Different Chromosome Segregation Patterns Coexist in the Tetraploid Adriatic Sturgeon Acipenser naccarii
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and DNA Purification
2.2. Selection of Loci and Genotyping
2.3. Data Analyses
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


References
- Ludwig, A. A Sturgeon View on Conservation Genetics. Eur. J. Wildl. Res. 2005, 52, 3–8. [Google Scholar] [CrossRef]
- Peng, Z.; Ludwig, A.; Wang, D.; Diogo, R.; Wei, Q.; He, S. Age and Biogeography of Major Clades in Sturgeons and Paddlefishes (Pisces: Acipenseriformes). Mol. Phylogenetics Evol. 2007, 42, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Congiu, L.; Mudrak, V.A.; Quattro, J.M.; Smith, T.I.; Ware, K.; Doroshov, S.I. Evidence of Hexaploid Karyotype in Shortnose Sturgeon. Genome 2008, 51, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Nam, Y.K.; Noh, J.K.; Park, C.H.; Chapman, F.A. Karyotype of North American Shortnose Sturgeon Acipenser Brevirostrum with the Highest Chromosome Number in the Acipenseriformes. Ichthyol. Res. 2005, 52, 94–97. [Google Scholar] [CrossRef]
- Fontana, F.; Zane, L.; Pepe, A.; Congiu, L. Polyploidy in Acipenseriformes: Cytogenetic and Molecular Approaches. In Fish Cytogenetics; Pisano, E., Ozouf-Costaz, C., Foresti, F., Kapoor, B.G., Eds.; Science Publisher, Inc.: Enfield, NH, USA, 2007; pp. 385–403. [Google Scholar]
- Fontana, F.; Lanfredi, M.; Chicca, M.; Congiu, L.; Tagliavini, J.; Rossi, R. Fluorescent in Situ Hybridization with rDNA Probes on Chromosomes of Acipenser ruthenus and Acipenser naccarii (Osteichthyes, Acipenseriformes). Genome 1999, 42, 1008–1012. [Google Scholar] [CrossRef]
- Boscari, E.; Vidotto, M.; Martini, D.; Papetti, C.; Ogden, R.; Congiu, L. Microsatellites from the Genome and the Transcriptome of the Tetraploid Adriatic Sturgeon, Acipenser naccarii (Bonaparte, 1836) and Cross-Species Applicability to the Diploid Beluga Sturgeon, Huso huso (Linnaeus, 1758). J. Appl. Icht. 2015, 31, 977–983. [Google Scholar] [CrossRef]
