Abstract
Decapods include species of economic importance, such as Achelata (lobsters) and Brachyura (true crabs), since they have aesthetic, commercial, gastronomic, and biomedical value. These groups exhibit a great variety of shapes, larval stages, habits, and sizes, making them difficult to recognize. In the Southern Mexican Caribbean (SMC), no taxonomic list or analysis of the biological diversity for the Achelata and Brachyura has been performed. Herein, the biological diversity of these groups was analyzed by reviewing the literature and collecting specimens in the SMC to obtain morphological, ecological, and molecular data. These results showed a total of 29 families, 67 genera, and 98 species recorded, of which, one is considered as a potentially new species, six are new records for the SMC, 12 expanded their distribution range, and 14 species names were updated. In addition, the BOLD system assigned 21 BINs supported with morphological identification. This work contributes positively to the knowledge of the marine and coastal decapods from the SCM as it represents the first effort to recognize their current biological diversity. This information will be used to develop adequate strategies for the conservation and management of marine and coastal natural resources of the SMC.
1. Introduction
The Order Decapoda belongs to the class Malacostraca (subphyla: Crustacea). It includes a wide variety of organisms: the true crabs (Brachyura), hermit and porcelanid crabs (Anomura), shrimps (Dendrobranchiata, Caridea, and Stenopodidea), and lobsters (Astacidae, Achelata) [1,2]. Decapods include species of economic importance, penaeid shrimps, palinurids, lobsters, portunids, and crabs since they have aesthetic, commercial, gastronomic, and biomedical value [3,4,5]. However, the most relevant decapods belong to the infraorders Achelata and Brachyura as they support some of the most remunerative fisheries worldwide [4,6].
These crustaceans live in marine and coastal ecosystems and fulfill different ecological functions. They are relevant in marine, pelagic, and benthic trophic networks due they serve as food for birds, marine mammals, sharks, turtles, starfish, cephalopods, and fish of commercial importance [7,8,9]. It has been documented that more than 50% of the diet of adult and juvenile snappers is based mainly on decapod crustaceans [9,10]. They also regulate the herbivore populations (e.g., sea urchins), favoring primary and secondary production and the trophic structure’s stability [7]. Furthermore, when some species of decapods (e.g., Rhithropanopeus harrisii) are introduced or invade a new region successfully, they become exotic species, and they can modify the native marine communities by altering habitat and ecosystem function [11,12].
Despite their economic and ecological importance, decapod species show severe taxonomic difficulties due to their high variety of shapes, larval stages, habits, and sizes. Martin & Davis [3] published an updated classification of the Crustacea where they highlighted the necessity for reaching a consensus on the relationships among the Decapoda because opinions and datasets remain sharply divided. According to Álvarez et al. [2], in Mexico, the species richness of decapods is about 1775 species classified in 537 genera and 115 families, representing 11.9% of the total species and 57.5% of the families in the world, respectively. Also, of the total number of these species, 1597 (89.9%) are marine, and 178 (10.1%) are freshwater; 46.7% of the marine species occur in the Mexican Pacific, 31.4% in the Gulf of Mexico, and 21.8% in the Mexican Caribbean.
In the Mexican Caribbean, some studies have been conducted to know the biological diversity of marine and coastal crustaceans, including the infraorders Achelata and Brachyura. Markham et al. [13], García-Madrigal et al. [14], and Álvarez [15] reported species lists for different orders of crustaceans (e.g., Stomatopoda, Peracarida, Decapoda) from the shallow Caribbean coast of Quintana Roo between 1990 and early 2000, since then, the information has not been updated. In the north coast of the Mexican Caribbean (Isla Mujeres and Puerto Morelos, Quintana Roo), Campos-Vázquez [16], González-Gómez et al. [17] and Briones-Fourzán et al. [18], conducted studies of decapods associated to coralline reefs and seagrass, respectively. In contrast, the Southern Mexican Caribbean has not been studied, and there is no taxonomic list of species or an analysis of the current biological diversity for the decapods Achelata and Brachyura.
