Museomics Provides Insights into Conservation and Education: The Instance of an African Lion Specimen from the Museum of Zoology “Pietro Doderlein”
Abstract
1. Introduction
2. Materials and Methods
2.1. The History of the Museum Collection and the Lion Sample
2.2. 3D Printing of Missing Skeletal Fragments and Restoration
2.3. Sampling and DNA Extraction
2.4. DNA Amplification and Sequencing
2.5. Data Analysis
3. Results
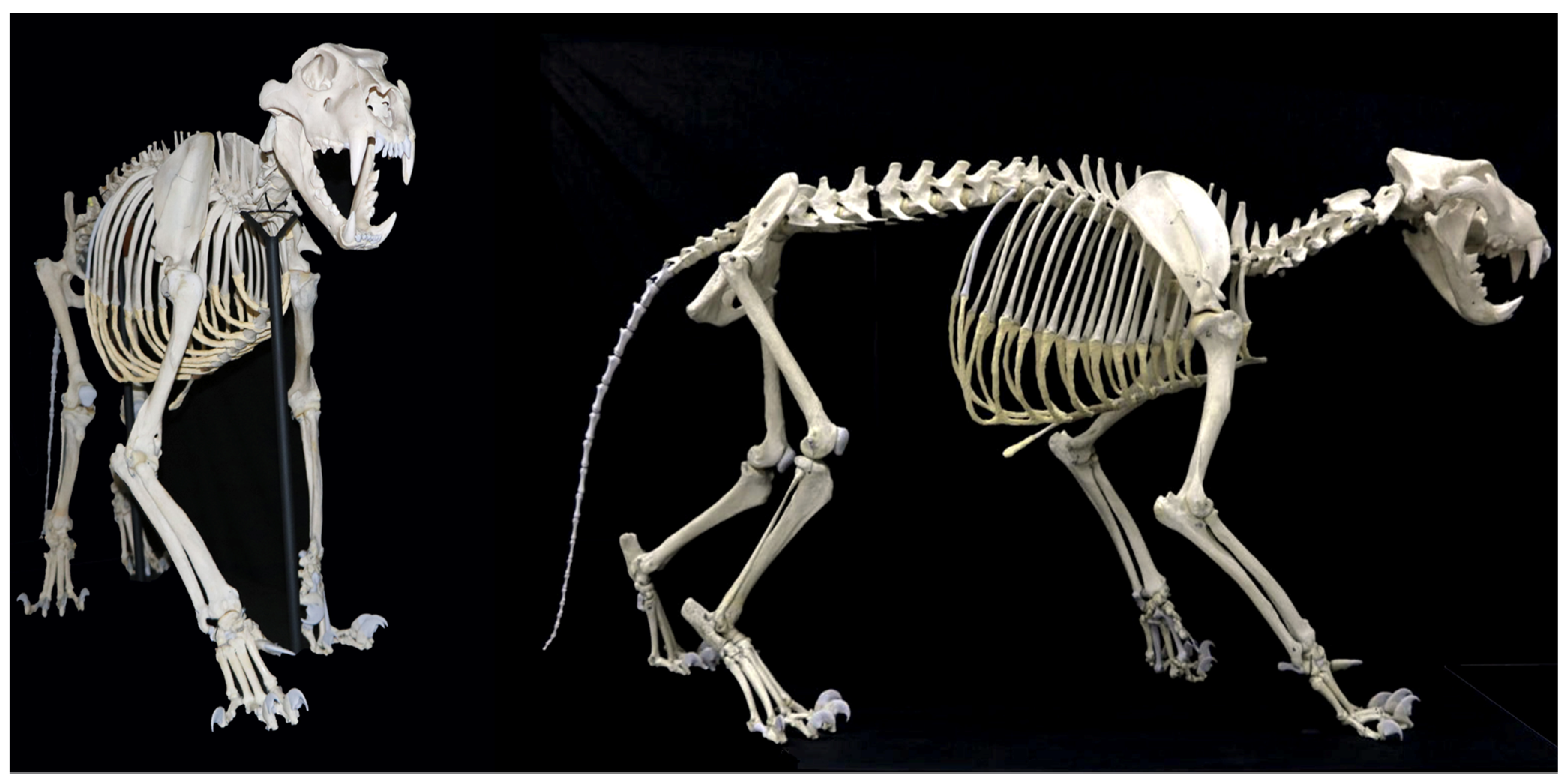
3.1. Skeletal Reconstruction and Exhibition
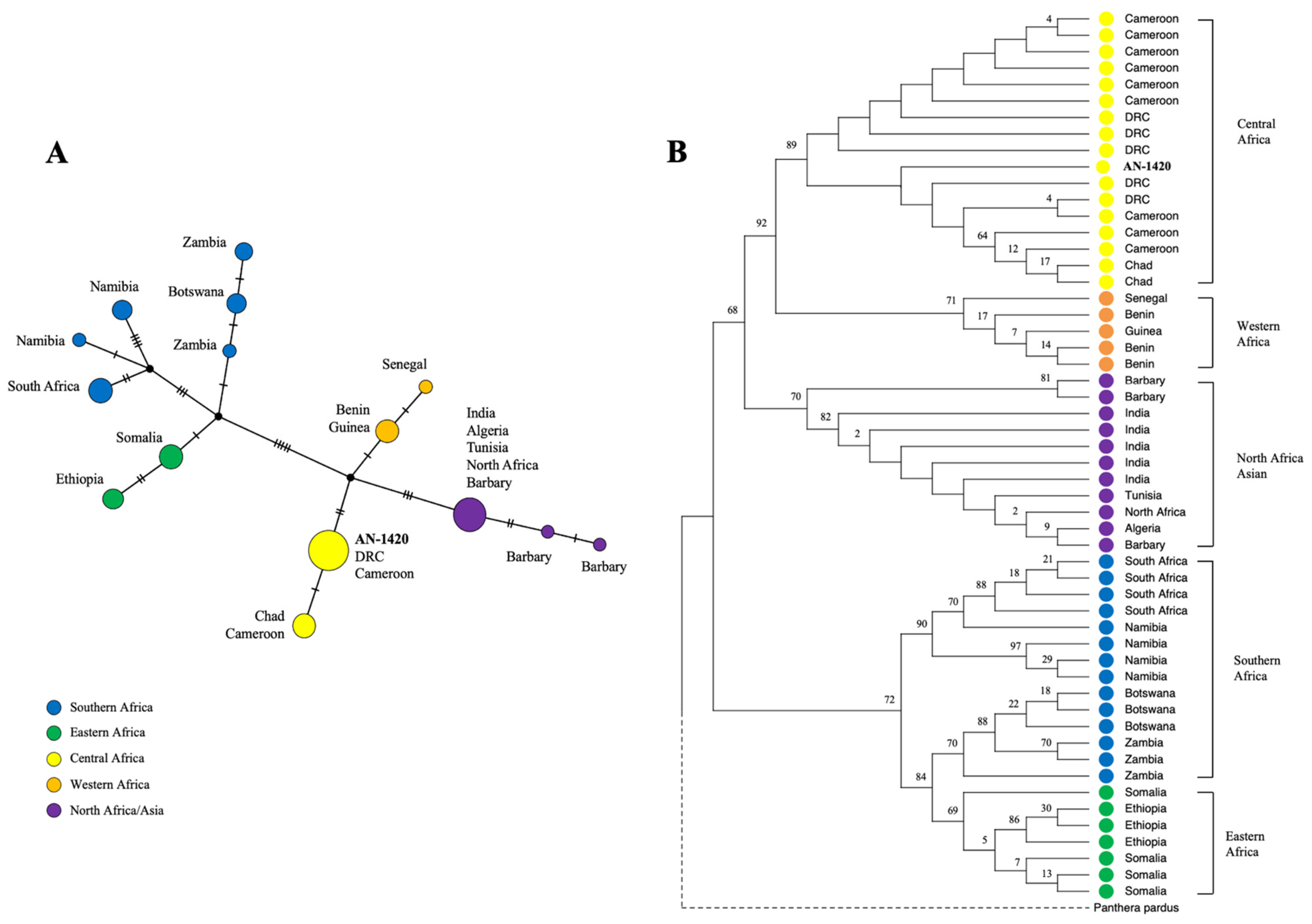
3.2. Phylogeographical Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Lo Brutto, S.; Puleio, R.; Vicari, D.; Calascibetta, A.; Bellia, E.; Bellante, A.; Giacalone, V.; Buffa, G. Dal Mare Al Museo: Il Recupero Di Uno Scheletro Di Stenella coeruleoalba (Meyen 1833). Museol. Sci. 2022, 16, 62–69. [Google Scholar]
- Movalli, P.; Koschorreck, J.; Treu, G.; Slobodnik, J.; Alygizakis, N.; Androulakakis, A.; Badry, A.; Baltag, E.; Barbagli, F.; Bauer, K.; et al. The Role of Natural Science Collections in the Biomonitoring of Environmental Contaminants in Apex Predators in Support of the EU’s Zero Pollution Ambition. Environ. Sci. Eur. 2022, 34, 1–7. [Google Scholar] [CrossRef]
- Tiralongo, F.; Badalamenti, R.; Arizza, V.; Prieto, L.; Lo Brutto, S. The Portuguese Man-of-War Has Always Entered the Mediterranean Sea—Strandings, Sightings, and Museum Collections. Front. Mar. Sci. 2022, 9, 377. [Google Scholar] [CrossRef]
- Curry, C.J.; Davis, B.W.; Bertola, L.D.; White, P.A.; Murphy, W.J.; Derr, J.N. Spatiotemporal Genetic Diversity of Lions Reveals the Influence of Habitat Fragmentation across Africa. Mol. Biol. Evol. 2021, 38, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Lo Brutto, S.; Calascibetta, A.; Pavan, G.; Buffa, G. Cetacean Strandings and Museum Collections: A Focus on Sicily Island Crossroads for Mediterranean Species. Diversity 2021, 13, 104. [Google Scholar] [CrossRef]
- Yeates, D.K.; Zwick, A.; Mikheyev, A.S. Museums Are Biobanks: Unlocking the Genetic Potential of the Three Billion Specimens in the World’s Biological Collections. Curr. Opin. Insect Sci. 2016, 18, 83–88. [Google Scholar] [CrossRef]
- Albert, C.; Luque, G.M.; Courchamp, F. The Twenty Most Charismatic Species. PLoS ONE 2018, 13, e0199149. [Google Scholar] [CrossRef]
