Abstract
Earthworms are an important ecological group, especially in agricultural regions in Northeast China. However, fewer studies focus on this group of organisms compared with other faunal groups. Here, we sequenced 15 new mitogenomes of Aporrectodea tuberculata Eisen, 1874, A. trapezoides Duges, 1828, Eisenia nordenskioldi Eisen, 1878 and Drawida ghilarovi Gates, 1969 in Northeast China using a high-throughput sequencing platform. These incomplete linear and double-stranded mitogenomes vary from 14,998 bp to 16,123 bp in size and include 37 genes and a putative control region. Intraspecific genetic divergence was quantified in the lumbricid species, and a control region in D. ghilarovi was reported for the first time by comparison to the mitogenomes of the congeners. Phylogenetic analysis based on coding genes and ribosomal DNA datasets using BI and ML inferences showed the non-monophyly of Aporrectodea and polyphyly of E. nordenskioldi. Future works should examine the taxonomy, phylogeny and population genetics not only of Lumbricidae but also the other earthworm families on the global scale using mitogenomic and nuclear data.
1. Introduction
Earthworms play an important role in many ecosystem processes, such as soil aeration, turnover and drainage, as well as the composition and decomposition of organic matter [1]. However, for such an important ecological group, the taxonomy and phylogeny of earthworms, particularly Lumbricidae and Drawida Michaelsen 1900 of Moniligastridae, which are widespread across the Holarctic, are inadequate [2,3,4]. The reasons for the taxonomic insufficiencies may be due to the structural simplicity, which lacks complex appendices or highly specialized copulatory apparatuses [5]. The lack of characters has led to many morphologically similar species being lumped into a single species with various morphotypes or as a species complex that includes various taxa of uncertain taxonomic category. This is especially true for the species of Aporrectodea Orley, 1885 and Eisenia Malm, 1877 of Lumbricidae, and Drawida [2,3,4,5,6,7,8,9,10,11,12].
Eisenia nordenskioldi, a species widespread in Northern Asia and adjacent regions, is known for its high morphological, karyotypic and genetic variation [8,13]. Its distribution extends from tundra to forest–steppe and broad-leaved forests. The diagnostic features of this species include the following: the body has no stripes; the clitellum is faint, yellow, and saddle-shaped in xxvii–xxxii; the spermathecal pores are paired in 9/10 and 10/11 ventrally; the spermathecae are round; and the setal arrangement is lumbricine. Aporrectodea trapezoides Dugés 1828 and A. tuberculata Eisen, 1874 belong to the Aporrectodea caliginosa species complex, the most abundant earthworm group from the Paleartic grassland and forest regions. One diagnostic feature of A. tuberculata is its lack of body pigmentation, while A. trapezoides is brown. The position of the clitellum for both of the species occurs within the same range of segments, but the form and position of the tubercula pubertatis differ—they appear as two protuberances in A. tuberculata, while they appear as two lateral bands in A. trapezoides [5]. Meanwhile, the vestiture of Drawida ghilarovi found in this region is characterized by color variations: light-bluish, brown and grey [13] and pitch-black [14,15]. The life-forms are considered as black-wetland worm and other colored forest worms.
The reported complete mitogenomes of earthworms range from 14,648 to 15,188 base pairs (bp), consisting of 37 genes, including 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs) and a putative control region (CR) [16,17,18,19,20,21,22,23,24,25]. Mitochondrial sequences are considered to be ideal genetic markers that are extensively employed for the study of systematics, phylogeography and species delimitation in animals [26,27,28,29,30,31,32]. In previous studies of the phylogeny in earthworms [5,33], the main clades were not robust, with low support values using single or several genetic makers. The advantages of using the mitogenome as a phylogenetic marker over other genes to infer an evolutionary relationship have been discussed in various works. For instance, mitogenomes have shown to provide resolution for an enormously broad range of phylogenetic depths, from shallow divergence times between populations of a single species to deep divergence within an entire phylum [34,35,36]. Mitogenomes have often proved to be useful in resolving formerly troublesome phylogenies, clarifying the relationships within phylogenetically difficult groups where rapid radiations made other markers ineffective [37,38]. Previously in earthworm mitogenomic studies, more attention was paid to the mitogenomic studies of pheretimoids (Megascolecidae) in the Oriental Realm [18,20,21], while only a few studies were completed on moniligastrids and lumbricids.
Here, 15 new mitogenomes of Aporrectodea tuberculata Eisen, 1874, A. trapezoides Duges, 1828, Eisenia nordenskioldi Eisen, 1878 and Drawida ghilarovi Gates, 1969 were sequenced, assembled and analyzed in order to shed light on the mitochondrial structures of these species to assess the phylogenetic relationships of the two genera of Lumbricidae and a genus of Moniligastridae.
2. Materials and Methods
2.1. Sample Preparation and DNA Extraction
Four earthworm species of Lumbricidae and Moniligastridae from Northeast China (Figure 1), namely: A. tuberculata (Heilongjiang: Anda: Renmin Park: 46.400° N, 125.330° E); A. trapezoides (Jilin: Jilin: 43.727° N, 126.628° E) and E. nordenskioldi (Jilin: Changchun: Nanhu Park: N43.859, E125.308; Heilongjiang: Bei’an: Renmin Park 48.2536° N, 126.5040° E; Heilongjiang: Wudalianchi: 48.5794° N, 125.9435° E) of Lumbricidae, and D. ghilarovi (Jilin: Changchun: Jingyuetan National Scenic Area: 43.490° N, 125.790° E) of Moniligastridae were collected by hand in July–August, 2021. See Table S1 for detailed information concerning the samples. All of the samples were preserved in 95% ethanol, which was replaced with new ethanol every week during the fieldwork, and stored at room temperature (15–25 °C) in the lab. One to six specimens of each species were taken for mitogenomic analysis. The genomic DNA was extracted using TIANamp Micro DNA Kit (TIANGEN, Beijing, China) according to the manufacturer’s protocol. The DNA concentrations were measured with a BIO DL MicroDrop spectrophotometer (Beijing, China). Table 1 shows the earthworm mitogenomes used in this study.
