Coprinus leucostictus Rediscovered after a Century, Epitypified, and Its Generic Position in Hausknechtia Resolved by Multigene Phylogenetic Analysis of Psathyrellaceae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Description of Sites
2.2. Sampling and Morphological Study
2.3. DNA Extraction and Sequencing
2.4. Data Analyses
3. Results
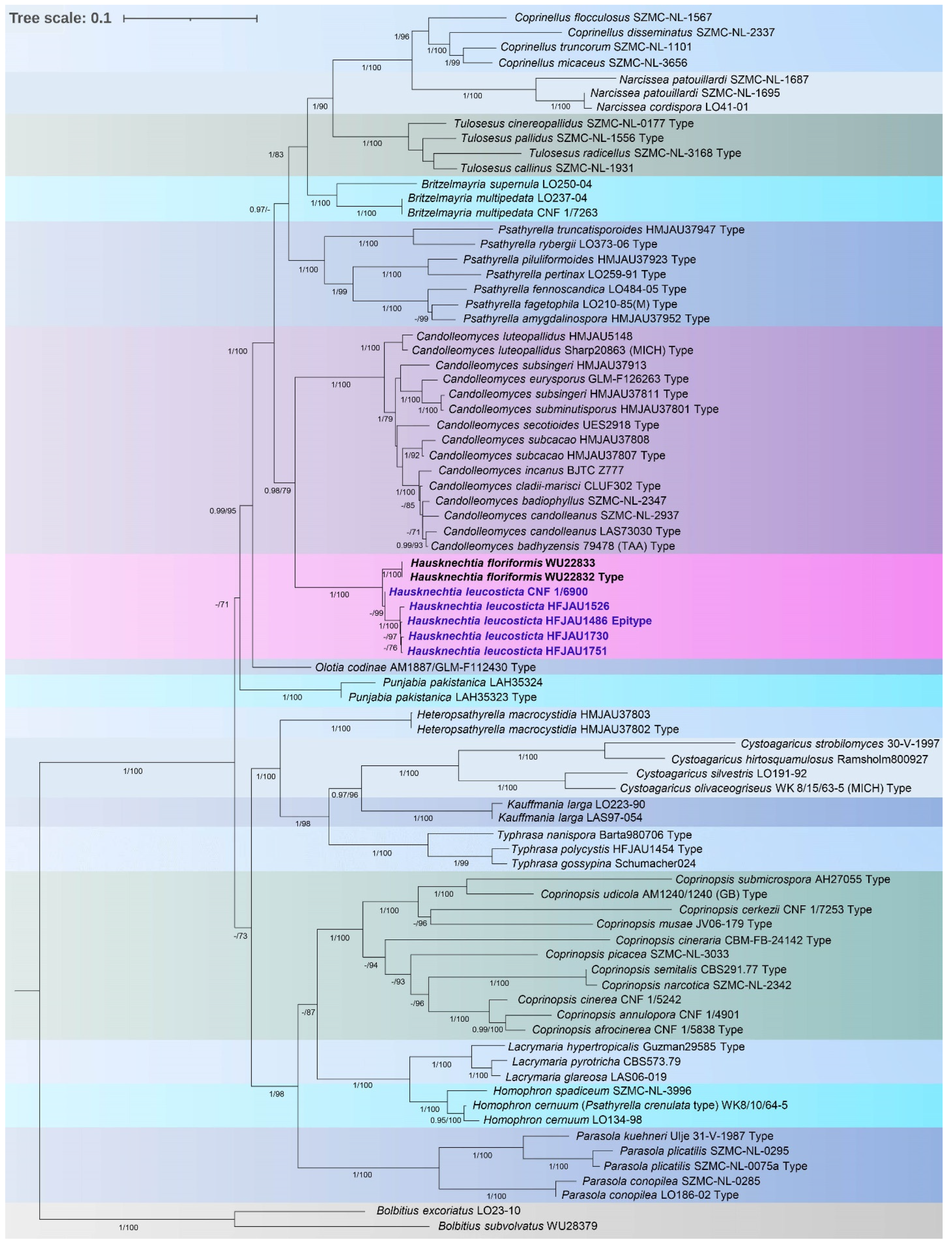
3.1. Molecular Phylogenetic Analysis
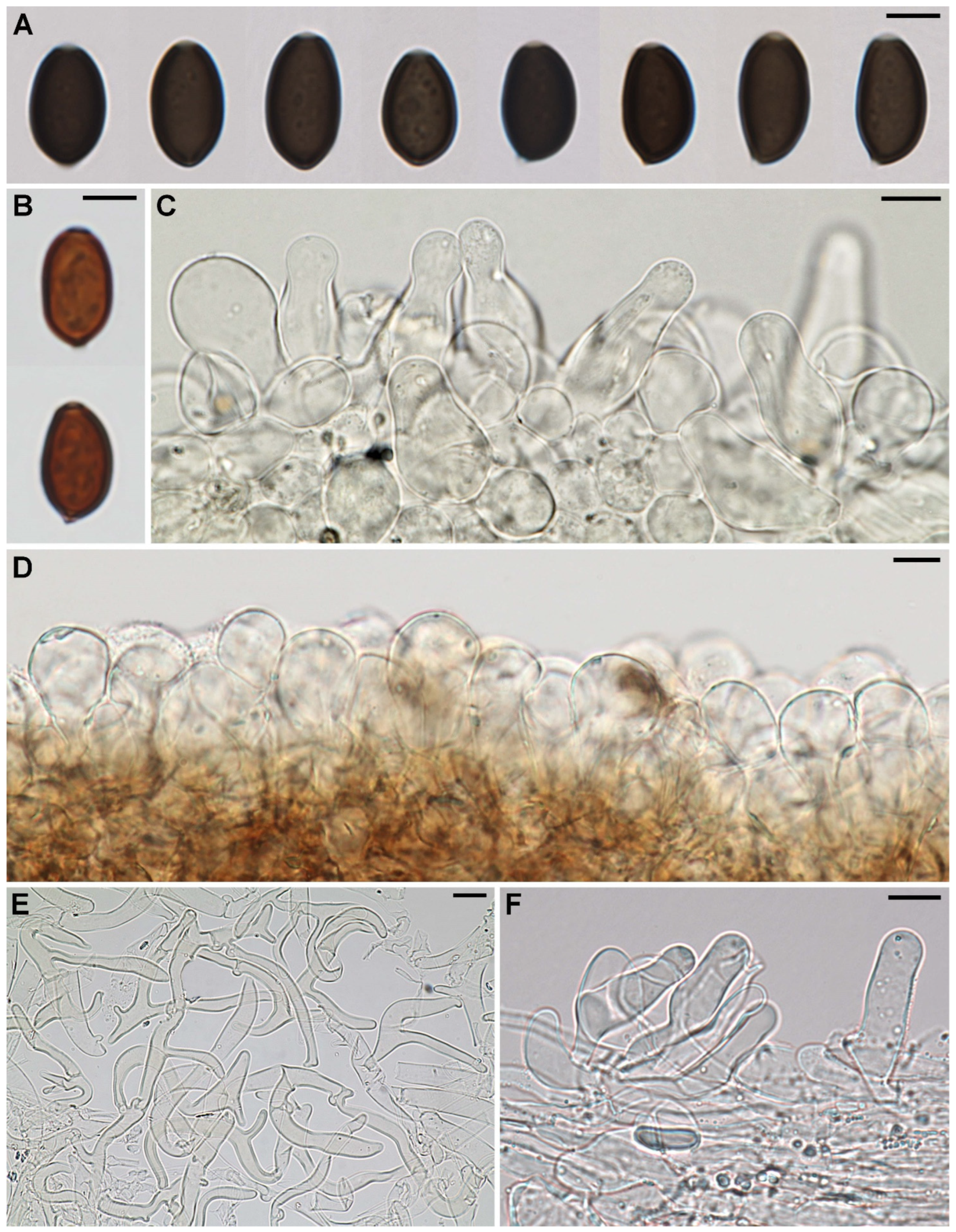
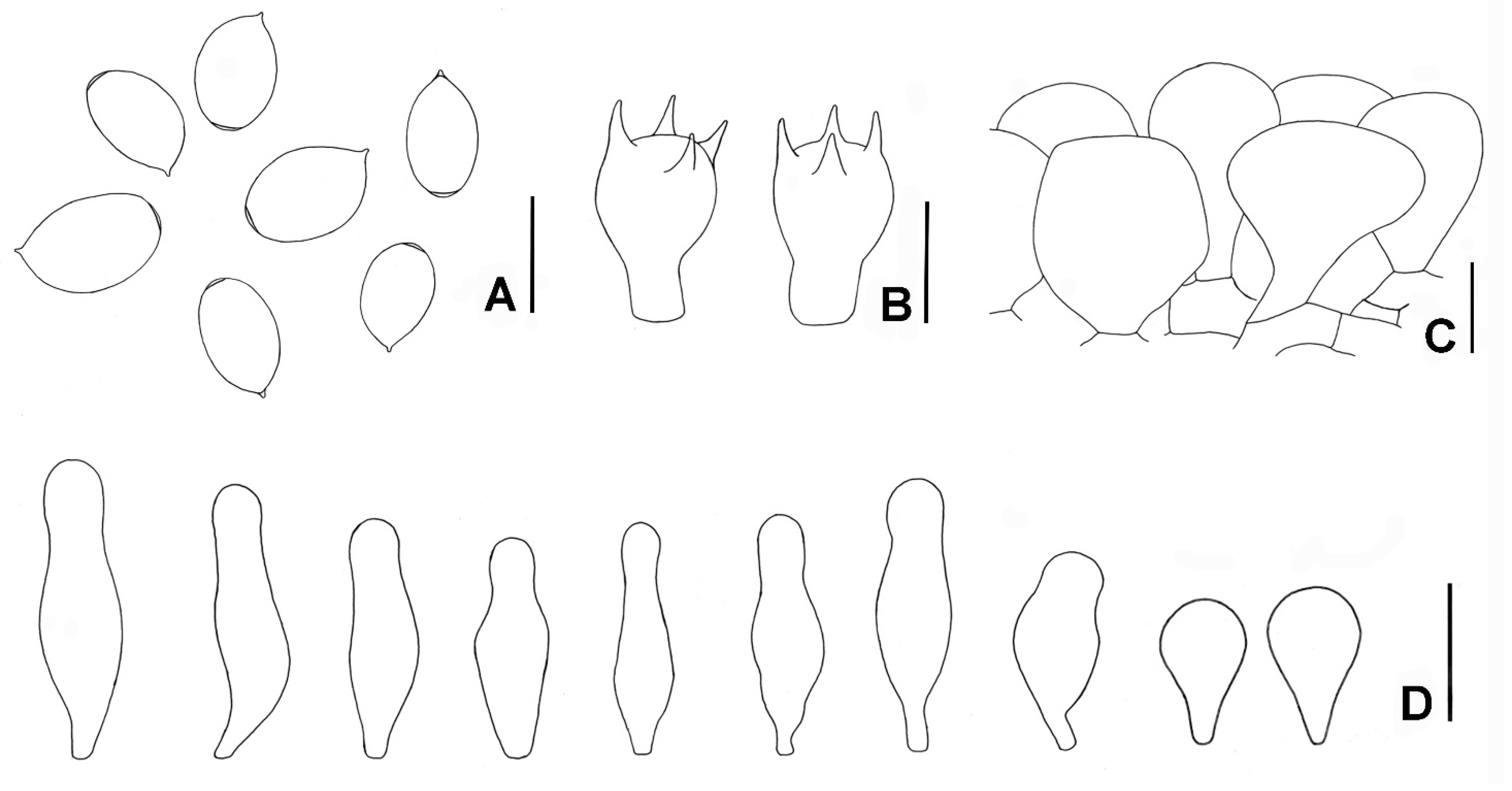
3.2. Taxonomy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wächter, D.; Melzer, A. Proposal for a subdivision of the family Psathyrellaceae based on a taxon-rich phylogenetic analysis with iterative multigene guide tree. Mycol. Prog. 2020, 19, 1151–1265. [Google Scholar] [CrossRef]
- Patouillard, N. Quelques champignons du Tonkin. Bull. Trimest. Soc. Mycol. Fr. 1917, 33, 50–63. [Google Scholar]
- Yang, Z.-L. Type studies on agarics described by N. Patouillard (and his co-authors) from Vietnam. Mycotaxon 2000, 75, 431–476. [Google Scholar]
- Redhead, S.A.; Vilgalys, R.; Moncalvo, J.M.; Johnson, J.; Hopple, J.S., Jr. Coprinus Pers. and the disposition of Coprinus species sensu lato. Taxon 2001, 50, 203–241. [Google Scholar] [CrossRef]
- Hausknecht, A.; Contu, M. The genus Galerella. A world-wide survey. Osterr. Z. Pilzkd. 2003, 12, 31–40. [Google Scholar]
- Tóth, A.; Hausknecht, A.; Krisai-Greilhuber, I.; Papp, T.; Vágvölgyi, C.; Nagy, L.G. Iteratively refined guide trees help improving alignment and phylogenetic inference in the mushroom family Bolbitiaceae. PLoS ONE 2013, 8, e56143. [Google Scholar] [CrossRef]
- Erb, B.; Matheis, W. Pilzmikroskopie, 1st ed.; Kosmos: Stuttgart, West Germany, 1983; pp. 1–166. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Hopple, J.S., Jr.; Vilgalys, R. Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: Divergent domains, outgroups, and monophyly. Mol. Phylogen. Evolut. 1999, 13, 1–19. [Google Scholar] [CrossRef]
- Matheny, P.B.; Wang, Z.; Binder, M.; Curtis, J.M.; Lim, Y.W.; Nilsson, R.H.; Hughes, K.W.; Hofstetter, V.; Ammirati, J.F.; Schoch, C.L.; et al. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol. Phylogen. Evol. 2007, 43, 430–451. [Google Scholar] [CrossRef]