- Bauer, H.; Packer, C.; Funston, P.F.; Henschel, P.; Nowell, K. Panthera leo. IUCN Red List Threat. Species 2016, e. T15951A115130419. [Google Scholar] [CrossRef]
- Henschel, P.; Coad, L.; Burton, C.; Chataigner, B.; Dunn, A.; MacDonald, D.; Saidu, Y.; Hunter, L.T.B. The Lion in West Africa Is Critically Endangered. PLoS ONE 2014, 9, e83500. [Google Scholar] [CrossRef]
- Straube, N.; Lyra, M.L.; Paijmans, J.L.A.; Preick, M.; Basler, N.; Penner, J.; Rödel, M.O.; Westbury, M.V.; Haddad, C.F.B.; Barlow, A.; et al. Successful Application of Ancient DNA Extraction and Library Construction Protocols to Museum Wet Collection Specimens. Mol. Ecol. Resour. 2021, 21, 2299–2315. [Google Scholar] [CrossRef]
- Roycroft, E.; Moritz, C.; Rowe, K.C.; Moussalli, A.; Eldridge, M.D.B.; Portela Miguez, R.; Piggott, M.P.; Potter, S. Sequence Capture From Historical Museum Specimens: Maximizing Value for Population and Phylogenomic Studies. Front. Ecol. Evol. 2022, 10, 667. [Google Scholar] [CrossRef]
- Kapp, J.D.; Green, R.E.; Shapiro, B. A Fast and Efficient Single-Stranded Genomic Library Preparation Method Optimized for Ancient DNA. J. Hered. 2021, 112, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Rohland, N.; Glocke, I.; Aximu-Petri, A.; Meyer, M. Extraction of Highly Degraded DNA from Ancient Bones, Teeth and Sediments for High-Throughput Sequencing. Nat. Protoc. 2018, 13, 2447–2461. [Google Scholar] [CrossRef] [PubMed]
- Puncher, G.N.; Cariani, A.; Cilli, E.; Massari, F.; Leone, A.; Morales-Muñiz, A.; Onar, V.; Toker, N.Y.; Bernal Casasola, D.; Moens, T.; et al. Comparison and Optimization of Genetic Tools Used for the Identification of Ancient Fish Remains Recovered from Archaeological Excavations and Museum Collections in the Mediterranean Region. Int. J. Osteoarchaeol. 2019, 29, 365–376. [Google Scholar] [CrossRef]
- Agne, S.; Naylor, G.J.P.; Preick, M.; Yang, L.; Thiel, R.; Weigmann, S.; Paijmans, J.L.A.; Barlow, A.; Hofreiter, M.; Straube, N. Taxonomic Identification of Two Poorly Known Lantern Shark Species Based on Mitochondrial DNA From Wet-Collection Paratypes. Front. Ecol. Evol. 2022, 10, 569. [Google Scholar] [CrossRef]
- Angelici, F.; Ciucani, M.; Angelini, S.; Annesi, F.; Caniglia, R.; Castiglia, R.; Fabbri, E.; Galaverni, M.; Palumbo, D.; Ravegnini, G.; et al. The Sicilian Wolf: Genetic Identity of a Recently Extinct Insular Population. Zoolog. Sci. 2019, 36, 189–197. [Google Scholar] [CrossRef]