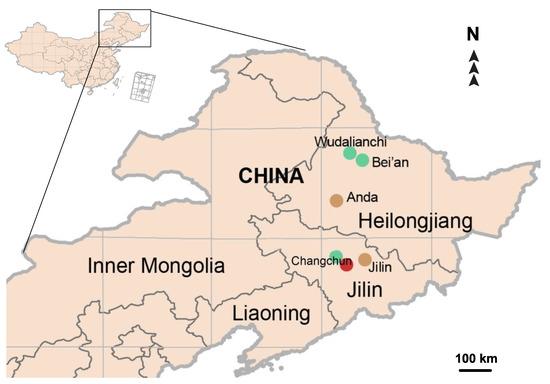
Figure 1.
The distribution of earthworm samples used in this study. The brown circles indicate the distribution of Aporrectodea species, the green circles indicate the distribution of Eisenia nordenskioldi and the red circle indicates the distribution of Drawida ghilarovi.

Table 1.
Mitogenomes used for the phylogeny of earthworms in this study.
2.2. Next Generation Sequencing
The genomic DNA was fragmented using the Covaris S220 Focused Ultrasonicator (Covaris, Woburn, MA, USA), and the 300–400 bp fragments were captured by magnetic beads. After repairing the blunt ends, adenylating 30 ends and ligating adapters, the fragmented DNA was enriched, and the PCR products were captured again by magnetic beads. The PCR products were sequenced by BGISEQ500 platform, and a sequence library was constructed. The raw data were cleaned by removing the following: (1) the adapters; (2) the reads that contained five and more Ns; (3) the reads that were shorter than 75 bp or the average Phred values lower than Q15; (4) the reads of the Phred values lower than Q20 with 4 bp slide windows; and (5) the reads that contained polyN (A, T, C, G) that were longer than 50 bp.
2.3. Data Assembly and Mitogenome Annotation
The clean reads were assembled by MitoZ v2.4 (Meng et al., Shenzhen, China) [45], NOVOplasty 4.3.1 (Dierckxsens et al., Brussels, Belgium) [46] and GetOrganelle 1.6.0 (Jin et al., Kunming, China) [47]. Annotation was performed using MitoZ and the MITOS2 webserver [48]. The mitochondrial genome was set as cox1 to begin with, following the other mitogenomes of earthworm with “Mitogenome_reorder.py” provided by MitoZ v2.4, and visualized by Circos 0.69 (Krzywinski et al., Vancouver, Canada) [49].
2.4. Statistics of the Earthworm Mitogenomes
The nucleotide composition of the whole mitogenome, which includes 13 PCGs, 22 tRNAs, 2 rRNAs and a putative control region (CR), were counted. The relative synonymous codon usage (RSCU) of the PCGs of the four earthworms were calculated in MEGA5 [50]. The nucleotide compositional skew was estimated according to the formula: AT skew = [A − T]/[A + T], GC skew = [G − C]/[G + C]) [51]. Genetic divergence within species, including genetic variants and indels, was detected by DNASP v5 (Librado, Barcelona, Spain) [52]. The tandem repeats were detected using the webserver “TANDEM REPEATS FINDER” (https://tandem.bu.edu/trf/trf.basic.submit.html, accessed on 3 January 2022).
2.5. Phylogenetic Analysis
To investigate the phylogeny of earthworms with the mitogenome data, we reconstructed the phylogenetic relationships among the earthworm species using two datasets (PCG: 13 PCGs, including all three codons, and PCGrRNA: 13 PCGs and 12S, 16S rRNAs) with Bayesian inference (BI) and maximum likelihood (ML) methods under a partitioned strategy. A total of 46 sequenced data are available online and 15 newly sequenced data are used here to perform the phylogenetic analysis. The details of the taxa are shown in Table 1. The mitogenomic phylogenetic trees of the earthworms were reconstructed representing 13 genera of earthworms belonging to three families: Megascolecidae, Lumbricidae and Moniligastridae. Tubifex tubifex Müller, 1774 and Nais communis Piguet, 1906 of Naididae (freshwater oligochaetes) were used as the outgroups in this study.
The best substitution models based on the Akaike information criterion (AIC) were selected by jModeltest 2.1.7 (Darriba et al., Vigo, Spain) [53] for the two datasets, and the selected models were listed in Table S2. BI analysis was performed with the chosen models above using MrBayes 3.2.6 (Ronquist et al., Florida, USA) [54]. To ensure that the average standard deviation of split frequencies was less than 0.01, two million generations were run with sampling every 1000 generations. All of the parameters achieved an effective sample size (ESS) larger than 200 that was checked by Tracer v1.7.2 (Rambaut et al., Edinburgh, UK) [55] after combining two runs and adjusting the burn-in. The consensus trees were calculated, including burning in the former 25% calculation. The ML trees were constructed by RAxML 8 (Stamatakis, Heidelberg, Germany) [56] using the default rapid hill-climbing algorithm, and 1000 replicates of bootstrap with GTRCAT model were generated. All of the phylogenetic trees were visualized using Figtree v1.4.4 (Rambaut, Edinburgh, UK) [57].
