Effects of Anthropogenic Habitat Fragmentation on the Genetic Connectivity of the Threatened and Endemic Campylorhynchus yucatanicus (Aves, Trogloditydae) in the Yucatan Peninsula, Mexico
Abstract
1. Introduction
2. Methods
2.1. Study Area and Sample Collection
2.2. Laboratory Process
2.3. Genetic Diversity Analysis
2.4. Genetic Structure Analysis
2.5. Landscape Composition and Genetic Diversity: Node Level
2.6. Genetic Distances and Landscape Resistance
3. Results
3.1. Genetic Diversity
3.2. Genetic Structure
3.3. Landscape Composition and Genetic Diversity: Node Level
3.4. Genetic Distances and Landscape Resistance
4. Discussion
4.1. Genetic Diversity
4.2. Genetic Structure
4.3. Landscape Composition and Genetic Diversity
5. Implications for Conservation
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| ID | Locus1 | Repetition Units | Primer Sequence (5′-3′) | AT | Length (pb) | TA (°C) |
|---|---|---|---|---|---|---|
| Locus1 | CACW3-01 | (ATT)5G(TTA)4(TTG)6TTATTG(TTGTTA)3(TCA)9 | F: ACTGTTCACCCTTGGACCTG R: TGTCTGGAAACCACTGAAGAAC | 6 | 168–188 | |
| Locus2 | CACW3-03 | (CTA)5CTG(CTA)8(ATA)10 | F: TCCTGAAATGTAATTCAGACACC | 5 | 259–279 | 57.6 |
| R: CAGAGTGCTACTTAAATTGATTCTTTC | ||||||
| Locus3 | CACW3-05 | (TGT)5 | F: GATGCATATTGTCAGAGTTCCAC | 5 | 131–149 | 57.6 |
| R: CTGGACTGAGCTAACAAATGATG | ||||||
| Locus4 | CACW3-11 | (ATA)5(AAC)6AAT(AAC)4(AAT)3AG(TAA)4 | F: TTCTCCTCCCTCTACCTCCTTT | 8 | 180–204 | 54 |
| R: GTGACAACAGAAAATTCCCTTTA | ||||||
| Locus5 | CACW4-01 | (GTAT)6GAATCTG(TCTA)11 | F: TTTTGCCTAATAAACTGGCTGAC | 3 | 122–133 | 54 |
| R: CACAGAACCACAACCTACATGG | ||||||
| Locus7 | CACW4-04 | (TCTA)14 | F: TCTCACGTCTTACCATCCTGTG | 5 | 241–257 | 57.6 |
| R: TTGATACTTGAAACTCTCCTTCTGTC | ||||||
| Locus9 | CACW4-09 | (GATG)22 | F: GCTAACTGAAAGGGATTGTTGG | 5 | 92–116 | 59 |
| R: TTTCTGGCATGTTTCCTGTC |
| Parameters | Type | Prior |
|---|---|---|
| N1 | N | UN~[10-100,000–50,000-0] |
| N2 | N | UN~[10-100,000–50,000-0] |
| N3 | N | UN~[10-100,000–50,000-0] |
| N4 | N | UN~[10-100,000–50,000-0] |
| N5 | N | UN~[10-100,000–50,000-0] |
| t1 | T | NO~[500–13,000–2500-1] |
| t2 | T | NO~[13,000–65,000–7,500-1] |
| ID | Sites | N | K1 | K2 | K3 | K4 |
|---|---|---|---|---|---|---|
| 1 | Southwest Celestún | 7 | 13.21 | 30.64 | 24.31 | 31.86 |
| 2 | Northeast Celestún | 15 | 10.87 | 30.36 | 27.31 | 31.44 |
| 3 | El Palmar | 10 | 19.81 | 27.72 | 24.89 | 27.62 |
| 4 | West Sisal | 10 | 9.21 | 30.36 | 29.61 | 30.85 |
| 5 | East Sisal | 13 | 15.69 | 28.04 | 28.95 | 27.34 |
| 6 | Chuburná | 15 | 11.36 | 31.49 | 24.71 | 32.44 |
| 7 | Capilla | 8 | 16.9 | 25.53 | 32.91 | 24.68 |
| 8 | Chixchulub | 4 | 26.12 | 24.16 | 26 | 23.7 |
| 9 | San Benito | 12 | 16.28 | 27.46 | 28.02 | 28.25 |
| 10 | Xcambó | 10 | 26.54 | 23.39 | 28.07 | 21.98 |
| 11 | Santa Clara | 10 | 18.62 | 25.37 | 31.42 | 24.61 |
| 12 | Dzilam | 12 | 46.11 | 17.8 | 18.71 | 17.37 |
| 13 | West Ría Lagartos | 4 | 68.63 | 10.57 | 11.27 | 9.53 |
| 14 | East Ría Lagartos | 10 | 72.37 | 9.33 | 9.4 | 8.91 |
| Source | Df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among Populations a priori | 3 | 38.837 | 12.946 | 0.190 | 12 |
| Among Sites | 10 | 20.629 | 2.063 | 0.043 | 3 |
| Among Indiv | 116 | 147.649 | 1.273 | 0.000 | 0 |
| Within Indiv | 130 | 179.000 | 1.377 | 1.377 | 86 |
| Total | 259 | 386.115 | 1.610 | 100 |
| Parameter | Mean | Median | Quantiles 0.05 | Quantiles 0.95 |
|---|---|---|---|---|
| N1 | 1293.24 | 1032.23 | 363.5 | 3056.45 |
| N2 | 44397.8 | 37826.6 | 15075.1 | 94751.9 |
| t1 | 7829.86 | 5750.76 | 2218.14 | 18108 |
| Model | |
|---|---|
| SEI | Shannon Equity Index |
| CA1 | Suitable habitat proportion |
| SDI | Fragments diversity index |
| CA2 | Disturbed habitat proportion |
| PA | Risk index for proximity of human settlements |
| ED | Fragments edge density of suitable habitat |
| MSI14 | Fragments average shape index of suitable habitat |