- Nagy, L.G.; Walther, G.; Házi, J.; Vágvölgyi, C.; Papp, T. Understanding the evolutionary processes of fungal fruiting bodies: Correlated evolution and divergence times in the Psathyrellaceae. Syst. Biol. 2011, 60, 303–317. [Google Scholar] [CrossRef]
- Bau, T.; Yan, J.-Q. A new genus and four new species in the /Psathyrella s.l. clade from China. MycoKeys 2021, 80, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Örstadius, L.; Larsson, E. Bolbitius excoriatus, flagnande guldskivling, funnen på spillning i Sverige. Sven. Mykol. Tidskr. 2013, 34, 2–6. [Google Scholar]
- Örstadius, L.; Ryberg, M.; Larsson, E. Molecular phylogenetics and taxonomy in Psathyrellaceae (Agaricales) with focus on psathyrelloid species: Introduction of three new genera and 18 new species. Mycol. Prog. 2015, 14, 25. [Google Scholar] [CrossRef]
- Mešić, A.; Šamec, D.; Jadan, M.; Bahun, V.; Tkalčec, Z. Integrated morphological with molecular identification and bioactive compounds of 23 Croatian wild mushrooms samples. Food Biosci. 2020, 37, 100720. [Google Scholar] [CrossRef]
- Sicoli, G.; Passalacqua, N.G.; De Giuseppe, A.B.; Palermo, A.M.; Pellegrino, G. A new species of Psathyrella (Psathyrellaceae, Agaricales) from Italy. MycoKeys 2019, 52, 89–102. [Google Scholar] [CrossRef]
- Büttner, E.; Karich, A.; Nghi, D.H.; Lange, M.; Liers, C.; Kellner, H.; Hofrichter, M.; Ullrich, R. Candolleomyces eurysporus—A new Psathyrellaceae (Agaricales) species from the tropical Cúc Phương National Park, Vietnam. Osterr. Z. Pilzkd. 2021, 28, 79–92. [Google Scholar]
- Zhou, H.; Cheng, G.Q.; Sun, X.M.; Cheng, R.Y.; Zhang, H.L.; Dong, Y.M.; Hou, C.L. Three new species of Candolleomyces (Agaricomycetes, Agaricales, Psathyrellaceae) from the Yanshan Mountains in China. MycoKeys 2022, 88, 109–121. [Google Scholar] [CrossRef]
- Yan, J.-Q.; Bau, T. The Northeast Chinese species of Psathyrella (Agaricales, Psathyrellaceae). MycoKeys 2018, 33, 85–102. [Google Scholar] [CrossRef]