3. Results
3.1. Mitogenome Organization and Nucleotide Composition of the Four Earthworm Species
All 15 mitogenomes assembled by the three bioinformatics tools are incomplete. The annotations of A. tuberculata, A. trapezoides, E. nordenskioldi and D. ghilarovi are listed in Tables S3 and S4, and the maps of mitogenomes of the four species are visualized in Figure 2. The ratio of AT, GC, AT skew and GC skew of whole mitogenomes, PCGs, tRNAs, rRNAs and CR are listed in Table S5. The accession numbers of the 15 new mitogenomes are OL840312–16 and OM687882–90 (see Table 1 for details). The total length of the six mitogenomes of A. tuberculata varies from 15,058 bp to 15,129 bp; that of A. trapezoides is 14,998 bp; that of the six mitogenomes of E. nordenskioldi vary between 15,290 bp and 16,123 bp; and that of the two mitogenomes of D. ghilarovi vary between 15,080 bp and 15,061 bp.
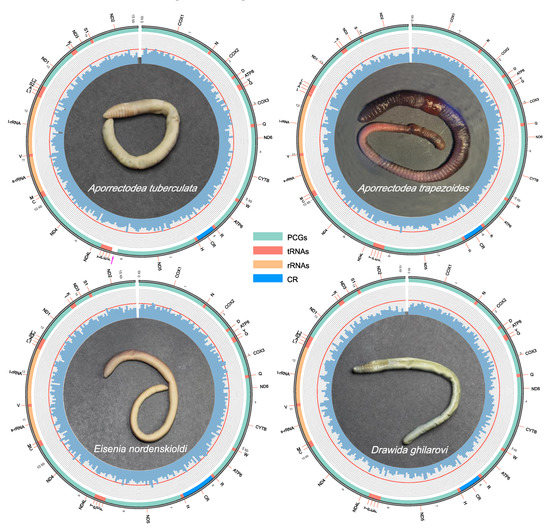
Figure 2.
Map of mitogenomes of A. tuberculata, A. trapezoides, E. nordenskioldi and D. ghilarovi. The inner circles indicate the GC content in every 50-site window, and the outer circle shows the arrangement of the genes; cox1 was set as the start of mitogenome. All genes are coded on the majority strand.
The intraspecies genetic divergence is quite high in the six mitogenomes from the same population of A. tuberculata, with 447 variants in 13 PCGs, 6 tRNAs, 2 rRNAs, CR and introns, and eight indel events were also detected in trnW, trnR, trnT, rrnL and CR (see Table S5). The genetic polymorphism of the two mitogenomes of E. nordenskioldi from Changchun (Nanhu Park) contains only five variants and three indels in cox1, cox2, nd4, nd5, rrnL and CR (see Table S5). Seven variants in atp8, cox1, cytb, rrnL and CR, and three indels in trnR and CR were also detected in the four mitogenomes within E. nordenskioldi lineages from Bei’an-Wudalianchi (see Table S5). Meanwhile, there was only one indel in the CR in the two mitogenomes of D. ghilarovi from Changchun (Jingyuetan National Scenic Area) (see Table S5).
The nucleotide compositions of the 15 new mitogenomes were similar within families while different between families (see Table S6 for details, Supplementary Materials). In Lumbricidae, the AT content of the whole mitogenomes of the lumbricids A. tuberculata, A. trapezoides and E. nordenskioldi ranged from 61.0% to 64.5%; that of the moniligastrid D. ghilarovi was 72.5%; the GC content of the three lumbricid species ranged from 35.5% to 39.0%; that of the D. ghilarovi was much lower (27.5%). The AT skews of the lumbricids ranged from −4.7% to 2.0%, while that of the moniligastrid was lower (−7.9%); the GC skews of the lumbricids ranged from −19.2% to −21.2%, and that of D. ghilarovi was much higher (−8.7%).
3.2. Protein-Coding Genes and Codon Usage in the Four Earthworm Species
In the 15 new mitogenomes, there was a large difference in the PCG length between families, a little difference between species and no difference within species. The lengths of the PCGs of the lumbricids A. tuberculata, A. trapezoides and E. nordenskioldi were 11,116 bp, 11,110 bp and 11,114 bp, respectively; that of the moniligastrid D. ghilarovi was 11,134 bp. The PCGs of the four earthworm species exhibited similar initiation and termination codons (Tables S3 and S4). All of the initiation codons of the four earthworms were ATG. Most of the termination codons were TAR, and atp8, cox1, cox3, nd2 and nd4l in all four species, nd1 in Aporrectodea, nd6 in E. nordenskioldi and nd4 in D. ghilarovi were incomplete stop codon T or TA (Tables S3 and S4). These truncated stop codons are commonly used in mitogenomes in Metazoa, and are modified to a complete stop codon by the post-transcriptional poly-adenylation [58]. The RSCU values of the four earthworm species are shown in Figure 3. The codons CGA-Arg and UCU/A-Ser2 in the four earthworm species and the UUA-Leu2 in D. ghilarovi were the most common.