- Ludwig, A.; Belfiore, N.M.; Pitra, C.; Svirsky, V.; Jenneckens, I. Genome Duplication Events and Functional Reduction of Ploidy Levels in Sturgeon (Acipenser, Huso and Scaphirhynchus). Genetics 2001, 158, 1203–1215. [Google Scholar] [CrossRef]
- Havelka, M.; Hulák, M.; Bailie, D.A.; Prodöhl, P.A.; Flajšhans, M. Extensive Genome Duplications in Sturgeons: New Evidence from Microsatellite Data. J. Appl. Ichthyol. 2013, 29, 704–708. [Google Scholar] [CrossRef]
- Stift, M.; Berenos, C.; Kuperus, P.; van Tienderen, P.H. Segregation Models for Disomic, Tetrasomic and Intermediate Inheritance in Tetraploids: A General Procedure Applied to Rorippa (Yellow Cress) Microsatellite Data. Genetics 2008, 179, 2113–2123. [Google Scholar] [CrossRef]
- Soares, N.R.; Mollinari, M.; Oliveira, G.K.; Pereira, G.S.; Vieira, M.L.C. Meiosis in Polyploids and Implications for Genetic Mapping: A Review. Genes 2021, 12, 1517. [Google Scholar] [CrossRef]
- Boscari, E.; Barbisan, F.; Congiu, L. Inheritance pattern of Microsatellite Loci in the Polyploid Adriatic Sturgeon (Acipenser naccarii). Aquaculture 2011, 321, 223–229. [Google Scholar] [CrossRef]
- Congiu, L.; Pujolar, J.M.; Forlani, A.; Cenadelli, S.; Dupanloup, I.; Barbisan, F.; Galli, A.; Fontana, F. Managing Polyploidy in Ex Situ Conservation Genetics: The Case of the Critically Endangered Adriatic Sturgeon (Acipenser naccarii). PLoS ONE 2011, 6, e18249. [Google Scholar] [CrossRef] [PubMed]
- Boscari, E.; Pujolar, J.M.; Dupanloup, I.; Corradin, R.; Congiu, L. Captive Breeding Programs Based on Family Groups in Polyploid Sturgeons. PLoS ONE 2014, 9, e110951. [Google Scholar] [CrossRef] [PubMed]
- May, B.; Krueger, C.C.; Kincaid, H.L. Genetic Variation at Microsatellite Loci in Sturgeon: Primer Sequence Homology in Acipenser and Scaphirhynchus. Can. J. Fish. Aquat. Sci. 1997, 54, 1542–1547. [Google Scholar] [CrossRef]