| log10(SA) | Log10 distance to human settlements |
| SC | Distance to road |
| CA1 + PA | Combined effect of Suitable habitat proportion and Risk index for proximity of human settlements |
| CA1 + MSI14 | Combined effect of Suitable habitat proportion and Fragments average shape index of suitable habitat |
| NumP14 + MedPS14 | Combined effect of fragments number of suitable habitat and average size of fragments of suitable habitat |

References
- Gentry, A.H. (Ed.) Four Neotropical Rainforests; Yale University Press: London, UK, 1991. [Google Scholar]
- Fahrig, L. Effect of habitat fragmentation on the extinction threshold: A synthesis. Ecol. Appl. 2002, 12, 346–353. [Google Scholar] [CrossRef]
- Trzcinski, M.K.; Fahrig, L.; Merriam, G. Independent effects of forest cover and fragmentation on the distribution of forest breeding birds. Ecol. Appl. 1999, 9, 586–593. [Google Scholar] [CrossRef]
- Li, Z.; Lu, Z.; Shu, X.; Jiang, G.; Xu, L.; Zhou, F. Nestedness of bird assemblages in the karst forest fragments of southwestern Guangxi, China. Chin. Birds 2013, 4, 170–183. [Google Scholar] [CrossRef]
- Connor, E.F.; Courtney, A.C.; Yoder, J.M. Individuals-area relationship: The relationship between animal population density and area. Ecology 2000, 81, 734–748. [Google Scholar] [CrossRef]
- Newmark, W.D. Tropical Forest Fragmentation and the Local Extinction of Understory Birds in the Eastern Usambara Mountains, Tanzania. Conserv. Biol. 1991, 5, 67–78. [Google Scholar] [CrossRef]
- Fahrig, L. Effects of habitat fragmentation on biodiversity. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 487–515. [Google Scholar] [CrossRef]
- Smith, A.L.; Salgado Ortiz, J.; Robertson, R.J. Distribution patterns of migrant and resident birds in successional forests of the Yucatan Peninsula, Mexico. Biotropica 2006, 33, 153–170. [Google Scholar] [CrossRef]
- Segelbacher, G.; Cushman, S.A.; Epperson, B.K.; Fortin, M.J.; Francois, O.; Hardy, O.J.; Holderegger, R.; Manel, S. Applications of landscape genetics in conservation biology: Concepts and challenges. Conservation Genetics 2010, 11, 375–385. [Google Scholar] [CrossRef]
- Luque, S.; Saura Santigo, S.; Fortin, M.J. Landscape connectivity analysis for conservation: Insights from combining new methods with ecological and genetic data. Landsc. Ecol. 2012, 27, 153–157. [Google Scholar] [CrossRef]
- Manel, S.; Holderegger, R. Ten years of landscape genetics. Trends Ecol. Evol. 2013, 28, 614–621. [Google Scholar] [CrossRef]
- Leberg, P.L.; Athrey, G.N.R.; Barr, K.R.; Lindsay, D.L.L.; Lance, R.F. Implications of landscape alteration for the conservation of genetic diversity of endangered species. In Molecular Approaches in Natural Resource Conservation and Management; De Woody, J., Bickham, J.W., Michler, C.H., Nichols, K.M., Rhodes, O.E., Woeste, K.E., Eds.; Cambridge University Press: New York, NY, USA, 2010; pp. 212–238. [Google Scholar] [CrossRef]
- Charlesworth, D.; Charlesworth, B. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 1987, 19, 237–268. [Google Scholar] [CrossRef]
- Hemmings, N.L.; Slate, J.; Birkhead, T.R. Inbreeding causes early death in a passerine bird. Nat. Commun. 2012, 3, 863. [Google Scholar] [CrossRef] [PubMed]
- Traill, L.W.; Brook, B.W.; Frankham, R.R.; Bradshaw, C.J.A. Pragmatic population viability targets in a rapidly changing world. Biol. Conserv. 2010, 143, 28–34. [Google Scholar] [CrossRef]
- Aguilar, C.; Martínez, E.; Arriaga, L. Deforestación y fragmentación de ecosistemas ¿Qué tan grave es el problema en México? Biodiversitas 2000, 30, 7–11. [Google Scholar]
- Paynter, R.A., Jr. The Ornithogeography of the Yucatan Peninsula. Peabody Mus. Nat. Hist. Bull. 1955, 9, 347. [Google Scholar]
- Abrams, E.M.; Rue, D.J. The causes and consequences of deforestation among the prehistoric Maya. Hum. Ecol. 1988, 16, 377–395. [Google Scholar] [CrossRef]