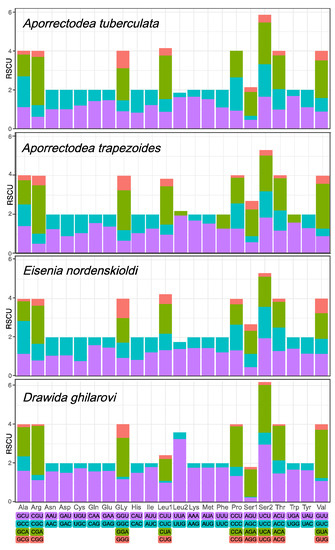
Figure 3.
Relative synonymous codon usage (RSCU) in the PCGs of the four newly sequenced earthworm species. Codon families are indicated below the X axis.
3.3. RNA Genes of the Four Earthworm Species
The secondary structures of 22 tRNAs of the 3 lumbricids, A. tuberculata (OL840316), A. trapezoides (OM687887) and E. nordenskioldi (OL840314) are illustrated in Figure 4. All of the tRNAs can fold as a typical cloverleaf structure, except the trnA, trnG and trnV whose lack the TψC loop and trnS1 which lacks the DHU stem. Figure 5 shows the secondary structures of the tRNAs of the moniligastrid D. ghilarovi: trnN, trnD, trnG, trnK, trnT, trnY and trnV lack the TψC loop, and trnS1 lacks the DHU loop. In addition, the identified wobble GT base pairs and unmatched base pairs AA, CA, CT and TT in different tRNA stems are shown in Figure 4 and Figure 5, and they might be restored during the post-transcriptional editing processes [59].
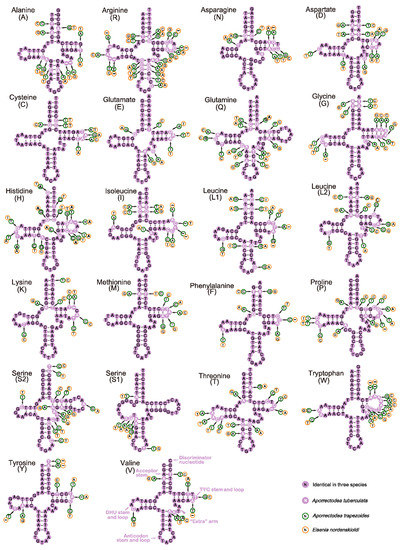
Figure 4.
Predicted secondary structure for the tRNAs of the three lumbricid species. The tRNAs are labeled with the abbreviations of their corresponding amino acids. Dashes indicate the Watson-Crick base pairs; dots indicate the wobble GT pairs and gaps indicate non-match bases.
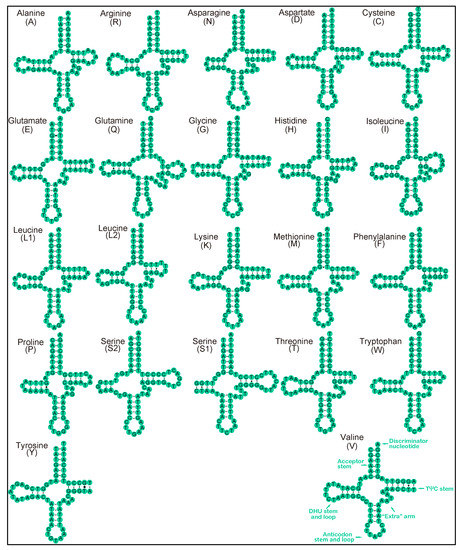
Figure 5.
Predicted secondary structure for the tRNAs of D. ghilarovi. The tRNAs are labeled with the abbreviations of their corresponding amino acids. Dashes indicate the Watson–Crick base pairs, dots indicate the wobble GT pairs and gaps indicate non-match bases.
The entire lengths of the rRNAs of the four species are similar, ranging from 2034 bp to 2044 bp. The rRNAs of the four species showed a positive AT skew and negative GC skew in the three lumbricids, which was positive in moniligastrid D. ghilarovi. The rRNAs A+T% was 62.4–65.6% in lumbricids and 72.9% in D. ghilarovi (see Table S3 for details).
3.4. Putative Control Region and Non-Coding Region
All of the putative CRs were located between trnR and trnH in the four earthworms. The lengths of CRs were different within and among species; the CR length varied from 374 to 444 bp among the individuals in A. tuberculata; 440 bp in A. trapezoides; 757 and 761bp in the E. nordenskioldi lineage from Changchun; 1569–1578 bp in the E. nordenskioldi lineage from Bei’an-Wudalianchi; and 468 and 487 bp in D. ghilarovi (see Table S3 for the details). There were three tandem repeats that were detected (AT, AATACA, ATACAAATATAT) in the CRs of A. tuberculata (OM687883 and/or OM687883 and/or OM687886); two copies of a 175 bp tandem repeat were detected in the Bei’an-Wudalianchi lineage of E. nordenskioldi; no tandem repeat was detected in the Changchun lineage of E. nordenskioldi; and 35–40 copies of AT were detected in D. ghilarovi.
3.5. Phylogenetic Analysis
The Aporrectodea tuberculata species in this work is similar to the lineages of North Europe (Denmark, Finland and Poland) (Figure S1) [5], while A. trapezoides belongs to the lineage of a parthenogenetic group that occurs in a large region in Europe (Serbia, France and Spain) [5] based on the cox2 gene data (Figure S1). On the other hand, Drawida ghilarovi in this study is separate from that of the cox1 data from Russia (Figure S2) [2,7].
The phylogenetic trees of BI and ML reveal that the three families are reciprocal monophyletic with high support (both posterior probabilities (PP) = 1 and bootstrap (BS) = 100, as shown in Figure 6). For Megascolecidae, the species of Amynthas and Metaphire are scattered throughout a multi-clade, which mostly agrees with Zhang et al.’s study [21]. For the details on the trees of Megascolecidae, see Figures S3–S6.