- Welsh, A.B.; Blumberg, M.; May, B. Identification of Microsatellite Loci in Lake Sturgeon, Acipenser Fulvescens, And Their Variability in Green Sturgeon, A. medirostris. Mol. Ecol. Notes 2002, 3, 47–55. [Google Scholar] [CrossRef]
- Zane, L.; Patarnello, T.; Ludwig, A.; Fontana, F.; Congiu, L. Isolation and Characterization of Microsatellites in the Adriatic Sturgeon (Acipenser naccarii). Mol. Ecol. Notes 2002, 2, 586–588. [Google Scholar] [CrossRef]
- Forlani, A.; Fontana, F.; Congiu, L. Isolation of Microsatellite Loci from the Endemic and Endangered Adriatic Sturgeon (Acipenser naccarii). Conserv. Genet. 2007, 9, 461–463. [Google Scholar] [CrossRef]
- Henderson-Arzapalo, A.; King, T.L. Novel Microsatellite Markers for Atlantic Sturgeon (Acipenser oxyrinchus) Population De-Lineation and Broodstock Management. Mol. Ecol. Notes 2002, 2, 437–439. [Google Scholar] [CrossRef]
- McQuown, E.C.; Sloss, B.L.; Sheehan, R.J.; Rodzen, J.; Tranah, G.J.; May, B. Microsatellite Analysis of Genetic Variation in Sturgeon: New Primer Sequences for Scaphirhynchus and Acipenser. Trans. Am. Fish. Soc. 2000, 129, 1380–1388. [Google Scholar] [CrossRef]
- Sybenga, J. Chromosome Pairing Affinity and Quadrivalent Formation in Polyploids: Do Segmental Allopolyploids Exist? Genome 1996, 39, 1176–1184. [Google Scholar] [CrossRef]
- Lloyd, A.; Bomblies, K. Meiosis in Autopolyploid and Allopolyploid Arabidopsis. Curr. Opin. Plant Biol. 2016, 30, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.; Wright, L.; Ponce, O.; Chen, J.; Prashar, A.; Sanchez-Moran, E.; Luo, Z.; Compton, L. Varietal Variation and Chromosome Behaviour During Meiosis in Solanum Tuberosum. Heredity 2020, 125, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.R. Analyzing tables of statistical tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.D.; Van de Velde, J.D.; Novikova, P.Y. Inference of Polyploidy Origin and Inheritance Mode from Population Genomic Data. BioRxiv. [CrossRef]