- Leone, A.; Puncher, G.N.; Ferretti, F.; Sperone, E.; Tripepi, S.; Micarelli, P.; Gambarelli, A.; Sarà, M.; Arculeo, M.; Doria, G.; et al. Pliocene Colonization of the Mediterranean by Great White Shark Inferred from Fossil Records, Historical Jaws, Phylogeographic and Divergence Time Analyses. J. Biogeogr. 2020, 47, 1119–1129. [Google Scholar] [CrossRef]
- Koupadi, K.; Fontani, F.; Ciucani, M.M.; Maini, E.; De Fanti, S.; Cattani, M.; Curci, A.; Nenzioni, G.; Reggiani, P.; Andrews, A.J.; et al. Population Dynamics in Italian Canids between the Late Pleistocene and Bronze Age. Genes 2020, 11, 1409. [Google Scholar] [CrossRef]
- Straube, N.; Preick, M.; Naylor, G.J.P.; Hofreiter, M. Mitochondrial DNA Sequencing of a Wet-Collection Syntype Demonstrates the Importance of Type Material as Genetic Resource for Lantern Shark Taxonomy (Chondrichthyes: Etmopteridae). R. Soc. Open Sci. 2021, 8, 210474. [Google Scholar] [CrossRef]
- Ciucani, M.M.; Palumbo, D.; Galaverni, M.; Serventi, P.; Fabbri, E.; Ravegnini, G.; Angelini, S.; Maini, E.; Persico, D.; Caniglia, R.; et al. Old Wild Wolves: Ancient DNA Survey Unveils Population Dynamics in Late Pleistocene and Holocene Italian Remains. PeerJ 2019, 7, e6424. [Google Scholar] [CrossRef]
- Ciucani, M.M.; Jensen, J.K.; Sinding, M.H.S.; Smith, O.; Lucenti, S.B.; Rosengren, E.; Rook, L.; Tuveri, C.; Arca, M.; Cappellini, E.; et al. Evolutionary History of the Extinct Sardinian Dhole. Curr. Biol. 2021, 31, 5571–5579.e6. [Google Scholar] [CrossRef] [PubMed]
- Andrews, A.J.; Di Natale, A.; Bernal-Casasola, D.; Aniceti, V.; Onar, V.; Oueslati, T.; Theodropoulou, T.; Morales-Muñiz, A.; Cilli, E.; Tinti, F. Exploitation History of Atlantic Bluefin Tuna in the Eastern Atlantic and Mediterranean—Insights from Ancient Bones. ICES J. Mar. Sci. 2022, 79, 247–262. [Google Scholar] [CrossRef]
- Lalueza-Fox, C. Museomics. Curr. Biol. 2022, 32, R1214–R1215. [Google Scholar] [CrossRef] [PubMed]
- Mondol, S.; Bruford, M.W.; Ramakrishnan, U. Demographic Loss, Genetic Structure and the Conservation Implications for Indian Tigers. Proc. R. Soc. B Biol. Sci. 2013, 280, 20130496. [Google Scholar] [CrossRef] [PubMed]