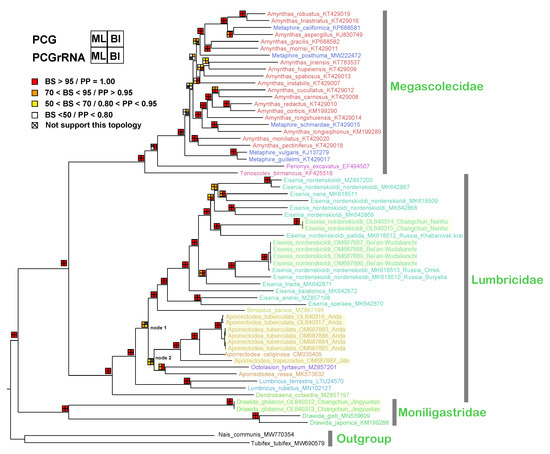
Figure 6.
Phylogenetic trees of earthworm using mitogenome data. The four squares near the node indicate the support value: top left, ML of PCG; bottom left, ML of PCGrRNA; top right, BI of PCG; bottom right, BI of PCGrRNA. The newly sequenced mitogenomes are highlighted, and genera delimited by the current taxonomic base are distinguished from each other by a particular color.
For Lumbricidae, the phylogenetic relationship between Aporrectodea, Octolasion and Lumbricus and the species within Eisenia is unclear. Clusters ((Eisenia, Bimastus) and (Aporrectodea, Octolasion)) formed with a low support value (node 1 in Figure 6). Aporrectodea is paraphyletic in both of the ML trees and the BI tree from the PCGrRNA dataset, as the topology shows (((A. tuberculata, A. caliginosa), A. trapezoides) (A. rosea, Octolasion)) (node 2 in Figure 6). Aporrectodea is also polyphyletic, as the topology shows ((Eisenia, Bimustus), (A. rosea, Octolasion)), ((A. tuberculata, A. caliginosa), A. trapezoides) in the BI tree of the PCG dataset with a low support value (PP = 0.66), as seen in the bottom right of Figure 6.
4. Discussion
4.1. Organization of a Partial Mitochondrial Genome
The 15 newly sequenced mitogenomes showed no variation in the number of genes, the strand location and the arrangement of genes (Table S3 and Figure 2) as compared with the mitogenomes of earthworm species in other works [13,14,15,16,17,18,19,20,21,22]. The 15 mitogenomic sequences that were assembled by the three tools failed to completely circularize, although lengths fell under the ranges of those circular mitogenomes [16,17,18,19,20,21,22,23,24,25]. As shown in Figure 2, the gap in the sequences was between nd2 and cox1. In the future, circular mitogenomes may be assembled using the Pacbio or Nanopore sequence platform that can produce reads longer than 10 kb [60], covering the polyNs or short tandem repeats of mitogenomes in earthworms.
The mitogenomes of A. tuberculata are distinguished from A. trapezoides and A. rosea [24] by containing a non-coding region (noted by the pink arrow on Figure 2). The mitogenome sequences of D. ghilarovi is differentiated from D. gisti [25] and D. japonica [21] by the presence of a putative CR (Figure 2). The putative control region is often absent in partial sequences due to certain technical difficulties in deciphering it by the traditional PCR method [21,25]. More mitogenomes of Drawida should be sequenced in the future by next generation sequencing to confirm the existence of CR in these earthworms.
4.2. Genetic Diversity of Lumbricids in Northeast China
Quantities of DNA polymorphs were detected in the mitogenomes of lumbricids that were collected from the gardens of downtown areas in Northeast China. This indicates that these gardens may be refuges for lumbricids. There were fewer earthworms collected in the farmland than in the gardens. This may be due the overuse of herbicides or pesticides that may have resulted in a reduction in earthworms in the farmland [61]. To test for the most common type of refuge for lumbricids in Northeast China, more earthworm samples need to be studied in the future.
4.3. Phylogenetic Inference
The paraphyly of E. nordenskioldi and the non-monophyly of Aporrectodea are supported by both BI and ML analyses. The non-monophyly may be explained by the following points: (1) the mitochondrial loci are a maternal inheritance, their rate of divergence is faster than that of the nuclear loci, and that may lead to the non-monophyletic topology of Aporrectodea and Octolasion; (2) taxonomical characters of E. nordenskioldi, Aporrectodea, Octolasion and Lumbricus are not synapomorphic, which results in the chaos of the taxonomy in Lumbricidae; (3) the number of species analyzed using mitogenomic data was inadequate for acquiring a clearer picture of the systematic relationships of Aporrectodea, Octolasion and Lumbricus. The non-monophyly of E. nordenskioldi agrees with the data from Shekhovtsov et al. [8], while the non-monophyly of Aporrectodea agrees with the data from Pop et al. [62], Pérez-Losada et al. [5] and Shekhovtsov et al. [4,63]. This suggests that certain morphological characters that are used to define these taxa could be homoplasious. A revision in the taxonomic identification and classification is needed, taking into consideration other morphological and ecological characteristics.
Our mitogenomic phylogenetic inference of Lumbricidae agrees with the study by Domínguez et al. [64], which contained the nuclear loci (18S and 28S rDNAs), the morphological characters and a large number of species that reach 76. The mitogenomic phylogeny of Lumbricidae here makes sense and aligns with the former attempts that have consistently indicated the need for extensive revision of the taxonomy of Lumbricidae.