- Pyatskowit, J.D.; Krueger, C.C.; Kincaid, H.L.; May, B. Inheritance of Microsatellite Loci in the Polyploid Lake Sturgeon (Acipenser fulvescens). Genome 2001, 44, 185–191. [Google Scholar] [CrossRef]
- Rodzen, J.A.; May, B. Inheritance of Microsatellite Loci in the White Sturgeon (Acipenser transmontanus). Genome 2002, 45, 1064–1076. [Google Scholar] [CrossRef]
- Schreier, A.D.; Van Eenennaam, J.P.; Anders, P.; Young, S.; Crossman, J. Spontaneous Autopolyploidy in the Acipenseriformes, with Recommendations for Management. Rev. Fish Biol. Fish. 2021, 31, 159–180. [Google Scholar] [CrossRef]
- Birstein, V.J. Sturgeon Species and Hybrids: Can Hybrids Produce Caviar. Environ. Policy Law 2002, 32, 210–214. [Google Scholar]

| Locus | Ref | Ta | Size | Family | Parents | Genotypes | Genotyped Fingerling (Y/N) | # |
|---|---|---|---|---|---|---|---|---|
| LS-39 | 15 | 52 °C | 116–155 | Not informative | - | - | N | - |
| AfuG113 | 16 | Td | 326–364 | Not informative | - | - | N | - |
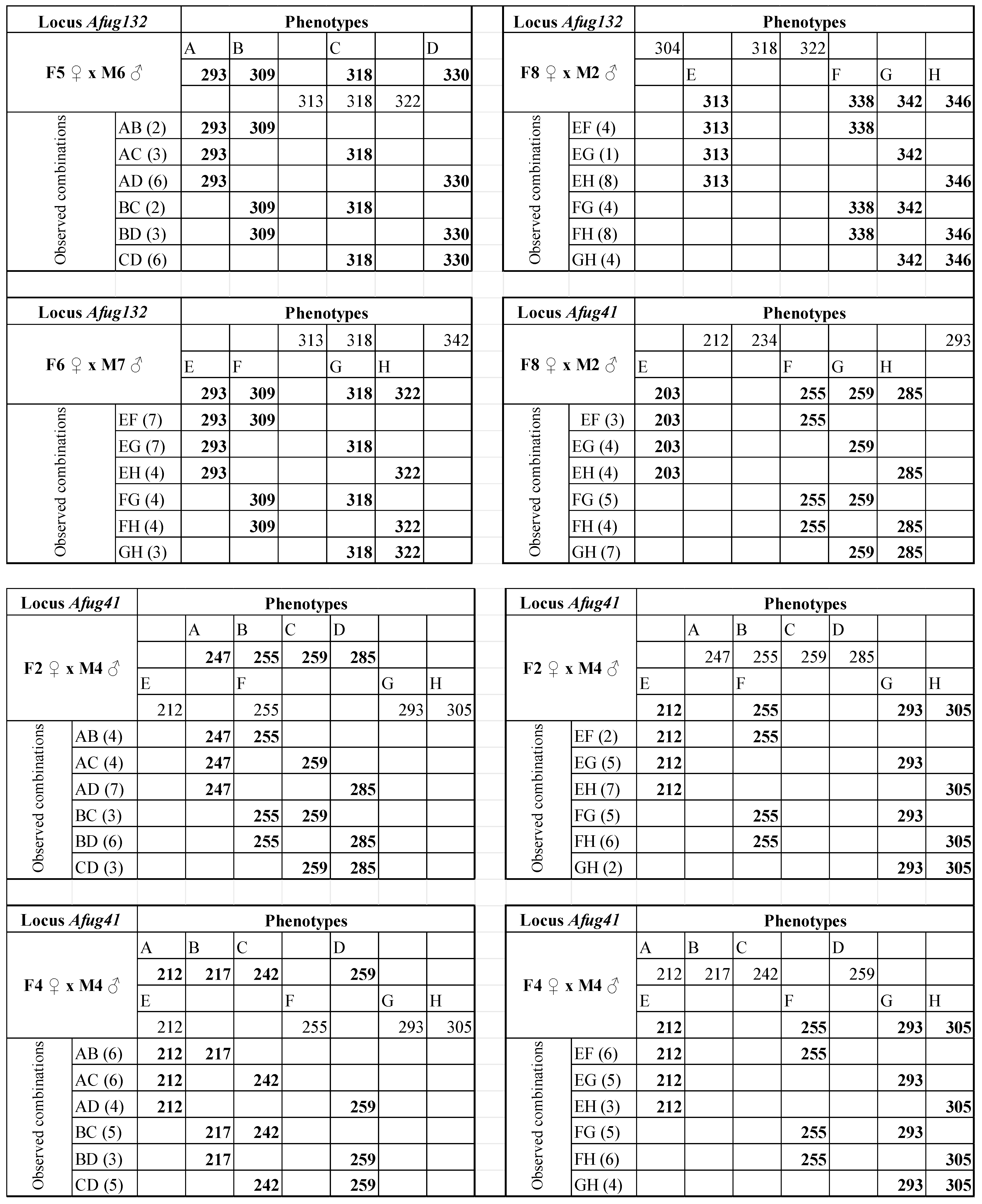
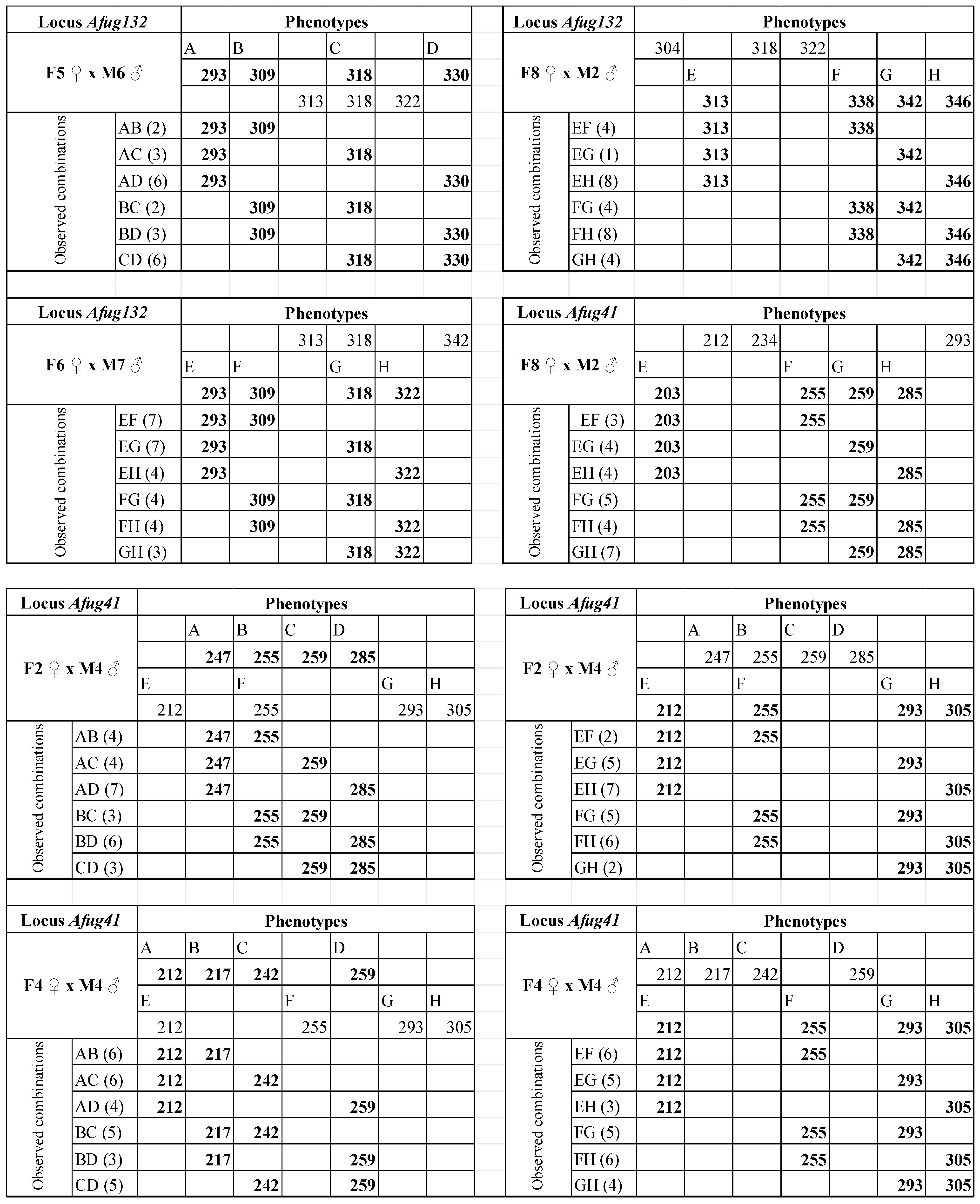
| AfuG132 | 16 | 61 °C | 259–346 | F5 ♀ × M6 ♂ | F5 ♀ M6 ♂ | 293/309/318/330 313/318/322 | Y | 32 |
| F6 ♀ × M7 ♂ | F6 ♀ M7 ♂ | 313/318/342 293/309/318/322 | Y | 32 | ||||
| F8 ♀ × M2 ♂ | F8 ♀ M2 ♂ | 304/318/322 313/338/342/346 | Y | 30 | ||||
| AfuG41 | 16 | 58 °C | 156–198 | F8 ♀ × M2 ♂ | F8 ♀ M2 ♂ | 212/234/293 203/255/259/285 | Y | 30 |
| F2 ♀ ×M4 ♂ | F2♀ M4♂ | 247/255/259/285 212/255/293/305 | Y | 30 | ||||
| F4 ♀ ×M4 ♂ | F4♀ M4♂ | 212/217/242/259 212/255/293/305 | Y | 30 | ||||
| F6 ♀ ×M7 ♂ | F6♀ M7♂ | 212/242/259/278 203/225/229/259 | Y | 32 | ||||
| Nacc7 ♀ ×Nacc5 ♂ | Nacc7♀ Nacc5♂ | 225/229/252/259 203/225/247/305 | Y | 32 | ||||
| Nacc8♀ ×Nacc 31♂ | Nacc8♀ Nacc31 ♂ | 212/242/293/305 203/234/305 | Y | 32 | ||||