- Geoghegan, J.; Cortina, S.V.; Klepeis, P.; Macario, P.M.; Ogneva-Himmelberger, Y.; Chowdhury, R.R. Modeling tropical deforestation in the southern Yucatán peninsular region: Comparing survey and satellite data. Agric. Ecosyst. Environ. 2001, 85, 25–46. [Google Scholar] [CrossRef]
- Porter-Bolland, L.; Bonilla-Moheno, M.; Garcia-Frapolli, E.; Morteo-Montiel, S. Forest ecosystems and conservation. In Biodiversity and Conservation of the Yucatán Peninsula; Springer: Cham, Swizerland, 2015; pp. 377–398. [Google Scholar]
- Barrera, N.; Toledo, V.M. Ethnoecology of the Yucatec Maya: Symbolism, knowledge and management of natural resources. J. Lat. Am. Geogr. 2005, 4, 9–41. [Google Scholar] [CrossRef]
- García, E.; Ayala, B.; Bonilla, M.; Espadas, C.; Ramos, G. Biodiversity conservation, traditional agriculture and ecotourism: Land cover/land-use change projections for a natural protected area in the northeastern Yucatan Peninsula, Mexico. Landsc. Urban. Plan. 2007, 83, 137–153. [Google Scholar] [CrossRef]
- Andrade, M. Transformación de los sistemas naturales por actividades antropogénicas. In Biodiversidad y Desarrollo Humano en Yucatán; Durán, R., Méndez, I., Eds.; CICY, PPD-FMAM, CONABIO, SEDUMA: Yucatán, México, 2010; pp. 316–319. [Google Scholar]
- Acosta-Lugo, E.; Alonzo-Parra, D.; Andrade-Hernández, M.; Castillo-Tzab, D.; Chablé-Santos, J.; Durán, R.; Espadas-Manrique, C.; Fernández-Stohanzlova, I.; Fraga, J.; Galicia, E.; et al. Plan de Conservación de la Eco-Región Petenes-Celestún-Palmar; Universidad Autónoma de Campeche, Pronatura Península de Yucatán, A.C.: Mérida, México, 2010. [Google Scholar]
- Lawrence, G.N. List of a Collection of Birds from Northern Yucatan. Ann. Lyceum Nat. Hist. N. Y. 1869, 9, 198–210. Available online: https://ia800702.us.archive.org/16/items/cbarchive_110228_listofacollectionofbirdsfromno1824/listofacollectionofbirdsfromno1824.pdf (accessed on 9 March 2022). [CrossRef]
- Zimmerman, D.A. Some remarks on the behavior of the Yucatan Cactus Wren. Condor 1957, 59, 53–58. [Google Scholar] [CrossRef]
- Ridgway, R. Birds of North and Middle America; U.S. National Museum Bulletin 50; Government Printing Office: Washington, DC, USA, 1904; 801p. [Google Scholar]
- Hellmayr, C.E. Catalogue of Americas and the Adjacent Islands. Field Mus. Nat. Hist. Zool. Ser. 1934, 7, 1–531. [Google Scholar]
- Selander, R.K. Speciation in wren sof the genus Campylorhynchus. Univ. Calif. Publ. Zool. 1964, 74, 1–375. [Google Scholar]
- Howell, S.; Webb, S. A Field Guide to the Birds of México and Northern Central America; Oxford University Press: New York, NY, USA, 1995. [Google Scholar]
- Vargas Soriano, J.; Salgado Ortíz, J.; Escalona Segura, G. Breeding phenology and nesting success of the Yucatan Wren in the Yucatan Peninsula, Mexico. Wilson J. Ornithol. 2010, 122, 439–446. [Google Scholar] [CrossRef]
- Flores, J.S.; Espejel, I. Tipos de Vegetación de la Península de Yucatán. Etnoflora Yucatanense; Fascículo 3; Universidad Autónoma de Yucatán: Yucatán, México, 1994; 136p. [Google Scholar]
- Herrera, J.A.; Comín, F.A.M.; Capurro, L. Los usos y abusos de la zona costera en la Península de Yucatán. In El Manejo Costero en México. Universidad Autónoma de Campeche; Rivera, E., Villalobos, G.J., Azuz, I., Rosado, F., Eds.; SEMARNAT, CETYS-Universidad, Universidad de Quintana Roo: Baja California, México, 2004; pp. 387–396. [Google Scholar]
- Norma Oficial Mexicana NOM-059-SEMARNAT-2010. Protección Ambiental. Especies Nativas de México de Flora y Fauna Silvestres. Categorías de Riesgo y Especificaciones Para su Inclusión, Exclusión o Cambio. Lista de Especies en Riesgo; SEMARNAT [Secretaría de Medio Ambiente y Recursos Naturales]: Ciudad de México, México, 2010. [Google Scholar]
- IUCN. The IUCN Red List of Threatened Species. Version 2015-4. 2015. Available online: www.iucnredlist.org (accessed on 30 May 2016).