5. Conclusions
Fifteen new mitogenomes from four Palearctic earthworm species are provided here. We reported the presence of CR in D. ghilarovi for the first time. Quantities of variants were detected among lumbricid species, particularly in the population of A. tuberculata. Phylogenetic analysis of BI and ML indicates the non-monophyletic taxa in Lumbricidae. Future works should examine the taxonomy, phylogeny and population genetics not only of Lumbricidae but also of the other earthworm families on a global scale using mitogenomic and nuclear data.
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/article/10.3390/d14090714/s1, Figure S1: NJ tree of COX2 of available online and this study of Aporrectodea; Figure S2: NJ tree of COX1 of all available online and this study of Drawida ghilarovi; Figure S3: ML tree of PCG dataset; Figure S4: ML tree of PCGrRNA dataset; Figure S5: BI tree of PCG dataset; Figure S6: BI tree of PCGrRNA dataset; Table S1: The information of specimens in the study; Table S2: The best substitute models were selected by jModeltest 2.1.7 for the genes in PCGrRNA and PCG datasets; Table S3: Organization of the mitogenomes of the lumbricid species A. tuberculata (OL840316), A. trapezoides (OM687887) and E. nordenskioldi (OL840314); Table S4: Organization of the mitogenomes of D. ghilarovi (OL840312; Table S5: The nucleotide sites of the variants and indels of earthworms in this study; Table S6: Statistics of mitogenomes of A. tuberculata (420Ra-f), A. trapezoides (DL377), E. nordenskioldi (374NHa-b, 451Ra-b, 456Ra-b) and D. ghilarovi (374JYa-b).
Author Contributions
Sampling, S.F., L.F. and Y.Z.; data curation and formal analysis, H.Z.; funding acquisition and investigation, Y.Z.; writing—original draft, H.Z. and Y.Z.; writing—review and editing, N.M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Science and Technology Project of Hebei Education Department (QN2021126), the project of Northeast Asia Biodiversity Research Center (2572022DS09) to H.Z., the National Science and Technology Fundamental Resources Investigation Program of China (2018FY100301) to Y.Z. and National Natural Scientific Fund of China (42071059).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Hebei Key Laboratory of Animal Diversity, Langfang Normal University.
Data Availability Statement
“MDPI Research Data Policies” at https://www.mdpi.com/ethics, accessed on 25 November 2021.
Acknowledgments
We are grateful to Hao Jiahua (Northeast Normal University, Changchun, China) for his assistance in the collection of specimens during the field work.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Edwards, C.A. Earthworm Ecology; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar] [CrossRef]
- Ganin, G.N.; Atopkin, D.M. Molecular differentiation of epigeic and anceic forms of Drawida ghilarovi Gates, 1969 (Moniligastridae, Clitellata) in the Russian Far East: Sequence data of two mitochondrial genes. Eur. J. Soil. Biol. 2018, 86, 1–7. [Google Scholar] [CrossRef]
- Shekhovtsov, S.V.; Berman, D.I.; Bulakhova, N.A.; Vinokurov, N.N.; Peltek, S.E. Phylogeography of Eisenia nordenskioldi nordenskioldi (Lumbricidae, Oligochaeta) from the north of Asia. Polar Biol. 2018, 41, 237–247. [Google Scholar] [CrossRef]
- Shekhovtsov, S.V.; Derzhinsky, Y.A.; Poluboyarova, T.V.; Golovanova, E.V.; Peltek, S.E. Phylogeography and genetic lineages of Aporrectodea rosea (Lumbricidae, Annelida). Eur. J. Soil. Biol. 2020, 99, 103191. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Ricoy, M.; Marshall, J.C.; Domínguez, J. Phylogenetic assessment of the earthworm Aporrectodea caliginosa species complex (Oligochaeta: Lumbricidae) based on mitochondrial and nuclear DNA sequences. Mol. Phylogenet. Evol. 2009, 52, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Ganin, G.; Anisimov, A.; Roslik, G.; Atopkin, D. The Russian Far East Endemic Drawida Ghilarovi (Oligochaeta, Moniligastridae): Polimorphism, Ecology Specifics And Karyotype. Russ. J. Zool. 2014, 93, 1070–1079. [Google Scholar] [CrossRef]
- Atopkin, D.M.; Ganin, G.N. Genetic differentiation of black and grey colored forms of the earthworm Drawida ghilarovi Gates, 1969 (Moniligastridae, Oligochaeta) on Russian Far East. Eur. J. Soil. Biol. 2015, 67, 12–16. [Google Scholar] [CrossRef]
- Shekhovtsov, S.V.; Golovanova, E.V.; Ershov, N.I.; Poluboyarova, T.V.; Berman, D.I.; Bulakhova, N.A.; Szederjesi, T.; Peltek, S.E. Phylogeny of the Eisenia nordenskioldi complex based on mitochondrial genomes. Eur. J. Soil. Biol. 2020, 96, 103137. [Google Scholar] [CrossRef]
- Shekhovtsov, S.V.; Golovanova, E.V.; Peltek, S.E. Cryptic diversity within the Nordenskiold’s earthworm, Eisenia nordenskioldi subsp. nordenskioldi (Lumbricidae, Annelida). Eur. J. Soil. Biol. 2013, 58, 13–18. [Google Scholar] [CrossRef]