| An20 * | 17 | 62 °C | 159–213 | F7 ♀ ×M8 ♂ | F7 ♀ M8 ♂ | 160/164/194 160/172/182/186 | Y | 30 |
| Anac-15214 | 7 | 61 °C | 259–285 | Not informative | - | - | N | - |
| Anac-2589 | 7 | 63 °C | 224–294 | Not informative | - | - | N | - |
| Anac-6784 | 7 | 62 °C | 311–346 | Nacc19 ♀ × Nacc17 ♂ | Nacc19♀ Nacc17 ♂ | 311/322/330/334 322/326 | Y | 32 |
| Anac-3133 | 7 | 56 °C | 164–178 | F7 ♀ × M8 ♂ | F7 ♀ M8 ♂ | 164/174 164/166/170/172 | Y | 30 |
| AnacA6 * | 18 | 62 °C | 289–313 | F5 ♀ × M6 ♂ | F5 ♀ M6 ♂ | 307/313 293/297/301/307 | Y | 32 |
| AnacB11 | 18 | 60 °C | 132–162 | F6 ♀ × M7 ♂ | F6 ♀ M7 ♂ | 148/150 138/144/148/162 | Y | 32 |
| Nacc19 ♀ × Nacc17 ♂ | Nacc19♀ Nacc17 ♂ | 132/136/138/144 132/150/162 | Y | 32 | ||||
| AnacB7 | 18 | 60 °C | 152–198 | F6 ♀ × M7 ♂ | F6♀ M7 ♂ | 166/172/174/176 154/156/164 | Y | 32 |
| Nacc19 ♀ × Nacc30 ♂ | Nacc19 ♀ Nacc30 ♂ | 156/166/174 154/170/174/176 | Y | 32 | ||||
| AnacC11 | 18 | 50 °C | 167–193 | Not informative | - | - | N | - |
| AnacE4 * | 18 | 58 °C | 326–354 | F5 ♀ × M6 ♂ | F5♀ M6 ♂ | 332/340/346/354 336/346 | Y | 32 |
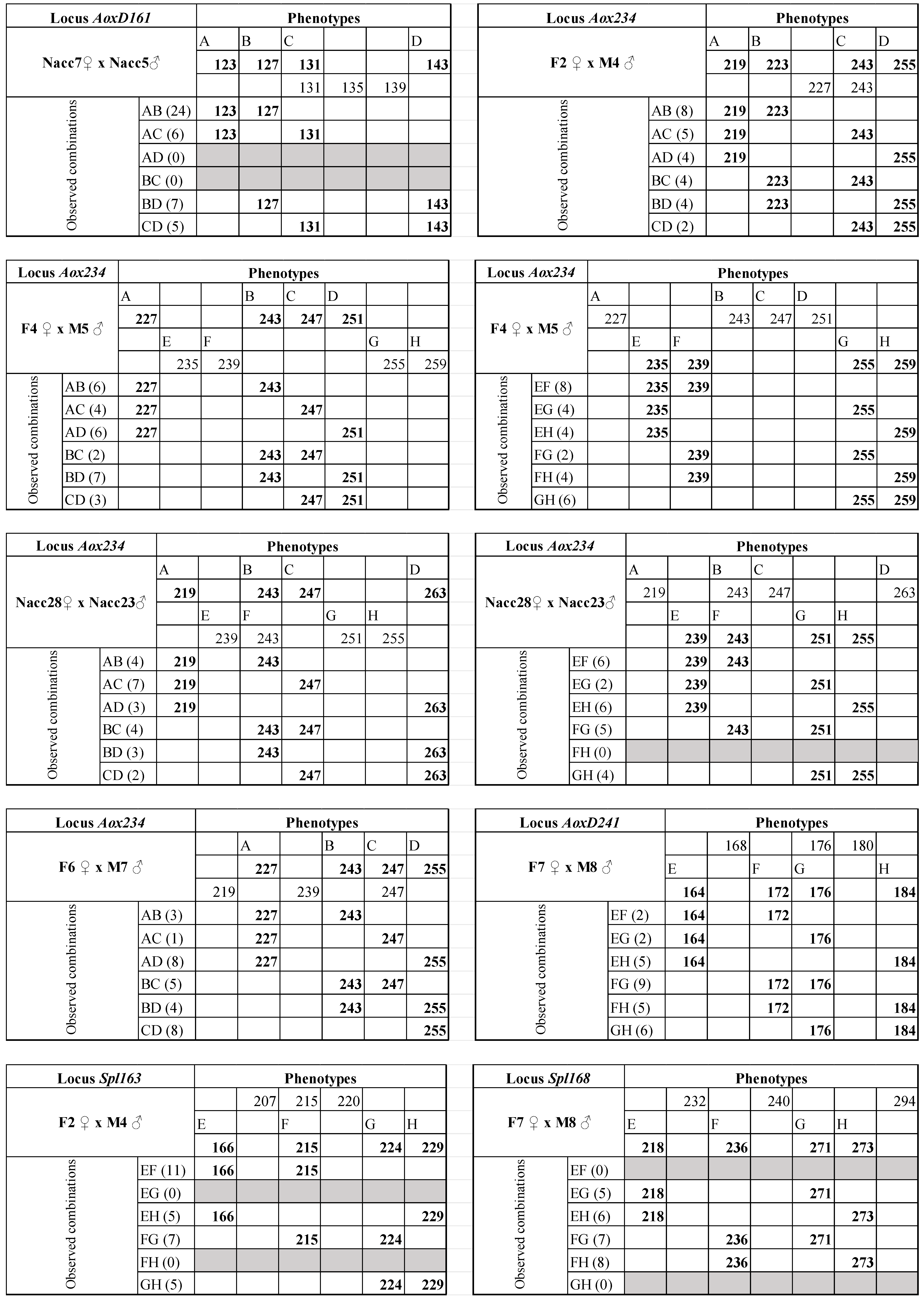
| AoxD161 | 19 | 60 °C | 111–155 | F4 ♀ × M5 ♂ | F4♀ M5 ♂ | 123/127/131/139 131/135/155 | Y | 30 |
| F8 ♀ × M2 ♂ | F8♀ M2 ♂ | 123/131/135/139 127/131 | Y | 30 | ||||
| Nacc7 ♀ × Nacc5 ♂ | Nacc7♀ Nacc5 ♂ | 123/127/131/143 131/135/139 | Y | 32 | ||||
| AoxD234 * | 19 | 52 °C | 215–275 | F2 ♀ × M4 ♂ | F2♀ M4 ♂ | 219/223/243/255 227/243 | Y | 30 |
| F4 ♀ × M5 ♂ | F4♀ M5♂ | 227/243/247/251 235/239/255/259 | Y | 30 | ||||
| Nacc28 ♀ × Nacc23 ♂ | Nacc28♀ Nacc23♂ | 219/243/247/263 239/243/251/255 | Y | 24 | ||||
| F6 ♀ × M7 ♂ | F6♀ M7 ♂ | 227/243/247/255 219/239/247 | Y | 32 | ||||
| AoxD241 * | 19 | 57 °C | 156–196 | F7 ♀ × M8 ♂ | F7 ♀ M8 ♂ | 168/176/180 164/172/176/184 | Y | 30 |
| AoxD64 * | 19 | 60 °C | 216–260 | Not informative | - | - | N | - |
| Spl-120 | 20 | 55 °C | 263–303 | Not informative | - | - | N | - |