The analysis of the biological diversity and the taxonomic and genetic status of different groups has been performed using molecular sequence data as the DNA barcodes [1,19,20,21,22,23]. This analysis mainly used mitochondrial gene cytochrome c oxidase subunit I (COI), providing a robust species-level resolution for different groups of animals, such as marine decapods [19,22,24]. As a result of the use of DNA barcodes and the inclusion of morphology description, Costa et al. [19] and Landschoff & Gouws [21] proposed the possible arbitrary threshold for genetic species delimitation in this group can be placed between 3.7% and 4.9%. In addition, DNA barcodes have proven to be a helpful tool in species differentiation in the last two decades, accelerating biodiversity inventories [24] to assign unknown specimens to already described and classified species, improving the discovery of new species, and facilitating their identification, particularly in cryptic, microscopic, and other organisms with complex morphology [25]. The Barcode of Life Data System (BOLD) is “an informatics workbench aiding the acquisition, storage, analysis and publication of DNA barcode records” [26]. This system allows the exchange of genetic and biological information and scientific collaborations. In addition, the genetic data can be associated with photographs, collection site metadata, life stage, and samples with vouchers in scientific collections [24]. Also, can assign a Barcode Index Number (BIN), equivalent to a Molecular Operating Taxonomic Unit (MOTU) for all samples that cover minimum information standards; with this, a standardized reference is made for unidentified organisms [27].
Assessing the current biological diversity correctly [28,29] and the ecological role of decapods in the SMC are priorities in understanding the geographic distribution patterns, taxonomic, genetic, and conservation status of these species. This knowledge is essential to make sustainable use of natural resources since there are currently various threats to public health, ecosystem health, fishing, and the economic development of the region, such as the growing arrival of Sargassum in the Mexican Caribbean, which generates hypoxia and deterioration of water quality, affecting individuals of a large number of species, mainly fish and crustaceans [30]. Therefore, the main objective of this study was to know the current state of the biological diversity of the Brachyura and Achelata decapods present in the SMC. The results will allow the development of instruments and strategies in favor of environmental sustainability and the conservation of the natural capital of the Southern Mexican Caribbean ecosystems.
2. Materials and Methods
2.1. Literature Review
The reports of Brachyura and Achelata species recorded in the Southern Mexican Caribbean (SMC) were analyzed to elaborate a taxonomic list. The Scopus, Google Academic, and Springer Link databases were consulted from October 2020 to November 2021. The specific search terms for revision were Decapoda, Brachyura, and Achelata (including the addition of the words COI, marine, native, exotic, and introduced), Mexican Caribbean, and the Atlantic Ocean. Additionally, to search the species records in the study area, the databases of the Reference Collection of Benthos of El Colegio de la Frontera Sur (ECOSUR), Chetumal, Mexico, and the online dataset of the National Collection of Crustaceans of Universidad Nacional Autónoma de Mexico (UNIBIO [31]), were visited and consulted. The online project “Stomatopod, amphipod, isopod and decapod crustaceans of the Quintana Roo coast” by Álvarez [15] was also included in the taxonomic revision. For each collection database, the search was restricted to localities from Punta Herrero (19.31157, −87.44517) to Xcalak (18.27001, −87.82649), Quintana Roo, Mexico. The BOLD Public Data Portal [32] (www.boldsystems.org, accessed on 13 July 2022), was consulted to obtain and compare molecular data. The taxonomic list includes the name of species, type locality, distribution, ecological notes, the process ID for the sequences obtained in this work, and BINs.