- Barr, K.R.; Lindsay, D.L.; Athrey, G.; Lance, R.F.; Hayden, T.J.; Tweddale, S.A.; Leberg, P.L. Population structure in an endangered songbird: Maintenance of genetic differentiation despite high vagility and significant population recovery. Mol. Ecol. 2008, 17, 3628–3639. [Google Scholar] [CrossRef]
- Lindsay, D.L.; Barr, K.R.; Lance, R.F.; Tweddale, S.A.; Hayden, T.J.; Leberg, P.L. Habitat fragmentation and genetic diversity of an endangered, migratory songbird, the golden-cheeked warbler (Dendroica chrysoparia). Mol. Ecol. 2008, 17, 2122–2133. [Google Scholar] [CrossRef]
- Athrey, G.N.R.; Barr, K.R.; Lance, R.F.; Leberg, P.L. Birds in space and time: Genetic changes accompanying anthropogenic habitat fragmentation in the endangered Blackcapped vireo (Vireo atricapilla). Evol. Appl. 2012, 5, 540–552. [Google Scholar] [CrossRef]
- American Ornithologists Union. Report of committee on the use of wild birds in research. Auk 1988, 105 (Suppl. 1), 1A–41A. [Google Scholar] [CrossRef]
- Bello, N.; Francino, O.; Sánchez, A. Isolation of genomic DNA from feathers. J. Vet. Diagn. Investig. 2001, 13, 162–164. [Google Scholar] [CrossRef]
- Barr, K.R.; Kus, B.E.; Preston, K.L.; Howell, S.; Perkins, E.; Vandergast, A.G. Habitat fragmentation in coastal southern California disrupts genetic connectivity in the cactus wren (Campylorhynchus brunneicapillus). Mol. Ecol. 2015, 24, 2349–2363. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Piry, S.; Luikart, G.; Cornuet, J.M. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Rousset, F. Genepop'007: A complete reimplementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.; vonHoldt, B. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Guillot, G.; Estoup, A.; Mortier, F.; Cosson, J.F. A spatial statistic model for landscape genetics. Genetics 2005, 170, 1261–1280. [Google Scholar] [CrossRef]
- Guillot, G.; Mortier, F.; Estoup, A. GENELAND: A computer package for landscape genetics. Mol. Ecol. Notes 2005, 5, 708–711. [Google Scholar] [CrossRef]
- Guillot, G. Inference of structure in subdivided populations at low levels of genetic differentiation—The correlated allele frequencies model revisited. Bioinformatics 2008, 24, 2222–2228. [Google Scholar] [CrossRef]
- Guillot, G.; Santos, F.; Estoup, A. Analysing georeferenced population genetics data with Geneland: A new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics 2008, 24, 1406–1407. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Rempel, R.S.; Kaukinen, D.; Carr, A.P. Patch analyst and patch grid. Ontario Ministry of Natural Resources; Centre for Northern Forest Ecosystem Research: Thunder Bay, ON, Canada, 2012. [Google Scholar]
- Raynal, L.; Marin, J.M.; Pudlo, P.; Ribatet, M.; Robert, C.P.; Estoup, A. ABC random forests for Bayesian parameter inference. Bioinformatics 2019, 35, 1720–1728. [Google Scholar] [CrossRef] [PubMed]
- Burnham, K.P.; Anderson, D.R. Model Selection and Inference: A practical Information-Theoretic Approach; Springer: New York, NY, USA, 2002. [Google Scholar]
- Zharikov, Y.; Milton, D.A. Valuing coastal habitats: Predicting high-tide roosts of nonbreeding migratory shorebirds from landscape composition. Emu 2009, 109, 107–120. [Google Scholar] [CrossRef]
- Rousset, F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 1997, 145, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- McRae, B.H.; Dickson, B.G.; Keitt, T.H.; Shah, V.B. Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology 2008, 89, 2712–2724. [Google Scholar] [CrossRef] [PubMed]
- McRae, B.H.; Shah, V.B. Circuitscape User’s Guide; The Univeristy of California: Santa Barbara, CA, USA, 2009; Available online: http://www.circuitscape.org (accessed on 17 December 2021).