- Moreno, G.; Heykoop, M.; Esqueda, M.; Olariaga, I. Another lineage of secotioid fungi is discovered: Psathyrella secotioides sp. nov. from Mexico. Mycol. Prog. 2015, 14, 34. [Google Scholar] [CrossRef]
- Bau, T.; Yan, J.-Q. Two new rare species of Candolleomyces with pale spores from China. MycoKeys 2021, 80, 149–161. [Google Scholar] [CrossRef]
- Nagy, L.G.; Házi, J.; Vágvölgyi, C.; Papp, T. Phylogeny and species delimitation in the genus Coprinellus with special emphasis on the haired species. Mycologia 2012, 104, 254–275. [Google Scholar] [CrossRef] [PubMed]
- Nagy, L.G.; Urban, A.; Örstadius, L.; Papp, T.; Larsson, E.; Vágvölgyi, C. The evolution of autodigestion in the mushroom family Psathyrellaceae (Agaricales) inferred from Maximum Likelihood and Bayesian methods. Mol. Phylogen. Evol. 2010, 57, 1037–1048. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.E.S.J.; Gené, J.; Guarro, J.; Baseia, I.G.; García, D.; Gusmão, L.F.P.; Souza-Motta, C.M.; et al. Fungal Planet description sheets: 716–784. Persoonia 2018, 40, 239–392. [Google Scholar] [CrossRef] [PubMed]
- Tibpromma, S.; Hyde, K.D.; Jeewon, R.; Maharachchikumbura, S.S.N.; Liu, J.K.; Bhat, D.J.; Jones, E.B.G.; McKenzie, E.H.C.; Camporesi, E.; Bulgakov, T.S.; et al. Fungal diversity notes 491–602: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017, 83, 1–261. [Google Scholar] [CrossRef]
- Nagy, L.G.; Kocsubé, S.; Papp, T.; Vágvölgyi, C. Phylogeny and character evolution of the coprinoid mushroom genus Parasola as inferred from LSU and ITS nrDNA sequence data. Persoonia 2009, 22, 28–37. [Google Scholar] [CrossRef]
- Vellinga, E.C. Genera in the family Agaricaceae: Evidence from nrITS and nrLSU sequences. Mycol. Res. 2004, 108, 354–377. [Google Scholar] [CrossRef]
- Larsson, E.; Örstadius, L. Fourteen coprophilous species of Psathyrella identified in the Nordic countries using morphology and nuclear rDNA sequence data. Mycol. Res. 2008, 112, 1165–1185. [Google Scholar] [CrossRef]
- Deschuyteneer, D.; Melzer, A.; Pérez-De-Gregorio, M.À. Psathyrella codinae, a new species from Spain. Bull. Assoc. Mycol. Francoph. Belg. 2018, 11, 4–8. [Google Scholar]