2.2. Material Collected
Punta Herrero, Punta Diamante, El Uvero, Mahahual, Bermejo River, North of Xahuayxol, Xahuayxol, and Huach River, in the Southern Mexican Caribbean were selected to collect individuals during 2021: January (27th to 29th), February (22nd to 24th), April 2021 (17th to 19th), October (27th to 29th), and November (10th to 12th) (Figure 1). At each site, samples of decapods were collected manually during free diving samplings, mainly of coralline rocks associated with algae at depths not exceeding 3 m (collection permit PPF/DGOPA-060/20). The sampling effort was 5 h by site and was performed by two persons. Individuals were photographed in situ with a digital camera (Sony DSC-H300) and fixed in 96% ethanol. Large specimens (more than 3 cm of carapace length) were injected with the same ethanol in the articulation between segments of the body. Subsequently, the samples were stored at −20 °C for a week to prevent DNA degradation [33]. The collected material was examined under a stereoscopic microscope Zeiss StemiDV4 and identified according to Rathbun [34,35,36], Williams [37,38,39], and Abele & Kim [40]. All material analyzed was then incorporated into the Reference Collection of Benthos (ECOSUR), Chetumal, Mexico. The species list was arranged alphabetically, and the nomenclatural status of each specie was assigned following the WoRMS Editorial Board [41].
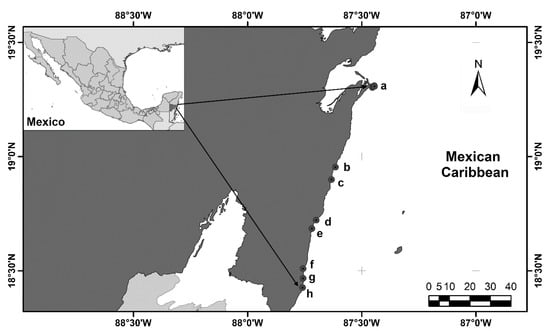
Figure 1.
Localities within the Southern Mexican Caribbean where decapods were collected. (a) Punta Herrero; (b) Punta Diamante; (c) El Uvero; (d) Mahahual; (e) Bermejo River; (f) North of Xahuayxol; (g) Xahuayxol; (h) Huach River. Figure edited by Jani Jarquín-González.
2.3. Molecular Analysis
For molecular analysis, 76 specimens were processed. A small piece of muscle (1–3 mm3) or 2–3 eggs (in the case of ovigerous females) were used to extract the DNA. The forceps and the material were sterilized using a solution of 1:5 chlorine/distiller water and subsequently neutralized with ethanol 96% between each tissue extraction.
A lysis buffer was used to digest each sample’s tissue with proteinase K, all samples were digested in an oven for 12 h. at 56 °C. The extraction was carried out through a 1.0 mm PALL glass fiber plate [42]. Cytochrome Oxidase I (COI) gene segment with an approximate length of 650 Bp [43] was amplified using the zooplankton primers [44]. Amplification was carried out with a final volume of 12.5 μL, prepared as follows: 6.5 μL of 10% trehalose, 2 μL of ultrapure water, 1.25 μL PCR buffer X10, 0.625 μL MgCl2 (50 mM), 0.125 μL of each Primer (0.01 mM), 0.06525 μL dNTP mix (10 mM), 0.625 μL Taq polymerase, and 2 μL of DNA template. The reactions were cycled at 94 °C for 1 min, followed by five cycles at 94 °C for 30 s, 45–50 °C for 40 s, and 72 °C for 1 min, followed by 35 cycles at 94 °C for 30 s, 51–54 °C for 40 s and 72 °C for 1 min, finally one last cycle of 72 °C for 10 min.
The PCR products were visualized in agarose gel InvitrogenTM with four μL of sample and 16 µL of water. The PCR products were sequenced at Macrogen (Seoul, Republic of Korea). Finally, the sequences were edited with Codon code v.3.0.1 and uploaded to BOLD. Specimen images, field data, and COI sequences obtained in this study can be consulted at BOLDSYSTEMS www.boldsystems.org within the dataset ID: DS-DECA01 Decapoda (Achelata and Brachyura) from Southern Mexican Caribbean (https://doi.org/10.5883/DS-DECA01, accessed on 13 July 2022).