- Taylor, P.D.; Fahrig, L.; Henein, K.; Merriam, G. Connectivity is a vital element of landscape structure. Oikos 1993, 68, 571–573. [Google Scholar] [CrossRef]
- Koen, E.L.; Garroway, C.J.; Wilson, P.J.; Bowman, J. The effect of map boundary on estimates of landscape resistance to animal movement. PLoS ONE 2010, 5, e11785. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Rodríguez, A.; Escalona-Segura, G.; Plasencia Vázquez, A.H.; Iñigo Elias, E.E.; Ruiz-Montoya, L. Distribución potencial y conectividad del paisaje: Criterios para reevaluar el grado de amenaza de Campylorhynchus yucatanicus (Aves: Troglodytidae). Rev. de Biol. Trop. (Int. J. Trop. Biol.) 2017, 65, 1554–1568. [Google Scholar] [CrossRef]
- Willi, Y.; Van Buskirk, J.; Hoffmann, A.A. Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 433–458. [Google Scholar] [CrossRef]
- Willi, Y.; Van Buskirk, J.; Schmid, B.; Fischer, M. Genetic isolation of fragmented populations is exacerbated by drift and selection. J. Evol. Biol. 2007, 20, 534–542. [Google Scholar] [CrossRef]
- Evans, S.R.; Sheldon, B.C. Interspecific patterns of genetic diversity in birds: Correlations with extinction risk. Conserv. Biol. 2008, 22, 1016–1025. [Google Scholar] [CrossRef]
- Bautista Vicente, F.; Carbajal, N.; Pineda Martínez, L.F. Estimation of total yearly CO2 emissions by wildfires in Mexico during the Period 1999–2010. Adv. Meteorol. 2014, 2014, 958457. [Google Scholar] [CrossRef]
- Sieving, K.E.; Wilson, M.F.; De Santo, T.L. Habitat barriers to movement of understory birds in fragmented South-Temperate Rainforest. Auk 1996, 113, 944–949. [Google Scholar] [CrossRef]
- Uezu, A.; Metzger, J.P.; Vielliard, J.M.E. Effects of structural and functional connectivity and patch size on the abundance of seven Atlantic Forest bird species. Biol. Conserv. 2005, 123, 507–519. [Google Scholar] [CrossRef]
- Crooks, K.R.; Sanjayan, M. Connectivity conservation: Maintaining connections for nature. In Connectivity Conservation; Crooks, K.R., Sanjayan, M., Eds.; Cambridge University Press: New York, NY, USA, 2006; 712p. [Google Scholar]
- INEGI México. Censo de Población y Vivienda 2010; MEX-INEGI.40.201.01-CPV-2010; Instituto Nacional de Estadística y Geografía Dirección General de Estadísticas Sociodemográficas Dirección General Adjunta del Censo General de Población y Vivienda: Aguascalientes, México, 2011. [Google Scholar]
- Chacea, J.F.; Walsh, J.J. Urban effects on native avifauna: A review. Landsc. Urban Plan. 2006, 74, 46–69. [Google Scholar] [CrossRef]