- Palkopoulou, E.; Lipson, M.; Mallick, S.; Nielsen, S.; Rohland, N.; Baleka, S.; Karpinski, E.; Ivancevic, A.M.; To, T.H.; Daniel Kortschak, R.; et al. A Comprehensive Genomic History of Extinct and Living Elephants. Proc. Natl. Acad. Sci. USA 2018, 115, E2566–E2574. [Google Scholar] [CrossRef]
- Andrews, A.J.; Puncher, G.N.; Bernal-Casasola, D.; Di Natale, A.; Massari, F.; Onar, V.; Toker, N.Y.; Hanke, A.; Pavey, S.A.; Savojardo, C.; et al. Ancient DNA SNP-Panel Data Suggests Stability in Bluefin Tuna Genetic Diversity despite Centuries of Fluctuating Catches in the Eastern Atlantic and Mediterranean. Sci. Rep. 2021, 11, 20744. [Google Scholar] [CrossRef] [PubMed]
- Pinsky, M.L.; Eikeset, A.M.; Helmerson, C.; Bradbury, I.R.; Bentzen, P.; Morris, C.; Gondek-Wyrozemska, A.T.; Baalsrud, H.T.; Brieuc, M.S.O.; Kjesbu, O.S.; et al. Genomic Stability through Time despite Decades of Exploitation in Cod on Both Sides of the Atlantic. Proc. Natl. Acad. Sci. USA 2021, 118, e2025453118. [Google Scholar] [CrossRef]
- Ciucani, M.M.; Ramos-Madrigal, J.; Hernández-Alonso, G.; Carmagnini, A.; Aninta, S.G.; Scharff-Olsen, C.H.; Lanigan, L.T.; Fracasso, I.; Clausen, C.G.; Aspi, J.; et al. Genomes of the Extinct Sicilian Wolf Reveal a Complex History of Isolation and Admixture with Ancient Dogs. bioRxiv 2022, 21, 477289. [Google Scholar] [CrossRef]
- Hempel, E.; Bibi, F.; Faith, J.T.; Koepfli, K.-P.; Klittich, A.M.; Duchêne, D.A.; Brink, J.S.; Kalthoff, D.C.; Dalén, L.; Hofreiter, M.; et al. Blue Turns to Grey—Palaeogenomic Insights into the Evolutionary History and Extinction of the Blue Antelope (Hippotragus Leucophaeus). Mol. Biol. Evol. 2022, 39, msac241. [Google Scholar] [CrossRef]
- Tedesco, M.R.; Mandalà, G.; Consentino, M.C.; Ienti, R.; Lo Valvo, F.; Movalli, P.; Sabella, G.; Spadola, F.; Spinnato, A.; Toscano, F.; et al. The First Inventory of Birds of Prey in Sicilian Museum Collections (Italy). Museol. Sci. 2020, 14, 67–80. [Google Scholar] [CrossRef]
- Llamas, B.; Valverde, G.; Fehren-Schmitz, L.; Weyrich, L.S.; Cooper, A.; Haak, W. From the Field to the Laboratory: Controlling DNA Contamination in Human Ancient DNA Research in the High-Throughput Sequencing Era. STAR Sci. Technol. Archaeol. Res. 2017, 3, 1–14. [Google Scholar] [CrossRef]