All the sequences obtained were compared with COI sequences previously published using the specimen identification tool in the Barcode of Life Data System (BOLD) [45]. Similarity values >98% were considered for all identified species, which confirmed their placement under different numbers of BINs [26,28]. The Kimura 2-parameter model (K2P) was used to calculate the genetic divergences between the species [46] and the maximum likelihood (ML) tree. The BOLD ID tree was simplified using the compression feature provided by MEGA X software [47]. The criteria to assign taxonomic level identification using BOLD was a similarity value ≥ 99% [45].
3. Results and Discussion
3.1. Literature Review
Results of the literature review showed that 91 species of decapods have been reported in the Southern Mexican Caribbean (Table S1). Of these, 78 species (86%) have type locality within Western Atlantic (e.g., Antilles, USA, Brazil); seven (8%) have type locality outside Western Atlantic (e.g., Africa, Australia, Indonesia, Chile, and the Mediterranean Sea); five (5%) with unknown type locality (e.g., Panulirus argus (Latreille, 1804) species of great economic importance); and one (1%) has type locality from the Mexican Caribbean (Parapinnixa bouvieri Rathbun, 1918 from Cape Catoche).
Regarding the habitat, 82% of the species were present in marine environments, including reefs, tide pools, rocky or sandy beaches, estuaries (15%), salt marshes (2%), and mudflats (1%) in a lesser proportion. The most important habitats reported for these species were coralline reefs (22%), sediments (sand, gravel, mud) (19%), and rocky bottoms (13%), followed by mollusk shells (10%), sponges (9%), seagrass (8%), algae (8%), other invertebrates (cnidarians, polychaetes, barnacles, echinoderms, foraminifera, and tunicates) (6%), mangrove roots (4%), and Sargassum (1%). Additionally, the species Calappa ocellata Holthuis, 1958, Euryplax nitida Stimpson, 1859, and Percnon gibbesi (H. Milne Edwards, 1853) was reported in stomach contents of bothids, holocentrids, diodontids, labrids, lutjanids, and serranids (Table S1).
Results of the number of records by locality showed that the localities with the highest number of recorded species were Mahahual (20%), Xahuayxol (17%), and El Placer (13%), followed by Banco Chinchorro and El Uvero (10%), Rio Indio (9%) and Chetumal Bay (5%). In contrast, the localities with the lowest number of records were Punta Herradura (4%), Xcalak (4%), and Huach River (3%). Of all species, 60% of species were recognized only in one specific location, the remaining 40% were recorded in more than one locality. Additionally, herein 14 names of the species found in the literature were updated (Table 1).

Table 1.
Update of the species names of Brachyura and Achelata found in the bibliographic review. SMC = Southern Mexican Caribbean.
3.2. Morphological Identification of Specimens Collected in the SMC
A total of 102 specimens were collected in the SMC. According to the morphology, they corresponded to 21 morphotypes, of which 20 were identified at the species level and one assigned to the genus Panopeus (Table 2). Six species represent new records for the SMC: Maguimithrax spinosissimus (Lamarck, 1818), Menippe nodifrons Stimpson, 1859, Mithraculus cinctimanus Stimpson, 1860, Mithrax tortugae Rathbun (1920), Plagusia immaculata Lamarck, 1818, and Portunus sayi (Gibbes, 1850).

Table 2.
Taxa of decapods (Achelata and Brachyura) recorded in this work and localities where they were found.
Considering the 91 records found in the literature review and the inclusion of the six new records and the morphotype, the result is a total of 98 species of decapods for the infraorders Achelata and Brachyura from the Southern Mexican Caribbean. Of these, the Achelata included two families, three genera, and four species; and Brachyura has 27 families, 64 genera, and 94 species (Table S1). Also, of the 98 species recorded only 2% (P. bouvieri and Panopeus sp.) are native to the region, while the rest of the species are shared with other places.