- Szarkándi, J.G.; Schmidt-Stohn, G.; Dima, B.; Hussain, S.; Kocsubé, S.; Papp, T.; Vágvölgyi, C.; Nagy, L.G. The genus Parasola: Phylogeny and the description of three new species. Mycologia 2017, 109, 620–629. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Usman, M.; Afshan, N.-ul-S.; Ahmad, H.; Khan, J.; Khalid, A.N. The genus Coprinellus (Basidiomycota; Agaricales) in Pakistan with the description of four new species. MycoKeys 2018, 39, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Házi, J.; Nagy, L.G.; Vágvölgyi, C.; Papp, T. Coprinellus radicellus, a new species with northern distribution. Mycol. Prog. 2010, 10, 363–371. [Google Scholar] [CrossRef]
- Wang, S.-N.; Hu, Y.-P.; Chen, J.-L.; Qi, L.-L.; Zeng, H.; Ding, H.; Huo, G.-H.; Zhang, L.-P.; Chen, F.-S.; Yan, J.-Q. First record of the rare genus Typhrasa (Psathyrellaceae, Agaricales) from China with description of two new species. MycoKeys 2021, 79, 119–128. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MrBayes: Bayesian inference of phylogeny. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Voto, P. Novelties in the Family Psathyrellaceae. Part VI. Mycol. Obs. 2022, 2, 9. [Google Scholar]

| Taxon | Voucher | Country | ITS | LSU | tef-1α | β-Tubulin | Refs. |
|---|---|---|---|---|---|---|---|
| Bolbitius excoriatus | LO23-10 | Sweden | KC456419 | KC456419 | KJ732834 | - | [1,16] |
| Bolbitius subvolvatus | WU28379 | Hungary | JX968248 | JX968365 | JX968454 | - | [6] |
| Britzelmayria multipedata | LO237-04 | Sweden | KC992888 | KC992888 | KJ732777 | KJ664867.1 | [17] |
| Britzelmayria multipedata | CNF 1/7263 | Croatia | MK169241 | - | - | - | [18] |
| Britzelmayria supernula | LO250-04 | Sweden | KC992867 | KC992867 | KJ732763 | KJ664849.1 | [17] |
| Candolleomyces badhyzensis (type) | 79478 (TAA) | Turkmenistan | KC992883 | KC992883 | - | - | [17] |
| Candolleomyces badiophyllus | SZMC-NL-2347 | - | FN430699 | FM876268 | FN396261 | FM897252 | [12] |
| Candolleomyces candolleanus (type) | LAS73030 | Hungary | KM030175 | KM030175 | - | - | [17] |
| Candolleomyces candolleanus | SZMC-NL-2937 | Hungary | FN396114 | FN396165 | FN396220 | FN396307 | [12] |
| Candolleomyces cladii-marisci (type) | CLUF302 | Italy | MK080112 | - | - | - | [19] |