- Delaney, K.S.; Riley, S.P.D.; Fisher, R.N. A rapid, strong and convergent genetic response to urban habitat fragmentation in four divergent and widespread vertebrates. PLoS ONE 2011, 5, e12767. [Google Scholar] [CrossRef]
- del Hoyo, J.; Elliott, A.; Christie, D. Handbook of the Birds of the World, Volume 10: Cuckoo-Shrikes to Thrushes; Lynx Editions: Barcelona, Spain, 2005. [Google Scholar]
- Martin-Albarracin, V.L.; Amico, G.C.; Simberloff, D.; Nuñez, M.A. Impact of non-native birds on native ecosystems: A global analysis. PLoS ONE 2015, 10, e0143070. [Google Scholar] [CrossRef]
- Programa de Ordenamiento Ecológico Territorial del Estado de Yucatán, México. 2021. Available online: https://bitacoraordenamiento.yucatan.gob.mx/index.php (accessed on 29 October 2021).
- DiLeo, M.F.; Wagner, H.H. A Landscape Ecologist’s Agenda for Landscape Genetics. Curr. Landsc. Ecol. Rep. 2016, 1, 115–126. [Google Scholar] [CrossRef]
- Íñigo-Elías, E.; Enkerlin-Hoeflich, E.C. Amenazas, estrategias e instrumentos para la conservación de las aves. In Conservación de Aves: Experiencias en México; Gómez de Silva, H., Oliveras de Ita, A., Eds.; Sección Mexicana del Consejo Internacional para la Preservación de las Aves, A.C., CIPAMEX: San Luis Potosi, México, 2003. [Google Scholar]
- Martínez-Morales, M.A.; Islas, V.M.; Zuria, I.; Hoffmann-Pinther, M.C.C.P.; Velasco, R.G.C. La conservación de las aves más allá de las áreas naturales protegidas: El caso de la avifauna del Rancho Santa Elena, Hidalgo. Huitzil 2013, 14, 87–100. [Google Scholar] [CrossRef]
- Bouzat, J.L.; Johnson, J.A.; Toepfer, J.E.; Simpson, S.A.; Esker, T.L.; Westemeier, R.L. Beyond the beneficial effects of translocations as an effective tool for the genetic restoration of isolated populations. Conserv. Genet. 2009, 10, 191–201. [Google Scholar] [CrossRef]
- Keller, L.F.; Waller, D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002, 17, 230–242. [Google Scholar] [CrossRef]
- Frankham, R. Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 2010, 143, 1919–1927. [Google Scholar] [CrossRef]
- Fahrig, L. How much habitat is enough? Biol. Conserv. 2001, 100, 65–74. [Google Scholar] [CrossRef]
- Balkenhol, N.; Waits, L.P. Molecular road ecology: Exploring the potential of genetics for investigating transportation impacts on wildlife. Mol. Ecol. 2009, 18, 4151–4164. [Google Scholar] [CrossRef] [PubMed]
- Minor, E.S.; Urban, D.L. A graph-theory framework for evaluating landscape connectivity and conservation planning. Conserv. Biol. 2008, 22, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Hortal, L.; Saura, S. Impact of spatial scale on the identification of critical habitat patches for the maintenance of landscape connectivity. Landsc. Urban Plan. 2007, 83, 176–186. [Google Scholar] [CrossRef]