| Spl-163 | 20 | 63 °C | 166–233 | F2 ♀ × M4 ♂ | F2 ♀ M4 ♂ | 207/215/220 166/215/224/229 | Y | 30 |
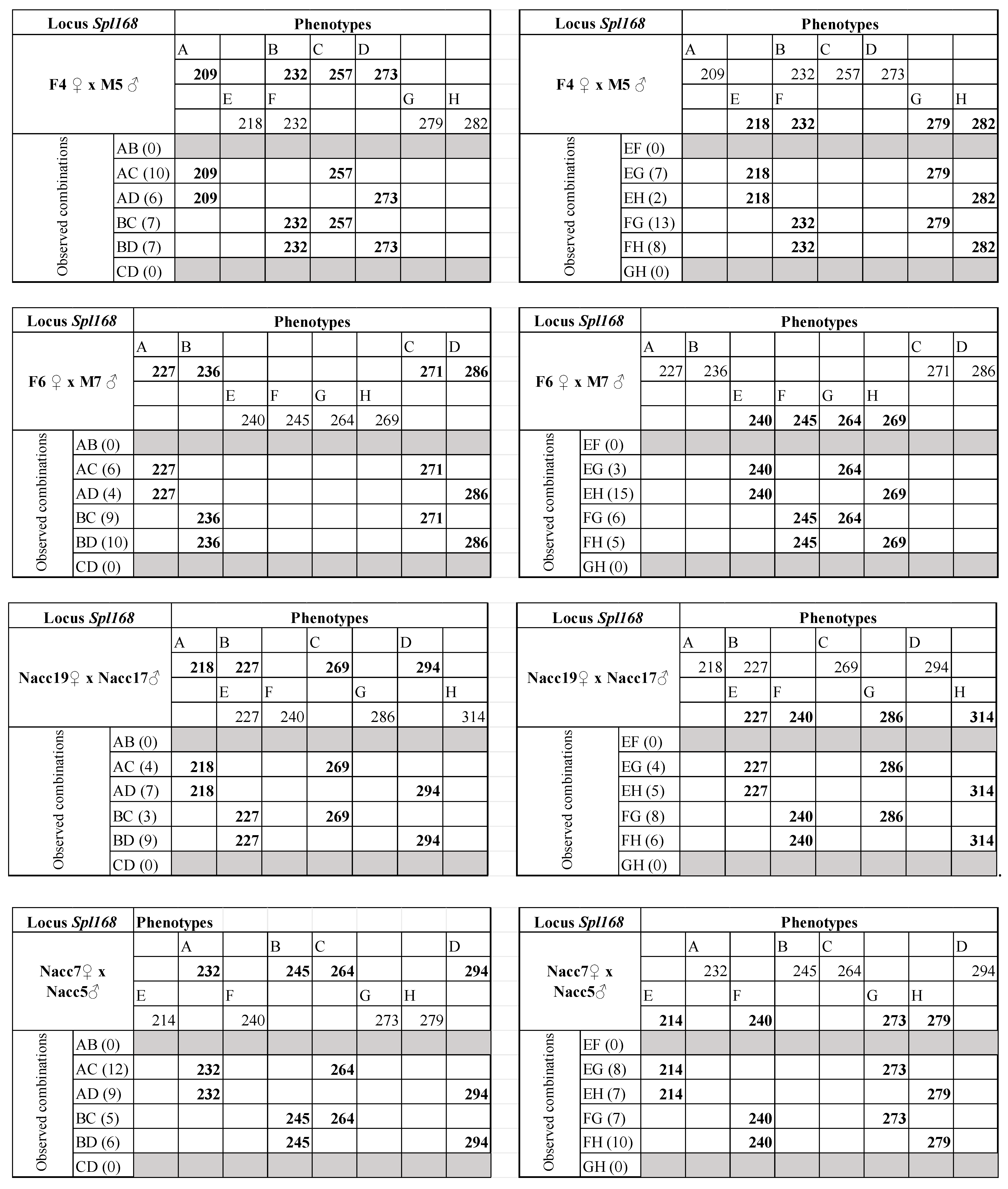
| Spl-168 | 10 | 63 °C | 200–314 | F7 ♀ × M8 ♂ | F7 ♀ M8 ♂ | 232/240/294 218/236/271/273 | Y | 30 |
| F4 ♀ × M5 ♂ | F4♀ M5♂ | 209/232/257/273 218/232/279/282 | Y | 30 | ||||
| F6 ♀ × M7 ♂ | F6♀ M7♂ | 227/236/271/286 240/245/264/269 | Y | 32 | ||||
| Nacc19 ♀ × Nacc17 ♂ | Nacc19♀ Nacc17♂ | 218/227/269/294 227/240/286/314 | Y | 32 | ||||
| Nacc7 ♀ × Nacc5 ♂ | Nacc7♀ Nacc5♂ | 232/245/264/294 214/240/273/279 | Y | 32 | ||||
| Nacc8 ♀ × Nacc31 ♂ | Nacc8♀ Nacc31♂ | 209/236/271/279 218/249/273/279 | Y | 32 |
| Locus | Family | Informative Parent | N (Nd) | Null Model (Τ = 1) Likelihood Obs | Best Intermediate Model | Model Comparison: LRT | p-Values | ||
|---|---|---|---|---|---|---|---|---|---|
| Pairing Alleles | Τ | Likelih-ood Obs | |||||||
| Afug132 | F5 ♀ × M6 ♂ | F5 ♀ | 22 (8) | 39.42 | AC/BD | 0.82 | 39.23 | 0.19 | 0.3322 |
| F6 ♀ × M7 ♂ | M7 ♂ | 29 (1) | 51.96 | AD/BC | 0.83 | 51.74 | 0.22 | 0.3185 | |
| F8 ♀ × M2 ♂ | M2 ♂ | 29 (1) | 51.96 | AB/CD | 0.83 | 51.74 | 0.22 | 0.3185 | |
| Afug41 | F8 ♀ × M2 ♂ | M2 ♂ | 27 (3) | 48.38 | AC/BD | 0.89 | 48.29 | 0.09 | 0.3853 |
| F2 ♀ × M4 ♂ | F2 ♀ | 27 (3) | 48.38 | AB/CD | 0.78 | 48.03 | 0.35 | 0.2776 | |
| M4 ♂ | 27 (3) | 48.38 | AB/CD | 0.44 | 45.98 | 2.39 | 0.0609 | ||
| F4 ♀ × M4 ♂ | F4 ♀ | 29 (1) | 51.96 | AC/BD and AD/BC | 0.93 | 51.93 | 0.03 | 0.4259 | |
| M4 ♂ | 29 (1) | 51.96 | AD/BC | 0.83 | 51.74 | 0.22 | 0.3185 | ||
| F6 ♀ × M7 ♂ | F6 ♀ | 28 (2) | 50.17 | AD/BC | 0.86 | 50.02 | 0.15 | 0.3509 | |
| M7 ♂ | 28 (2) | 50.17 | AB/CD | 0.86 | 50.02 | 0.15 | 0.3509 | ||
| Nacc7 ♀ × Nacc5 ♂ | Nacc7 ♀ | 28 (4) | 50.17 | AD/BC | 0.64 | 49.21 | 0.96 | 0.1631 | |
| Nacc5 ♂ | 28 (4) | 50.17 | AD/BC | 0.75 | 49.71 | 0.46 | 0.2489 | ||
| Nacc8 ♀ × Nacc 31♂ | Nacc8 ♀ | 31 (1) | 55.54 | AD/BC | 0.77 | 55.13 | 0.41 | 0.2603 | |