| Candolleomyces eurysporus (type) | GLM-F126263 | Germany | MT651560 | MT651560 | - | - | [20] |
| Candolleomyces incanus | BJTC Z777 | China, Beijing | ON042759 | ON042766 | ON098508 | ON098513 | [21] |
| Candolleomyces luteopallidus (type) | Sharp20863 (MICH) | USA | KC992885 | KC992885 | KJ732775 | KJ664865 | [17] |
| Candolleomyces luteopallidus | HMJAU5148 | China, Jilin | MG734736 | MW301084 | MW314056 | MW314073 | [22] |
| Candolleomyces secotioides (type) | UES2918 | Mexico | KR003281 | KR003282 | - | KR003283 | [23] |
| Candolleomyces subcacao (type) | HMJAU37807 | China, Henan | NR_173317 | NG_079683 | MW314081 | MW314063 | [24] |
| Candolleomyces subcacao | HMJAU37808 | China, Henan | MW301065 | MW301093 | MW314082 | MW314064 | [24] |
| Candolleomyces subminutisporus | HMJAU37916 | China, Henan | MW301067 | MW301095 | MW314084 | MW314066 | [24] |
| Candolleomyces subsingeri (type) | HMJAU37811 | China, Jilin | MG734715 | MW301097 | MW314085 | MW314067 | [22,24] |
| Candolleomyces subsingeri | HMJAU37913 | China, Jilin | MG734725 | MW301098 | MW314086 | MW314067 | [22,24] |
| Coprinellus disseminatus | SZMC-NL-2337 | Hungary | FM878017 | FM876274 | - | FN396282 | [12] |
| Coprinellus flocculosus | SZMC-NL-1567 | - | FN430683 | JN159593 | - | FN396345 | [12] |
| Coprinellus micaceus | SZMC-NL-3656 | - | JN159567 | JN159588 | - | JN159644 | [25] |
| Coprinellus truncorum | SZMC-NL-1101 | Sweden | JN159562 | FM876262 | FM897225 | JN159629 | [12,26] |
| Coprinopsis afrocinerea (type) | CNF 1/5838 | Nigeria | MG662162 | MG662158 | - | - | [27] |
| Coprinopsis annulopora | CNF 1/4901 | Croatia | MG662170 | - | - | - | [27] |
| Coprinopsis cerkezii (type) | CNF 1/7253 | Croatia | NR_173361 | NG_068782 | - | - | [28] |
| Coprinopsis cineraria (type) | CBM-FB-24142 | Japan | KC992962 | - | - | - | [17] |
| Coprinopsis cinerea | CNF 1/5242 | Croatia | MG662167 | - | - | - | [27] |
| Coprinopsis musae (type) | JV06-179 | Denmark | NR_148070 | KC992965 | - | KJ664920 | [17] |
| Coprinopsis narcotica | SZMC-NL-2342 | Hungary | FM163180 | FM160729 | FN396244 | FN396290 | [29] |