- Higgins, D.; Austin, J.J. Teeth as a Source of DNA for Forensic Identification of Human Remains: A Review. Sci. Justice 2013, 53, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Allentoft, M.E.; Sikora, M.; Sjogren, K.-G.; Rasmussen, S.; Rasmussen, M.; Stenderup, J.; Damgaard, P.B.; Schroeder, H.; Ahlstrom, T.; Vinner, L.; et al. Population Genomics of Bronze Age Eurasia. Nature 2015, 522, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.; Poinar, H.N. Ancient DNA: Do It Right or Not at All. Science 2000, 289, 1139. [Google Scholar] [CrossRef] [PubMed]
- Fulton, T.L.; Shapiro, B. Setting Up an Ancient DNA Laboratory. Methods Mol. Biol. 2019, 1963, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cilli, E.; Gabanini, G.; Ciucani, M.M.; De Fanti, S.; Serventi, P.; Bazaj, A.; Sarno, S.; Ferri, G.; Fregnani, A.; Cornaglia, G.; et al. A Multifaceted Approach towards Investigating Childbirth Deaths in Double Burials: Anthropology, Paleopathology and Ancient DNA. J. Archaeol. Sci. 2020, 122, 105219. [Google Scholar] [CrossRef]
- Dabney, J.; Knapp, M.; Glocke, I.; Gansauge, M.-T.; Weihmann, A.; Nickel, B.; Valdiosera, C.; Garcia, N.; Paabo, S.; Arsuaga, J.-L.; et al. Complete Mitochondrial Genome Sequence of a Middle Pleistocene Cave Bear Reconstructed from Ultrashort DNA Fragments. Proc. Natl. Acad. Sci. USA 2013, 110, 15758–15763. [Google Scholar] [CrossRef]
- Barnett, R.; Yamaguchi, N.; Shapiro, B.; Ho, S.Y.; Barnes, I.; Sabin, R.; Werdelin, L.; Cuisin, J.; Larson, G. Revealing the Maternal Demographic History of Panthera leo Using Ancient DNA and a Spatially Explicit Genealogical Analysis. BMC Evol. Biol. 2014, 14, 70. [Google Scholar] [CrossRef]
- Castresana, J. Cytochrome b Phylogeny and the Taxonomy of Great Apes and Mammals. Mol. Biol. Evol. 2001, 18, 465–471. [Google Scholar] [CrossRef]
- Merheb, M.; Matar, R.; Hodeify, R.; Siddiqui, S.S.; Vazhappilly, C.G.; Marton, J.; Azharuddin, S.; Zouabi, H.A.L. Mitochondrial DNA, a Powerful Tool to Decipher Ancient Human Civilization from Domestication to Music, and to Uncover Historical Murder Cases. Cells 2019, 8, 433. [Google Scholar] [CrossRef]
- Burger, J.; Rosendahl, W.; Loreille, O.; Hemmer, H.; Eriksson, T.; Götherström, A.; Hiller, J.; Collins, M.J.; Wess, T.; Alt, K.W. Molecular Phylogeny of the Extinct Cave Lion Panthera leo Spelaea. Mol. Phylogenet. Evol. 2004, 30, 841–849. [Google Scholar] [CrossRef]