| An20 * | F7 ♀ × M8 ♂ | M8 ♂ | 30 (0) | 53.75 | AD/BC | 0.70 | 53.03 | 0.72 | 0.1984 |
| Anac6784 | Nacc19 ♀ × Nacc17 ♂ | Nacc19 ♀ | 30 (2) | 53.75 | AC/BD and AD/BC | 0.91 | 53.68 | 0.08 | 0.3912 |
| Anac3133 | F7 ♀ × M8 ♂ | M8 ♂ | 29 (1) | 51.96 | AB/CD | 0.41 | 49.06 | 2.90 | 0.0444 ª |
| AnacA6 * | F5 ♀ × M6 ♂ | M6 ♂ | 29 (1) | 51.96 | AB/CD and AC/BD | 0.72 | 51.38 | 0.58 | 0.2225 |
| AnacB11 | F6 ♀ × M7 ♂ | M7 ♂ | 26 (4) | 46.59 | AB/CD | 0.58 | 45.31 | 1.28 | 0.1290 |
| Nacc19 ♀ × Nacc17 ♂ | Nacc19 ♀ | 26 (6) | 46.59 | AB/CD | 0.81 | 46.34 | 0.25 | 0.3088 | |
| AnacB7 | F6 ♀ × M7 ♂ | F6 ♀ | 27 (3) | 48.38 | AB/CD | 0.67 | 47.57 | 0.80 | 0.1849 |
| Nacc19 ♀ × Nacc30 ♂ | Nacc30 ♂ | 32 (0) | 57.34 | AD/BC | 0.94 | 57.30 | 0.03 | 0.4295 | |
| AnacE4 * | F5 ♀ × M6 ♂ | F5 ♀ | 28 (2) | 50.17 | AB/CD and AD/BC | 0.96 | 50.16 | 0.01 | 0.4622 |
| AoxD161 | F4 ♀ × M5 ♂ | F4 ♀ | 29 (1) | 51.96 | AB/CD | 0.00 | 40.20 | 11.76 | 0.0003 |
| F8 ♀ × M2 ♂ | F8 ♀ | 30 (0) | 53.75 | AC/BD | 0.10 | 45.28 | 8.47 | 0.0018 ª | |
| Nacc7 ♀ × Nacc5 ♂ | Nacc7 ♀ | 32 0) | 57.34 | AD/BC | 0.00 | 44.36 | 12.97 | 0.0002 | |
| AoxD234 * | F2 ♀ × M4 ♂ | F2 ♀ | 27 (3) | 48.38 | AD/BC | 0.89 | 48.29 | 0.09 | 0.3853 |
| F4 ♀ × M5 ♂ | F4 ♀ | 28 (2) | 50.17 | AD/BC | 0.86 | 50.02 | 0.15 | 0.3509 | |
| M5 ♂ | 28 (2) | 50.17 | AD/BC | 0.64 | 49.21 | 0.96 | 0.1631 | ||
| Nacc28 ♀ × Nacc23 ♂ | Nacc28 ♀ | 23 (1) | 41.21 | AB/CD | 0.78 | 40.93 | 0.28 | 0.2972 | |
| Nacc23 ♂ | 23 (1) | 41.21 | AC/BD | 0.26 | 37.29 | 3.92 | 0.0239 ª | ||
| F6 ♀ × M7 ♂ | F6 ♀ | 29 (1) | 51.96 | AC/BD | 0.52 | 50.07 | 1.89 | 0.0844 | |
| AoxD241 * | F7 ♀ × M8 ♂ | M8 ♂ | 29 (1) | 51.96 | AC/BD | 0.72 | 51.38 | 0.58 | 0.2225 |
| Spl163 | F2 ♀ × M4 ♂ | M4 ♂ | 28 (2) | 50.17 | AC/BD | 0.00 | 38.82 | 11.35 | 0.0004 |
| Spl168 | F7 ♀ × M8 ♂ | M8 ♂ | 26 (4) | 46.59 | AB/CD | 0.00 | 36.04 | 10.54 | 0.0006 |