| Coprinopsis picacea | SZMC-NL-3033 | - | FN396119 | FN396168 | FN396223 | FN396312 | [12] |
| Coprinopsis semitalis (type) | CBS291.77 | - | GQ249278 | GQ249287 | GQ249270 | GQ249262 | [12] |
| Coprinopsis submicrospora (type) | AH27055 | Spain | KC992959 | KC992959 | - | KJ664918 | [17] |
| Coprinopsis udicola (type) | AM1240 | Germany | NR_148071 | KC992967 | KJ732831 | KJ664922 | [17] |
| Cystoagaricus hirtosquamulosus | Ramsholm800927 | Finland | KC992945 | KC992945 | - | - | [17] |
| Cystoagaricus olivaceogriseus (type) | WK 8/15/63-5 | USA | KC992948 | KC992948 | - | - | [17] |
| Cystoagaricus silvestris | LO191-92 | Sweden | KC992949 | KC992949 | - | - | [17] |
| Cystoagaricus strobilomyces | 30-V-1997 | Japan | AY176347 | AY176348 | - | - | [30] |
| Hausknechtia floriformis (type) | WU22832 | Vanuatu | ON745613 | ON745616 | ON746007 | ON746008 | This study |
| Hausknechtia floriformis | WU22833 | Vanuatu | ON745619 | ON745615 | ON746009 | ON746010 | This study |
| Hausknechtia leucosticta | CNF 1/6900 | India | ON745618 | ON745617 | ON746005 | ON746006 | This study |
| Hausknechtia leucosticta (epitype) | HFJAU1486 | China, Anhui | OL435561 | OL435565 | OL439896 | ON677539 | This study |
| Hausknechtia leucosticta | HFJAU1526 | China, Fujian | OL435563 | 0L435566 | OL439897 | ON677541 | This study |
| Hausknechtia leucosticta | HFJAU1730 | China, Jiangxi | OL435562 | - | OL439898 | ON677540 | This study |
| Hausknechtia leucosticta | HFJAU1751 | China, Jiangxi | OL435564 | OL435567 | OL439899 | ON677542 | This study |
| Heteropsathyrella macrocystidia (type) | HMJAU37802 | China, Fujian | MW405102 | MW413359 | MW411004 | - | [24] |
| Heteropsathyrella macrocystidia | HMJAU37803 | China, Fujian | MW405101 | MW413358 | MW411003 | - | [24] |
| Homophron cernuum | LO134-98 | Sweden | DQ389726 | DQ389726 | KJ732828 | KJ664915 | [17,31] |
| Homophron cernuum (Psathyrella crenulata type) | WK8/10/64-5 | USA | KC992957 | - | - | . | [17] |
| Homophron spadiceum | SZMC-NL-3996 | Hungary | FN396132 | FN396180 | FN396231 | FN396333.1 | [12] |
| Kauffmania larga | LO223-90 | Sweden | DQ389694 | DQ389694 | KJ732824 | KJ664912.1 | [17] |