- Bertola, L.D.; Jongbloed, H.; Van Der Gaag, K.J.; De Knijff, P.; Yamaguchi, N.; Hooghiemstra, H.; Bauer, H.; Henschel, P.; White, P.A.; Driscoll, C.A.; et al. Phylogeographic Patterns in Africa and High Resolution Delineation of Genetic Clades in the Lion (Panthera leo). Sci. Rep. 2016, 6, 30807. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. Popart: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More Models, New Heuristics and High-Performance Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- White, P.A.; Belant, J.L. Individual Variation in Dental Characteristics for Estimating Age of African Lions. Wildlife Biol. 2016, 22, 71–77. [Google Scholar] [CrossRef]
- Naples, V.L.; Rothschild, B.M. Sex Determination in Lions (Panthera leo, Felidae): A Novel Method of Distinguishing Male and Female Skulls. Mammalia 2012, 76, 99–103. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Singh, H.S.; Gibson, L. A Conservation Success Story in the Otherwise Dire Megafauna Extinction Crisis: The Asiatic Lion (Panthera leo Persica) of Gir Forest. Biol. Conserv. 2011, 144, 1753–1757. [Google Scholar] [CrossRef]
- Kitchener, A.C.; Breitenmoser-Würsten, C.; Eizirik, E.; Gentry, A.; Werdelin, L.; Wilting, A.; Yamaguchi, N.; Abramov, A.V.; Christiansen, P.; Driscoll, C.; et al. A Revised Taxonomy of the Felidae: The Final Report of the Cat Classification Task Force of the IUCN Cat Specialist GroupCAT News Spec. Issue 2017, 11, 71–73. [Google Scholar]
- de Manuel, M.; Barnett, R.; Sandoval-Velasco, M.; Yamaguchi, N.; Vieira, F.G.; Lisandra Zepeda Mendoza, M.; Liu, S.; Martin, M.D.; Sinding, M.H.S.; Mak, S.S.T.; et al. The Evolutionary History of Extinct and Living Lions. Proc. Natl. Acad. Sci. USA 2020, 117, 10927–10934. [Google Scholar] [CrossRef] [PubMed]
- Lo Brutto, S.; Battaglia, F.; Biundo, G.; Calascibetta, A.; Conti, F.; Nuccio, A.; Pellerito, F. Un’esperienza Di Divulgazione Condotta Da Studenti Universitari. Museol. Sci. 2020, 14, 133–138. [Google Scholar] [CrossRef]
- Lo Brutto, S. The Case of a Rudderfish Highlights the Role of Natural History Museums as Sentinels of Bio-Invasions. Zootaxa 2017, 4254, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Lo Brutto, S. A Finding at the Natural History Museum of Florence Affords the Holotype Designation of Orchestia stephenseni Cecchini, 1928 (Crustacea: Amphipoda: Talitridae). Zootaxa 2017, 