2023. "Gamete Recognition Gene Divergence Yields a Robust Eutherian Phylogeny across Taxonomic Levels" Diversity 15, no. 11: 1145. https://doi.org/10.3390/d15111145
APA StyleRoberts, E. K., Wright, E. A., Worsham, A. E., Hardy, D. M., & Bradley, R. D. (2023). Gamete Recognition Gene Divergence Yields a Robust Eutherian Phylogeny across Taxonomic Levels. Diversity, 15(11), 1145. https://doi.org/10.3390/d15111145