| F4 ♀ × M5 ♂ | F4 ♀ | 30 (0) | 53.75 | AB/CD | 0.00 | 41.59 | 12.16 | 0.0002 | |
| M5 ♂ | 30 (0) | 53.75 | AB/CD | 0.00 | 41.59 | 12.16 | 0.0002 | ||
| F6 ♀ × M7 ♂ | F6 ♀ | 29 (1) | 51.96 | AB/CD | 0.00 | 40.20 | 11.76 | 0.0003 | |
| M7 ♂ | 29 (1) | 51.96 | AB/CD | 0.00 | 40.20 | 11.76 | 0.0003 | ||
| Nacc19 ♀ × Nacc17 ♂ | Nacc19 ♀ | 23 (9) | 41.21 | AB/CD | 0.00 | 31.88 | 9.33 | 0.0011 | |
| Nacc17 ♂ | 23 (9) | 41.21 | AB/CD | 0.00 | 31.88 | 9.33 | 0.0011 | ||
| Nacc7 ♀ × Nacc5 ♂ | Nacc7 ♀ | 32 (0) | 57.34 | AB/CD | 0.00 | 44.36 | 12.97 | 0.0002 | |
| Nacc5 ♂ | 32 (0) | 57.34 | AB/CD | 0.00 | 44.36 | 12.97 | 0.0002 | ||
| Nacc8 ♀ × Nacc31 ♂ | Nacc8 ♀ | 29 (3) | 51.96 | AB/CD | 0.00 | 40.20 | 11.76 | 0.0003 | |
| Nacc31 ♂ | 29 (3) | 51.96 | AB/CD | 0.10 | 43.86 | 8.10 | 0.0022 ª | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dalle Palle, S.; Boscari, E.; Bordignon, S.G.; Muñoz-Mora, V.H.; Bertorelle, G.; Congiu, L. Different Chromosome Segregation Patterns Coexist in the Tetraploid Adriatic Sturgeon Acipenser naccarii. Diversity 2022, 14, 745. https://doi.org/10.3390/d14090745
Dalle Palle S, Boscari E, Bordignon SG, Muñoz-Mora VH, Bertorelle G, Congiu L. Different Chromosome Segregation Patterns Coexist in the Tetraploid Adriatic Sturgeon Acipenser naccarii. Diversity. 2022; 14(9):745. https://doi.org/10.3390/d14090745
Chicago/Turabian StyleDalle Palle, Stefano, Elisa Boscari, Simone Giulio Bordignon, Víctor Hugo Muñoz-Mora, Giorgio Bertorelle, and Leonardo Congiu. 2022. "Different Chromosome Segregation Patterns Coexist in the Tetraploid Adriatic Sturgeon Acipenser naccarii" Diversity 14, no. 9: 745. https://doi.org/10.3390/d14090745
APA StyleDalle Palle, S., Boscari, E., Bordignon, S. G., Muñoz-Mora, V. H., Bertorelle, G., & Congiu, L. (2022). Different Chromosome Segregation Patterns Coexist in the Tetraploid Adriatic Sturgeon Acipenser naccarii. Diversity, 14(9), 745. https://doi.org/10.3390/d14090745