4231, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Movalli, P.; Duke, G.; Ramello, G.; Dekker, R.; Vrezec, A.; Shore, R.F.; García-Fernández, A.; Wernham, C.; Krone, O.; Alygizakis, N.; et al. Progress on Bringing Together Raptor Collections in Europe for Contaminant Research and Monitoring in Relation to Chemicals Regulation. Environ. Sci. Pollut. Res. 2019, 26, 20132–20136. [Google Scholar] [CrossRef]
- Suarez, A.V.; Tsutsui, N.D. The Value of Museum Collections for Research and Society. Bioscience 2004, 54, 66–74. [Google Scholar] [CrossRef]
- Cook, J.A.; Light, J.E. The Emerging Role of Mammal Collections in 21st Century Mammalogy. J. Mammal. 2019, 100, 733–750. [Google Scholar] [CrossRef]
- Bradley, R.D.; Bradley, L.C.; Garner, H.J.; Baker, R.J. Assessing the Value of Natural History Collections and Addressing Issues Regarding Long-Term Growth and Care. Bioscience 2014, 64, 1150–1158. [Google Scholar] [CrossRef]
- Mannino, A.M.; Balistreri, P.; Iaciofano, D.; Galil, B.S.; Lo Brutto, S. An Additional Record of Kyphosus vaigiensis (Quoy & Gaimard, 1825) (Osteichthyes, Kyphosidae) from Sicily Clarifies the Confused Situation of the Mediterranean Kyphosids. Zootaxa 2015, 3963, 45–54. [Google Scholar] [CrossRef]
- Jensen, E.L.; Díez-del-Molino, D.; Gilbert, M.T.P.; Bertola, L.D.; Borges, F.; Cubric-Curik, V.; de Navascués, M.; Frandsen, P.; Heuertz, M.; Hvilsom, C.; et al. Ancient and Historical DNA in Conservation Policy. Trends Ecol. Evol. 2022, 37, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Pales, L.; Lambert, C. Atlas Ostéologique Pour Servir à l’identification Des Mammifères Du Quaternaire; Editions du CNRS: Paris, France, 1971. [Google Scholar]
- Bertola, L.D.; van Hooft, W.F.; Vrieling, K.; Uit de Weerd, D.R.; York, D.S.; Bauer, H.; Prins, H.H.T.; Funston, P.J.; Udo de Haes, H.A.; Leirs, H.; et al. Genetic Diversity, Evolutionary History and Implications for Conservation of the Lion (Panthera leo) in West and Central Africa. J. Biogeogr. 2011, 38, 1356–1367. [Google Scholar] [CrossRef]