| Kauffmania larga | LAS97-054 | Sweden | DQ389695 | DQ389695 | - | [17] | |
| Lacrymaria glareosa | LAS06-019 | Sweden | KC992954 | KC992954 | KJ732827 | KJ664914 | [17] |
| Lacrymaria hypertropicalis (type) | Guzman29585 | Mexico | KC992958 | KC992958 | - | KJ664916 | [17] |
| Lacrymaria pyrotricha | CBS573.79 | - | GQ249280 | GQ249289 | GQ249272 | GQ249264 | [12] |
| Narcissea cordispora | LO41-01 | Sweden | DQ389723 | DQ389723 | - | KJ664910 | [17,31] |
| Narcissea patouillardi | SZMC-NL-1687 | Hungary | FM878009 | FM876265 | FM897238 | FN396257 | [26] |
| Narcissea patouillardi | SZMC-NL-1695 | Hungary | FN430685 | FN396196 | - | FN396258 | [26] |
| Olotia codinae (type) | AM1887/GLM-F112430 | Spain | MG696611 | MG674714 | - | - | [32] |
| Parasola conopilea (type) | LO186-02 | Sweden | DQ389725 | DQ389725 | - | - | [31] |
| Parasola conopilea | SZMC-NL-0285 | Hungary | FM163225 | FM160684 | FM897237 | FN396247 | [29] |
| Parasola kuehneri (type) | Ulje 31-V-1987 | Netherlands | KY928608 | KY928633 | - | - | [33] |
| Parasola plicatilis | SZMC-NL-0295 | Hungary | FM163216 | FM160693 | FM897242 | FN396253 | [29] |
| Parasola plicatilis (type) | SZMC-NL-0075a | Hungary | FM163213 | FM160696 | - | - | [29] |
| Psathyrella amygdalinospora (type) | HMJAU37952 | China, Sichuan | NR_173320 | MW413361 | MW410999 | MW410991 | [24] |
| Psathyrella fagetophila (type) | LO210-85 (M) | Sweden | NR_167955 | KC992902 | - | KJ664879 | [17] |
| Psathyrella fennoscandica (type) | LO484-05 | Sweden | KC992903 | KC992903 | KJ732790 | KJ664881 | [17] |
| Psathyrella pertinax (type) | LO259-91 | Sweden | DQ389701 | DQ389701 | KJ732809 | - | [17] |
| Psathyrella piluliformoides (type) | HMJAU37923 | China, Zhejiang | MW405106 | MW413362 | MW411002 | - | [24] |
| Psathyrella rybergii (type) | LÖ373-06 | Sweden | KC992893 | KC992893 | KJ732781 | KJ664872 | [17] |
| Psathyrella truncatisporoides (type) | HMJAU37947 | China, Zhejiang | MW405107 | MW413363 | MW410990 | MW410998 | [24] |
| Punjabia pakistanica (type) | LAH35323 | Pakistan | MH366736 | - | - | - | [34] |
| Punjabia pakistanica | LAH35324 | Pakistan | MH366737 | - | - | - | [34] |