- Bernuésa, A.; Ruiz, R.; Olaizola, A.; Villalba, D.; Casasús, I. Sustainability of pasture-based livestock farming systems in the European Mediterranean context: Synergies and trade-offs. Livest. Sci. 2011, 139, 44–57. [Google Scholar] [CrossRef]
- Cantú, J.C.; García De la Puente, E.; González, G.M.; Sánchez., M.E. Riqueza Alada: El Crecimiento del Aviturismo en México; Defenders of Wildlife, UABCS, ENESUM, Teyeliz, A.C.: Washington, DC, USA, 2020; 40p. [Google Scholar]

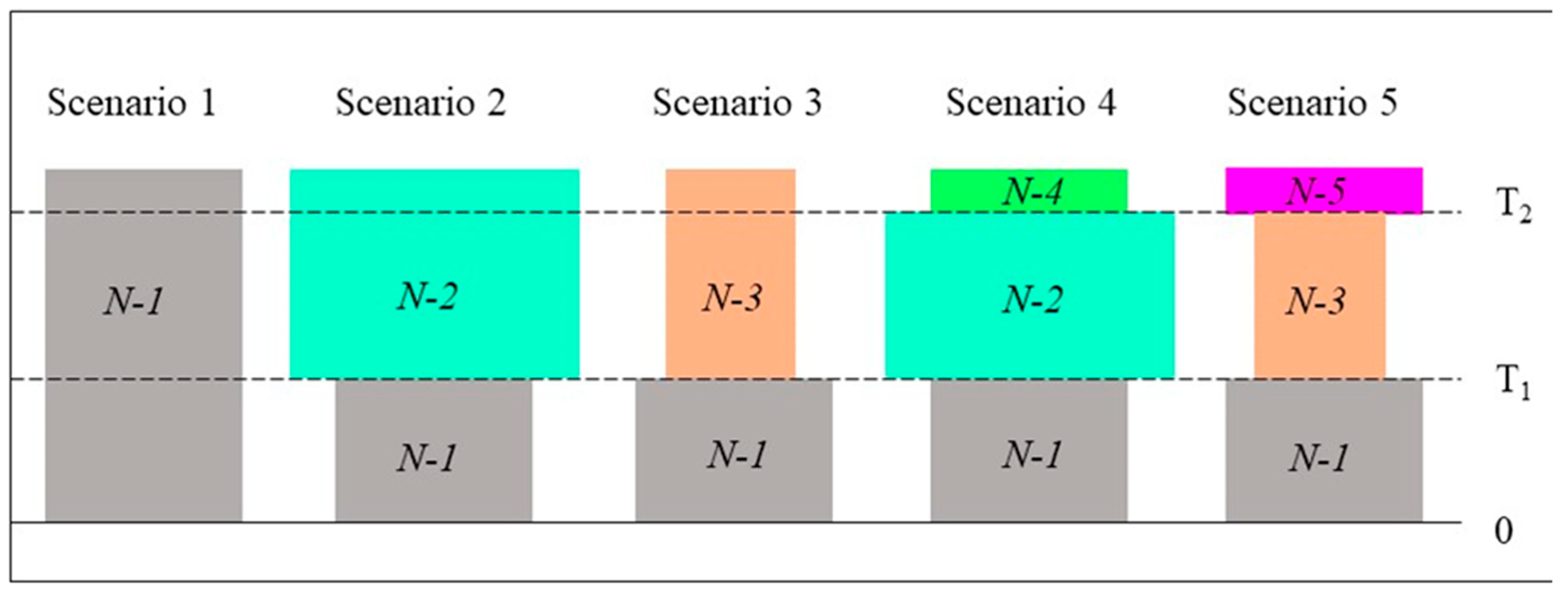
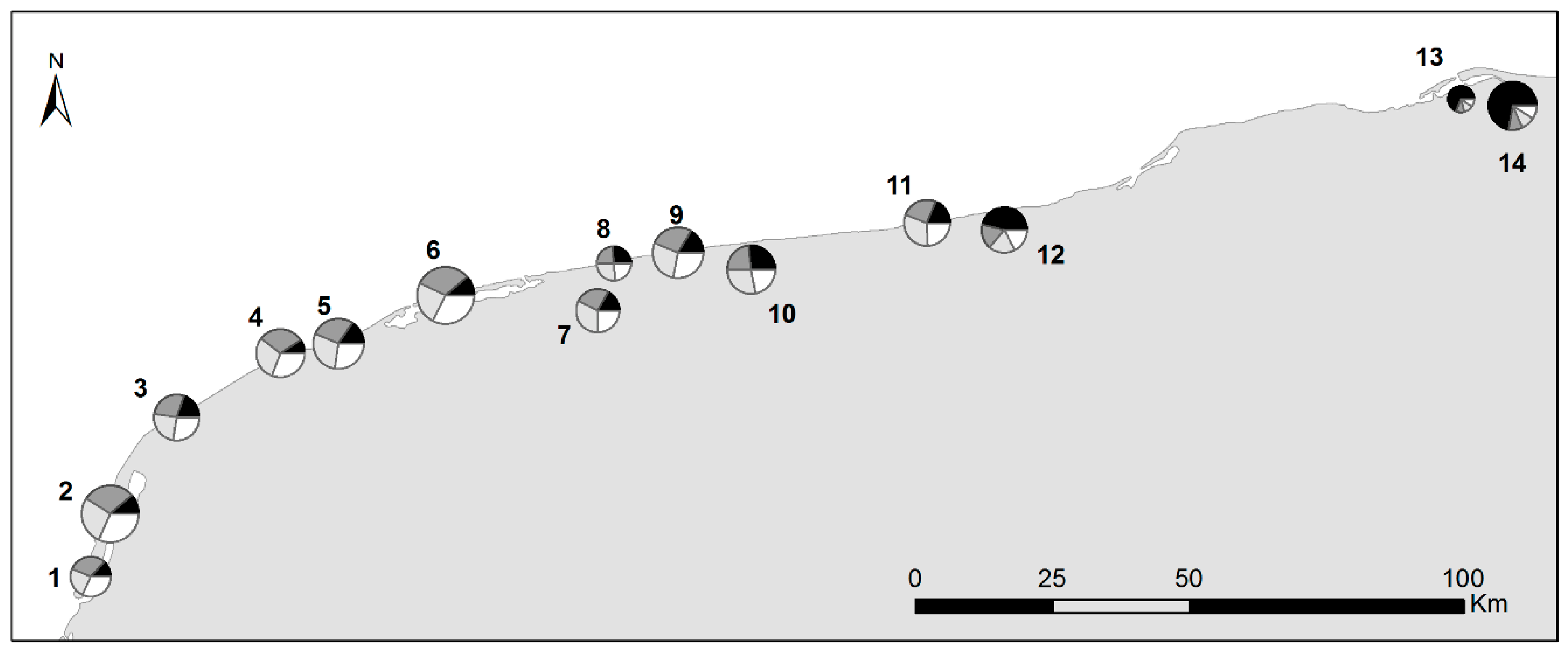


| ID | Sites | N | Na ± SE | F ± SE | I ± SE | He ± SE |
|---|---|---|---|---|---|---|
| 1 | Southwest Celestún | 14 | 2.71 ± 0.28 | 0.08 ± 0.13 | 0.65 ± 0.13 | 0.37 ± 0.07 |
| 2 | Northeast Celestún | 7 | 3.57 ± 0.29 | 0.16 ± 0.07 | 0.72 ± 0.10 | 0.39 ± 0.06 |
| 3 | El Palmar | 9 | 3.14 ± 0.26 | 0.27 ± 0.16 | 0.89 ± 0.05 | 0.52 ± 0.02 |
| 4 | West Sisal | 10 | 2.71 ± 0.42 | 0.21 ± 0.15 | 0.59 ± 0.15 | 0.33 ± 0.08 |
| 5 | East Sisal | 11 | 3.71 ± 0.28 | −0.10 ± 0.05 | 0.89 ± 0.03 | 0.49 ± 0.01 |
| 6 | Chuburná | 14 | 4.14 ± 0.45 | −0.06 ± 0.07 | 0.95 ± 0.12 | 0.50 ± 0.05 |
| 7 | Capilla | 8 | 3.57 ± 0.57 | 0.02 ± 0.09 | 0.81 ± 0.16 | 0.43 ± 0.08 |
| 8 | Chixchulub | 5 | 2.57 ± 0.29 | −0.27 ± 0.11 | 0.73 ± 0.14 | 0.43 ± 0.08 |
| 9 | San Benito | 11 | 3.85 ± 0.49 | −0.06 ± 0.05 | 1.04 ± 0.16 | 0.57 ± 0.06 |
| 10 | Xcambó | 10 | 3.57 ± 0.48 | 0.02 ± 0.16 | 0.95 ± 0.12 | 0.53 ± 0.05 |
| 11 | Santa Clara | 9 | 3.00 ± 0.31 | −0.23 ± 0.04 | 0.75 ± 0.11 | 0.43 ± 0.06 |
| 12 | Dzilam | 9 | 3.71 ± 0.28 | −0.07 ± 0.10 | 0.88 ± 0.10 | 0.48 ± 0.06 |
| 13 | West Ría Lagartos | 3 | 2.42 ± 0.20 | −0.23 ± 0.04 | 0.61 ± 0.09 | 0.36 ± 0.05 |
| 14 | East Ría Lagartos | 10 | 2.85 ± 0.26 | −0.17 ± 0.09 | 0.73 ± 0.11 | 0.41 ± 0.06 |
| Genetic Population (Sites) | N | Na ± SE | F ± SE | I ± SE | He ± SE |
|---|---|---|---|---|---|
| Celestún-Chuburná group (1–6) | 65 | 4.17 ± 0.60 | 0.05 ± 0.03 | 0.81 ± 0.13 | 0.43 ± 0.07 |
| Chixchulub group (7–9) | 24 | 3.83 ± 0.79 | 0.10 ± 0.08 | 0.89 ± 0.19 | 0.48 ± 0.08 |
| Xcambó-Dzilam group (10–12) | 28 | 3.33 ± 0.49 | 0.11 ± 0.05 | 0.93 ± 0.16 | 0.52 ± 0.08 |
| Ría Lagartos group (13–14) | 13 | 2.50 ± 0.43 | 0.07 ± 0.01 | 0.56 ± 0.19 | 0.31 ± 0.11 |
| Model | N | K | logLik | AIC | AICc | ΔAICc | LIKAIC | ωi | R | R2 | F | p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SEI | 14 | 3 | 14.79 | −23.59 | −21.19 | 0.00 | 1.00 | 0.35 | 0.30 | 0.24 | 5.19 | 0.04 * |
| CA1 | 14 | 3 | 14.19 | −22.37 | −19.97 | 1.22 | 0.54 | 0.19 | 0.24 | 0.18 | 3.76 | 0.04 * |