- Bertola, L.D.; Tensen, L.; Van Hooft, P.; White, P.A.; Driscoll, C.A.; Henschel, P.; Caragiulo, A.; Dias-Freedman, I.; Sogbohossou, E.A.; Tumenta, P.N.; et al. Autosomal and MtDNA Markers Affirm the Distinctiveness of Lions in West and Central Africa. PLoS ONE 2015, 10, e0137975. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Davis, B.W.; Eizirik, E.; Murphy, W.J. Phylogenomic Evidence for Ancient Hybridization in the Genomes of Living Cats (Felidae). Genome Res. 2016, 26, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Paijmans, J.L.A.; Fickel, J.; Courtiol, A.; Hofreiter, M.; Förster, D.W. Impact of Enrichment Conditions on Cross-Species Capture of Fresh and Degraded DNA. Mol. Ecol. Resour. 2016, 16, 42–55. [Google Scholar] [CrossRef]
- Ma, Y.P.; Wang, S. Mitochondrial Genome of the African Lion Panthera Leo Leo. Mitochondrial DNA 2015, 26, 951–952. [Google Scholar] [CrossRef]
- Bruche, S.; Gusset, M.; Lippold, S.; Barnett, R.; Eulenberger, K.; Junhold, J.; Driscoll, C.A.; Hofreiter, M. A Genetically Distinct Lion (Panthera Leo) Population from Ethiopia. Eur. J. Wildl. Res. 2013, 59, 215–225. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cilli, E.; Fontani, F.; Ciucani, M.M.; Pizzuto, M.; Di Benedetto, P.; De Fanti, S.; Mignani, T.; Bini, C.; Iacovera, R.; Pelotti, S.; et al. Museomics Provides Insights into Conservation and Education: The Instance of an African Lion Specimen from the Museum of Zoology “Pietro Doderlein”. Diversity 2023, 15, 87. https://doi.org/10.3390/d15010087
Cilli E, Fontani F, Ciucani MM, Pizzuto M, Di Benedetto P, De Fanti S, Mignani T, Bini C, Iacovera R, Pelotti S, et al. Museomics Provides Insights into Conservation and Education: The Instance of an African Lion Specimen from the Museum of Zoology “Pietro Doderlein”. Diversity. 2023; 15(1):87. https://doi.org/10.3390/d15010087
Chicago/Turabian StyleCilli, Elisabetta, Francesco Fontani, Marta Maria Ciucani, Marcella Pizzuto, Pierangelo Di Benedetto, Sara De Fanti, Thomas Mignani, Carla Bini, Rocco Iacovera, Susi Pelotti, and et al. 2023. "Museomics Provides Insights into Conservation and Education: The Instance of an African Lion Specimen from the Museum of Zoology “Pietro Doderlein”" Diversity 15, no. 1: 87. https://doi.org/10.3390/d15010087
APA StyleCilli, E., Fontani, F., Ciucani, M. M., Pizzuto, M., Di Benedetto, P., De Fanti, S., Mignani, T., Bini, C., Iacovera, R., Pelotti, S., Spadola, F., Luiselli, D., & Lo Brutto, S. (2023). Museomics Provides Insights into Conservation and Education: The Instance of an African Lion Specimen from the Museum of Zoology “Pietro Doderlein”. Diversity, 15(1), 87. https://doi.org/10.3390/d15010087