| Tulosesus callinus | SZMC-NL-1931 | Hungary | FN396105 | FN396158 | FN396213 | FN396299 | [12] |
| Tulosesus cinereopallidus (type) | SZMC-NL-0177 | Hungary | HQ847001 | HQ847090 | - | HQ847149 | [25] |
| Tulosesus pallidus (type) | SZMC-NL-1556 | Hungary | HQ846989 | HQ847076 | - | HQ847155 | [25] |
| Tulosesus radicellus (type) | SZMC-NL-3168 | Sweden | GU227719 | HQ847077 | - | GU227737 | [25,35] |
| Typhrasa gossypina | Schumacher024 | Germany | KC992946 | KC992946 | KJ732825 | - | [17] |
| Typhrasa nanispora (type) | Barta980706 | Austria | KC992947 | KC992947 | - | - | [17] |
| Typhrasa polycystis (type) | HFJAU1454 | China, Zhejiang | MW466538 | MW466544 | MW475280 | - | [36] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, C.; Wang, S.-N.; Tkalčec, Z.; Yan, J.-Q.; Hu, Y.; Ge, Y.; Na, Q.; Zeng, H.; Ding, H.; Huo, G.; et al. Coprinus leucostictus Rediscovered after a Century, Epitypified, and Its Generic Position in Hausknechtia Resolved by Multigene Phylogenetic Analysis of Psathyrellaceae. Diversity 2022, 14, 699. https://doi.org/10.3390/d14090699
Nie C, Wang S-N, Tkalčec Z, Yan J-Q, Hu Y, Ge Y, Na Q, Zeng H, Ding H, Huo G, et al. Coprinus leucostictus Rediscovered after a Century, Epitypified, and Its Generic Position in Hausknechtia Resolved by Multigene Phylogenetic Analysis of Psathyrellaceae. Diversity. 2022; 14(9):699. https://doi.org/10.3390/d14090699
Chicago/Turabian StyleNie, Chengfeng, Sheng-Nan Wang, Zdenko Tkalčec, Jun-Qing Yan, Yaping Hu, Yupeng Ge, Qin Na, Hui Zeng, Hui Ding, Guanghua Huo, and et al. 2022. "Coprinus leucostictus Rediscovered after a Century, Epitypified, and Its Generic Position in Hausknechtia Resolved by Multigene Phylogenetic Analysis of Psathyrellaceae" Diversity 14, no. 9: 699. https://doi.org/10.3390/d14090699
APA StyleNie, C., Wang, S.-N., Tkalčec, Z., Yan, J.-Q., Hu, Y., Ge, Y., Na, Q., Zeng, H., Ding, H., Huo, G., Pošta, A., Pradeep, C. K., & Mešić, A. (2022). Coprinus leucostictus Rediscovered after a Century, Epitypified, and Its Generic Position in Hausknechtia Resolved by Multigene Phylogenetic Analysis of Psathyrellaceae. Diversity, 14(9), 699. https://doi.org/10.3390/d14090699