- Shekhovtsov, S.V.; Berman, D.I.; Peltek, S.E. Phylogeography of the earthworm Eisenia nordenskioldi nordenskioldi (Lumbricidae, Oligochaeta) in northeastern Eurasia. Dokl. Biol. Sci. 2015, 461, 85–88. [Google Scholar] [CrossRef]
- Shekhovtsov, S.V.; Berman, D.I.; Bazarova, N.E.; Bulakhova, N.A.; Porco, D.; Peltek, S.E. Cryptic genetic lineages in Eisenia nordenskioldi pallida (Oligochaeta, Lumbricidae). Eur. J. Soil. Biol. 2016, 75, 151–156. [Google Scholar] [CrossRef]
- Shekhovtsov, S.; Berman, D.; Golovanova, E.; Peltek, S. Genetic diversity of the earthworm Eisenia nordenskioldi (Lumbricidae, Annelida). Vavilov J. Genet. Breed. 2017, 21, 589–595. [Google Scholar] [CrossRef]
- Vsevolodova-Perel, T.S. The Earthworms of the Fauna of Russia; Nauka: Moscow, Russia, 1997. [Google Scholar]
- Ganin, G. Structural and Functional Organization of Mezopedobiont Communities of the Southern Russian Far East; Russian Academy of Sciences Far Eastern Banch, Institute of Water and Ecological Problems: Vladivostok, Russia, 2011; p. 380. [Google Scholar]
- Ganin, G.; Anisimov, A.; Roslik, G.; Atopkin, D. Earthworms Drawida ghilarovi Gates, 1969 (Oligochaeta, Moniligastridae): 1. Polimorphism, geographic range, ecology specifics. Amurian Zool. J. 2013, 4, 401–404. [Google Scholar] [CrossRef]
- Boore, J.L.; Brown, W.M. Complete sequence of the mitochondrial DNA of the annelid worm Lumbricus terrestris. Genetics 1995, 141, 305–319. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.R.; Hong, Y.; Win, T.M.; Kim, I. Complete mitochondrial genome of the Burmese giant earthworm, Tonoscolex birmanicus (Clitellata: Megascolecidae). Mitochondrial DNA 2015, 26, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, J.; Dong, Y.; Qiu, J. Complete mitochondrial genome of four pheretimoid earthworms (Clitellata: Oligochaeta) and their phylogenetic reconstruction. Gene 2015, 574, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, J.; Dong, Y.; Qiu, J. Complete mitochondrial genome of an Amynthas earthworm, Amynthas aspergillus (Oligochaeta: Megascolecidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 1876–1877. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, J.; Dong, Y.; Qiu, J. Complete mitochondrial genome of a Pheretimoid earthworm Metaphire vulgaris (Oligochaeta: Megascolecidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2014, 27, 297–298. [Google Scholar] [CrossRef]
- Zhang, L.; Sechi, P.; Yuan, M.; Jiang, J.; Dong, Y.; Qiu, J. Fifteen new earthworm mitogenomes shed new light on phylogeny within the Pheretima complex. Sci. Rep. 2016, 6, 20096. [Google Scholar] [CrossRef]
- Conrado, A.C.; Arruda, H.; Stanton, D.W.G.; James, S.W.; Peter, K.; Brown, G.; Silva, E.; Dupont, L.; Taheri, S.; Morgan, A.J.; et al. The complete mitochondrial DNA sequence of the pantropical earthworm Pontoscolex corethrurus (Rhinodrilidae, Clitellata): Mitogenome characterization and phylogenetic positioning. ZooKeys 2017, 688, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Liu, H.; Zhang, Y.; Ruan, H. The complete mitochondrial genome of Lumbricus rubellus (Oligochaeta, Lumbricidae) and its phylogenetic analysis. Mitochondrial DNA B Resour. 2019, 4, 2677–2678. [Google Scholar] [CrossRef]
- Shekhovtsov, S.V.; Peltek, S.E. The complete mitochondrial genome of Aporrectodea rosea (Annelida: Lumbricidae). Mitochondrial DNA B Resour. 2019, 4, 1752–1753. [Google Scholar] [CrossRef]
- Liu, H.; Xu, N.; Zhang, Q.; Wang, G.; Xu, H.; Ruan, H. Characterization of the complete mitochondrial genome of Drawida gisti (Metagynophora, Moniligastridae) and comparison with other Metagynophora species. Genomics 2020, 112, 3056–3064. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Zhao, H.; Yu, F.; Zhang, A.; Li, H. The First Mitogenomes of the Subfamily Odontiinae (Lepidoptera, Crambidae) and Phylogenetic Analysis of Pyraloidea. Insects 2021, 12, 486. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-García, M.; Pinilla-Beltrán, D.; Murillo-García, O.E.; Pinto, C.M.; Brito, J.; Shostell, J.M. Comparative mitogenome phylogeography of two anteater genera (Tamandua and Myrmecophaga; Myrmecophagidae, Xenarthra): Evidence of discrepant evolutionary traits. Zool. Res. 2021, 42, 525–547. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Hasegawa, H.; Cooley, J.R.; Simon, C.; Yoshimura, J.; Cai, W.; Sota, T.; Li, H. Mitochondrial Genomics Reveals Shared Phylogeographic Patterns and Demographic History among Three Periodical Cicada Species Groups. Mol. Biol. Evol. 2019, 36, 1187–1200. [Google Scholar] [CrossRef]
- Hou, Q.; Tu, F.; Liu, Y.; Liu, S. Characterization of the mitogenome of Uropsilus gracilis and species delimitation. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 1836–1837. [Google Scholar] [CrossRef]