According to Álvarez et al. [2], the Mexican Caribbean ranks third place in the biological diversity of marine decapods compared to the Mexican Pacific and the Gulf of Mexico. This low decapod diversity can be explained by few taxonomic studies performed in the region, the sampling of decapods has not been intense and continuous, and the collecting methodologies have been different between the studies carried out [2]. For this reason, it is recommended to increase the sampling in the SMC since, as indicated by Briones et al. [18] and Vargas-Castillo & Vargas-Zamora [47], increasing the sampling frequency during different times of the year, the number of collecting sites, and the different types of substrates are excellent strategies to know the biological diversity of the decapods.
3.3. Identified Species and Sampled Sites
After reviewing the faunal composition of each sampled site, it was found that the locality with the highest number of species was Punta Herrero with 10 species (23%), followed by El Uvero with seven species (16%), Mahahual with six species (14%), south of Xahuayxol with five species (12%), Punta Diamante, Huach River, and north of Xahuayxol with four species (9%) each, and Bermejo River with three species (7%). With respect to species frequency, Mithraculus sculptus (Lamarck, 1818) and Omalacantha bicornuta (Latreille, 1825) were the most predominant species in six of the eight sampled sites; followed by Cataleptodius floridanus (Gibbes, 1850) found in five sites; Mithraculus coryphe (Herbst, 1801), and Pachygrapsus transversus (Gibbes, 1850) in three places; M. nodrifrons, Panulirus argus Latreille, 1804, and Pitho lherminieri (Desbonne, 1867) in two locations; and the rest of the species were recognized once (Table 2).
Punta Herrero was the locality with the highest number of species because unlike the central zone of the SMC (e. g., Mahahual), it has not experienced important urban, tourist, and economic development [48]. Personal observations of the authors demonstrated that this locality showed a low accumulation of Sargassum, contrasting with the impact that the high concentration of Sargassum is creating between Mahahual and Xcalak localities. As the massive influx of Sargassum can influence the transformation of the ecosystems and fauna composition of the region [17,18,49], the abundance and diversity of crustaceans will be modified. According to Vargas-Castillo & Vargas-Zamora [49], the species lists are the first step for evaluating temporal changes in the composition and abundance of decapods due to coastal development, pollution, and climate change. To date, there are 15 sites where Achetala and Brachyura crustaceans have been collected and reported for the SMC (including localities from the literature and those studied in this work). These sites are found between Punta Herrero and Xcalak. This area corresponds to approximately a quarter of the total extension of the Mexican Caribbean. Therefore, updating the lists of decapod species, inclusion of other localities, and continuous monitoring of the populations must be priorities for the SMC.
3.4. Molecular Analysis
From 21 morphotypes identified, between one and five specimens were selected for molecular analysis. In total, 76 samples were processed and 65 (86%) were amplified correctly. No insertions, deletions, or stop codons were observed in the sequences and the lengths ranged between 608 and 665 base pairs (bp). The average K2P distance between barcode sequences within species was 0.38%, whereas interspecific divergences were 15.18%. These values are within the range reported by Raupach et al. [25] for crustaceans, including the decapod group from the North Sea.
The 65 sequences matched with sequences in the BOLD library with similarity values ≥99%. Based on these results, 10 families with 15 genera and 20 species were identified, and only one specimen was identified to genus level (Figure 2; Table S1; Figure S1). The BOLD system assigned 21 BINs to the sequences (see project DS-DECA01 in www.boldsystems.org). This result is consistent with the morphological identification of the 20 species and one morphotype (Panopeus sp.).

Figure 2.
ID tree showing the clustering of the 21 morphotypes. The numbers on the branches are the bootstrap support after 500 replicates. Numbers after the names are the BINs.