| SDI | 14 | 3 | 13.48 | −20.97 | −18.57 | 2.62 | 0.27 | 0.09 | 0.16 | 0.09 | 2.25 | 0.16 |
| CA2 | 14 | 3 | 12.82 | −19.64 | −17.24 | 3.95 | 0.14 | 0.05 | 0.07 | 0.00 | 0.96 | 0.35 |
| PA | 14 | 3 | 12.44 | −18.87 | −16.47 | 4.72 | 0.09 | 0.03 | 0.02 | −0.06 | 0.28 | 0.61 |
| ED1 | 14 | 3 | 12.39 | −18.79 | −16.39 | 4.80 | 0.09 | 0.03 | 0.02 | −0.07 | 0.20 | 0.66 |
| MSI14 | 14 | 3 | 12.33 | −18.67 | −16.27 | 4.92 | 0.09 | 0.03 | 0.01 | −0.07 | 0.10 | 0.76 |
| log10(SA) | 14 | 3 | 12.33 | −18.66 | −16.26 | 4.93 | 0.08 | 0.03 | 0.01 | −0.08 | 0.09 | 0.77 |
| SC | 14 | 3 | 12.30 | −18.60 | −16.20 | 4.99 | 0.08 | 0.03 | 0.00 | −0.08 | 0.03 | 0.86 |
| CA1 + PA | 14 | 4 | 12.83 | −17.66 | −13.21 | 7.98 | 0.02 | 0.01 | 0.08 | 0.00 | 0.98 | 0.34 |
| CA1 + MSI14 | 14 | 4 | 12.75 | −17.50 | −13.05 | 8.14 | 0.02 | 0.01 | 0.06 | −0.01 | 0.83 | 0.38 |
| NumP14 + MedPS14 | 14 | 4 | 12.33 | −16.66 | −12.21 | 8.98 | 0.01 | 0.00 | 0.01 | −0.08 | 0.09 | 0.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serrano-Rodríguez, A.; Escalona-Segura, G.; Rodríguez, A.G.; Machkour-M’Rabet, S.; Ruiz-Montoya, L.; Elias, E.E.I.; Plasencia-Vázquez, A.H. Effects of Anthropogenic Habitat Fragmentation on the Genetic Connectivity of the Threatened and Endemic Campylorhynchus yucatanicus (Aves, Trogloditydae) in the Yucatan Peninsula, Mexico. Diversity 2022, 14, 1108. https://doi.org/10.3390/d14121108
Serrano-Rodríguez A, Escalona-Segura G, Rodríguez AG, Machkour-M’Rabet S, Ruiz-Montoya L, Elias EEI, Plasencia-Vázquez AH. Effects of Anthropogenic Habitat Fragmentation on the Genetic Connectivity of the Threatened and Endemic Campylorhynchus yucatanicus (Aves, Trogloditydae) in the Yucatan Peninsula, Mexico. Diversity. 2022; 14(12):1108. https://doi.org/10.3390/d14121108
Chicago/Turabian StyleSerrano-Rodríguez, Anay, Griselda Escalona-Segura, Antonio González Rodríguez, Salima Machkour-M’Rabet, Lorena Ruiz-Montoya, Eduardo E. Iñigo Elias, and Alexis Herminio Plasencia-Vázquez. 2022. "Effects of Anthropogenic Habitat Fragmentation on the Genetic Connectivity of the Threatened and Endemic Campylorhynchus yucatanicus (Aves, Trogloditydae) in the Yucatan Peninsula, Mexico" Diversity 14, no. 12: 1108. https://doi.org/10.3390/d14121108
APA StyleSerrano-Rodríguez, A., Escalona-Segura, G., Rodríguez, A. G., Machkour-M’Rabet, S., Ruiz-Montoya, L., Elias, E. E. I., & Plasencia-Vázquez, A. H. (2022). Effects of Anthropogenic Habitat Fragmentation on the Genetic Connectivity of the Threatened and Endemic Campylorhynchus yucatanicus (Aves, Trogloditydae) in the Yucatan Peninsula, Mexico. Diversity, 14(12), 1108. https://doi.org/10.3390/d14121108