- Sun, X. Divergence across the mitogenomes of Branchinella kugenumaensis (Anostraca: Thamnocephalidae) with implications for species delimitation. Mitochondrial DNA B Resour. 2021, 6, 631–633. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Y.; Wang, Z.; Chen, H.; Qin, Y. Two Complete Mitogenomes of Chalcididae (Hymenoptera: Chalcidoidea): Genome Description and Phylogenetic Implications. Insects 2021, 12, 1049. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, H.; Li, Y. A report of the complete mitochondrial genome of Bisetocreagris titanium (Arachnida: Pseudoscorpiones: Neobisiidae) from Yunnan Province, China. Mitochondrial DNA Part B Resour. 2021, 6, 3212–3213. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Ganin, G.N.; Atopkin, D.M.; Wu, D.H. Earthworm Drawida (Moniligastridae) Molecular phylogeny and diversity in Far East Russia and Northeast China. Eur. Zool. J. 2020, 87, 180–191. [Google Scholar] [CrossRef]
- Saccone, C.; De Giorgi, C.; Gissi, C.; Pesole, G.; Reyes, A. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene 1999, 238, 195–209. [Google Scholar] [CrossRef]
- Ramesh, A.; Small, S.T.; Kloos, Z.A.; Kazura, J.W.; Nutman, T.B.; Serre, D.; Zimmerman, P.A. The complete mitochondrial genome sequence of the filarial nematode Wuchereria bancrofti from three geographic isolates provides evidence of complex demographic history. Mol. Biochem. Parasitol. 2012, 183, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Kern, E.M.A.; Kim, T.; Park, J.-K. The Mitochondrial Genome in Nematode Phylogenetics. Front. Ecol. Evol. 2020, 8, 250. [Google Scholar] [CrossRef]
- Yu, L.; Li, Y.-W.; Ryder, O.A.; Zhang, Y.-P. Analysis of complete mitochondrial genome sequences increases phylogenetic resolution of bears (Ursidae), a mammalian family that experienced rapid speciation. BMC Evol. Biol. 2007, 7, 198. [Google Scholar] [CrossRef] [PubMed]
- Miya, M.; Nishida, M. The mitogenomic contributions to molecular phylogenetics and evolution of fishes: A 15-year retrospect. Ichthyol. Res. 2015, 62, 29–71. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Y.; Xu, W.; Fang, Y.; Ruan, H. Characterization of Five New Earthworm Mitogenomes (Oligochaeta: Lumbricidae): Mitochondrial Phylogeny of Lumbricidae. Diversity 2021, 13, 580. [Google Scholar] [CrossRef]
- Hong, Y.; Kim, M.J.; Wang, A.R.; Kim, I. Complete mitochondrial genome of the earthworm, Amynthas jiriensis (Clitellata: Megascolecidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2017, 28, 163–164. [Google Scholar] [CrossRef]
- Yu, X.; Yang, H.; Liu, J.; Qi, Y.; Sun, L.; Tian, X. A strategy for a high enrichment of insect mitochondrial DNA for mitogenomic analysis. Gene 2022, 808, 145986. [Google Scholar] [CrossRef]
- Kim, D.W.; Lee, K.S.; Jee, S.H.; Seo, S.B.; Park, S.C.; Choo, J.K. Complete sequence analysis of the mitochondrial genome in the earthworm, Perionyx excavatus. Integr. Biosci. 2005, 9, A705. [Google Scholar]
- Lee, J.; Jung, J. First record of the complete mitochondrial genome of Tubifex tubifex (Müller) 1774 (Annelida; Clitellata; Oligochaeta) and phylogenetic analysis. Mitochondrial DNA B Resour. 2022, 7, 1208–1210. [Google Scholar] [CrossRef]
- Lee, J.; Jung, J. The complete mitochondrial genome of Nais communis Piguet, 1906 (Annelida; Clitellata; Naididae). Mitochondrial DNA B Resour. 2022, 7, 199–201. [Google Scholar] [CrossRef] [PubMed]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Yu, W.; Yang, J.; Song, Y.; dePamphilis, C.W.; Yi, T.; Li, D. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Donath, A.; Jühling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 53942. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. Figtree 1.4.0. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 July 2022).
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. USA 2000, 97, 13738–13742. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef]
- Hu, F.; Piao, Y.; Wang, H.; Pan, Y.; Fu, Y.; Song, W. The Investigation of the Usage of Herbicide in Heilongjiang Province. J. Agric. 2015, 5, 25–31. [Google Scholar]
- Pop, A.A.; Csuzdi, C.; Wink, M.; Pop, V.V. An Attempt to Reconstruct the Molecular Phylogeny of the Genus Allolobophora (sensu lato, Pop, 1941) Using 16S rDNA and COI Sequences (Oligochaeta, Lumbricidae); University Press: Cluj-Napoca, Romania, 2006. [Google Scholar]
- Shekhovtsov, S.; Ermolov, S.; Poluboyarova, T.; Kim-Kashmenskaya, M.; Derzhinsky, Y.; Peltek, S. Morphological differences between genetic lineages of the peregrine earthworm: Aporrectodea caliginosa (Savigny, 1826). Acta Zool. Acad. Sci. Hung. 2021, 67, 235–246. [Google Scholar] [CrossRef]
- Domínguez, J.; Aira, M.; Breinholt, J.W.; Stojanovic, M.; James, S.W.; Pérez-Losada, M. Underground evolution: New roots for the old tree of lumbricid earthworms. Mol. Phylogenet. Evol. 2015, 83, 7–19. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).