The genus Panopeus has 16 species reported in the literature, three for the Eastern Pacific (P. chilensis, P. convexus, P. diversus), one from the Eastern Atlantic (P. africanus), and 12 for the Western Atlantic (P. americanus, P. austrobesus, P. boekei, P. harttii, P. herbstii, P. lacustris, P. meridionalis, P. obesus, P. occidentalis, P. purpureus, P. rugosus, P. simpsoni). In the Bold database, there were 12 species with sequences (except for P. boekei, P. convexus, P. diversus, and P. occidentalis). The comparison of the sequence of the specimen of Panopeus sp., with the genetic material available in BOLD, showed that it did not match any of the species. Regarding the morphology, Panopeus sp. is similar to P. obesus since both have rounded lateral teeth in the carapace and the distribution of the dark color on the palm of the fixed finger. However, they differ in the body, Panopeus sp. showed a brown-white coloring pattern and a marked granular pattern on the carapace, while in P. obesus the color of the body is dark purple to russet and has few or no granular patterns on the carapace. Thoma et al. [50], settled that the traditional morphological characters used to assign members of the genus Panopeus, have not proven useful, and additional studies (including morphological and genetic data) are necessary to clarify the taxonomic status of the species and their evolutionary relationships. Thus, the authors will continue working with the DNA sequencing of the Panopeus sp. found in this work to contrast with morphological characters and assign the correct species or propose a new one for the SMC.
4. Conclusions
The revision of the Achelata and Brachyura species and the inclusion of one potential new species and six new records found in this work resulted in the increase to 98 species in the SMC. The analysis allowed the identification of 12 species that expanded their geographic distribution range, and 14 species were updated to a current scientific name. These results highlight the importance of natural protected areas in the preservation of diversity and abundance of species. The area with less anthropogenic disturbance “Punta Herrero” showed the highest number of species, contrasting with the low diversity in the southern locality of Mahahual, probably related to an increase in the residual waters and accumulation of plastic debris and Sargasso. Finally, these results contribute to the knowledge of the Caribbean crustaceans and are useful for adequate strategies for conservation and management of the regional fauna. It is important to continue the monitoring of the biological diversity of decapods in the Southern Mexican Caribbean, since they are an essential part of the ecosystems, and some of them are food resources appreciated by Mexican society.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14080649/s1, Table S1: List of current Achelata and Brachyura decapods recorded in the Southern Mexican Caribbean. Figure S1: Full BOLD Taxon ID Tree. References [51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79] are cited in Supplementary Materials.
Author Contributions
Conceptualization, J.J.-G., M.V.-M. and R.R.-L.; methodology, J.J.-G., M.V.-M. and R.R.-L.; Software, J.J.-G. and M.V.-M.; Resources, R.R.-L. and M.V.-M.; writing—original draft preparation, J.J.-G. Writing—Review & Editing, R.R.-L. and M.V.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by CONACyT with the project “Isotopic niches of key marine invertebrates to understanding the degradation of coral reefs in the Caribbean (ORDECYT-PRONACES/1312440/2020)”, and the postdoctoral research grant “Estancias Posdoctorales por México 2020–2021” (J.J.-G.). The Mexican Barcode of Life (MEXBOL) node Chetumal, assisted with DNA extraction, PCR reactions, and sequence edition of all material presented here.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Manuel Elías Gutiérrez and Alma Estrella García Morales (ECOSUR) for helping us in the processing of DNA Barcoding, as part of the CONACyT network Mexican Barcode of Life (MEXBOL). Special thanks are given to Nancy Cabanillas Terán (ECOSUR), Isabella Pérez Posada (ECOSUR), Víctor Conde Vela (UANL), Javier Nolasco Tinoco (I.T. de Chetumal) for helping us with the collection of biological material. Thanks to Sara Covarrubias (UMSNH) who elaborated on the localities map. This work was supported partially by the projects “Cephalopods and Crustaceans of the Mexican Caribbean Sea” and “Isotopic niches of key marine invertebrates to understanding the degradation of coral reefs in the Caribbean (ORDECYT-PRONACES/1312440/2020)”. J.J.-G. would like to thank CONACyT for the postdoctoral research grant belonging to the program “Estancias Posdoctorales por México 2020–2021”.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Matzen da Silva, J.; Creer, S.; dos Santos, A.; Costa, A.C.; Cunha, M.R.; Costa, F.O.; Carvalho, G.R. Systematic and Evolutionary Insights Derived from MtDNA COI Barcode Diversity in the Decapoda (Crustacea: Malacostraca). PLoS ONE 2011, 6, e19449. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, F.; Villalobos, J.L.; Hendrickx, M.E.; Escobar-Briones, E.; Rodríguez-Almaraz, G.; Campos, E. Biodiversidad de crustáceos decápodos (Crustacea: Decapoda) en México. Rev. Mex. Biodivers. 2014, 85, 208–219. [Google Scholar] [CrossRef]
- Martin, J.W.; Davis, G.E. An Updated Classification of the Recent CRUSTACEA; Natural History Museum of Los Angeles County: Los Angeles, CA, USA, 2001; Volume 39, pp. 1–124. [Google Scholar]
- Rana, S. Multiformity and Economic Importance of True Brachyuran Crabs. Edelweiss Appl. Sci. Technol. 2018, 2, 253–259. [Google Scholar] [CrossRef]
- Susanto, G.N. Crustacea: The Increasing Economic Importance of Crustaceans to Humans. In Arthropods—Are They Beneficial for Mankind? Ranz, R.E.R., Ed.; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Radhakrishnan, E.V.; Phillips, B.F.; Achamveetil, G. (Eds.) Lobsters: Biology, Fisheries and Aquaculture; Springer: Singapore, 2019. [Google Scholar] [CrossRef]
- Boudreau, S.; Worm, B. Ecological Role of Large Benthic Decapods in Marine Ecosystems: A Review. Mar. Ecol. Prog. Ser. 2012, 469, 195–213. [Google Scholar] [CrossRef]
- Szaniawska, A. Function and Importance of Crustaceans. In Baltic Crustaceans; Springer International Publishing: Cham, Switzerland, 2018; pp. 185–188. [Google Scholar] [CrossRef]
- Rosas-Luis, R.; Elias-Valdez, A. Trophic Resource Partitioning among Four Sympatric Lutjanid Species in the Southern Mexican Caribbean Sea. Mar. Biol. Res. 2021, 17, 615–624. [Google Scholar] [CrossRef]
- Santamaría-Miranda, A.; Saucedo-Lozano, M.; Herrera-Moreno, M.N.; Apún-Molina, J.P. Hábitos alimenticios del pargo amarillo Lutjanus argentiventris y del pargo rojo Lutjanus colorado (Pisces: Lutjanidae) en el norte de Sinaloa, México. Rev. Biol. Mar. Oceanogr. 2005, 40, 33–34. [Google Scholar] [CrossRef]
- Projecto-Garcia, J.; Cabral, H.; Schubart, C.D. High Regional Differentiation in a North American Crab Species throughout Its Native Range and Invaded European Waters: A Phylogeographic Analysis. Biol. Invasions 2010, 12, 253–263. [Google Scholar] [CrossRef]
- Stasolla, G.; Tricarico, E.; Vilizzi, L. Risk Screening of the Potential Invasiveness of Non-Native Marine Crustacean Decapods and Barnacles in the Mediterranean Sea. Hydrobiologia 2021, 848, 1997–2009. [Google Scholar] [CrossRef]
- Markham, J.C.; Donath-Hernández, F.E.; Villalobos-Hiriart, J.L.; Cantú Díaz-Barriga, A. Notes on the Shallow-Water Marine Crustacea of the Caribbean Coast of Quintana Roo, Mexico. An. Inst. Biol. Univ. Nac. México 1990, 61, 405–446. [Google Scholar]
- García-Madrigal, M.; del, S.G.; González, N.E.; Vázquez, C.C. Sección De Crustáceos De La Colección De Referencia De Bentos Costero De Ecosur. Univ. Cienc. 2002, 36, 140–148. [Google Scholar]
- Álvarez, F.Á. Crustáceos Estomatópodos, Anfípodos, Isópodos y Decápodos del Litoral de Quintana Roo; CONABIO: Tialpan, Mexico, 2021. [Google Scholar] [CrossRef]
- Campos-Vázquez, C. Crustáceos asociados a macroalgas en Bajo Pepito, Isla Mujeres, Caribe mexicano. Rev. Biol. Trop. 2000, 48, 361–364. [Google Scholar] [CrossRef]
- González-Gómez, R.; Briones-Fourzán, P.; Álvarez-Filip, L.; Lozano-Álvarez, E. Diversity and Abundance of Conspicuous Macrocrustaceans on Coral Reefs Differing in Level of Degradation. PeerJ 2018, 6, e4922. [Google Scholar] [CrossRef]
- Briones-Fourzán, P.; Monroy-Velázquez, L.V.; Estrada-Olivo, J.; Lozano-Álvarez, E. Diversity of Seagrass-Associated Decapod Crustaceans in a Tropical Reef Lagoon Prior to Large Environmental Changes: A Baseline Study. Diversity 2020, 12, 205. [Google Scholar] [CrossRef]
- Costa, F.O.; deWaard, J.R.; Boutillier, J.; Ratnasingham, S.; Dooh, R.T.; Hajibabaei, M.; Hebert, P.D. Biological Identifications through DNA Barcodes: The Case of the Crustacea. Can. J. Fish. Aquat. Sci. 2007, 64, 272–295. [Google Scholar] [CrossRef]
- Mantelatto, F.L.; Terossi, M.; Negri, M.; Buranelli, R.C.; Robles, R.; Magalhães, T.; Tamburus, A.F.; Rossi, N.; Miyazaki, M.J. DNA Sequence Database as a Tool to Identify Decapod Crustaceans on the São Paulo Coastline. Mitochondrial DNA Part A 2018, 29, 805–815. [Google Scholar] [CrossRef]
- Landschoff, J.; Gouws, G. DNA Barcoding as a Tool to Facilitate the Taxonomy of Hermit Crabs (Decapoda: Anomura: Paguroidea). J. Crustac. Biol. 2018, 38, 780–793. [Google Scholar] [CrossRef]
- Venera-Pontón, D.; Driskell, A.; De Grave, S.; Felder, D.; Scioli, J.; Collin, R. Documenting Decapod Biodiversity in the Caribbean from DNA Barcodes Generated during Field Training in Taxonomy. Biodivers. Data J. 2020, 8, e47333. [Google Scholar] [CrossRef]
- Marco-Herrero, E.; Cuesta, J.A.; González-Gordillo, J.I. DNA Barcoding Allows Identification of Undescribed Crab Megalopas from the Open Sea. Sci. Rep. 2021, 11, 20573. [Google Scholar] [CrossRef]
- Elías-Gutiérrez, M.; Hubert, N.; Collins, R.A.; Andrade-Sossa, C. Aquatic Organisms Research with DNA Barcodes. Diversity 2021, 13, 306. [Google Scholar] [CrossRef]
- Raupach, M.J.; Barco, A.; Steinke, D.; Beermann, J.; Laakmann, S.; Mohrbeck, I.; Neumann, H.; Kihara, T.C.; Pointner, K.; Radulovici, A.; et al. The Application of DNA Barcodes for the Identification of Marine Crustaceans from the North Sea and Adjacent Regions. PLoS ONE 2015, 10, e0139421. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. Bold: The Barcode of Life Data System (Http://Www.Barcodinglife.Org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef] [PubMed]
- Pozo, C.; Armijo, N.; Calmé, S. (Eds.) Riqueza Biológica de Quintana Roo: Un Análisis Para su Conservación, 1st ed.; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO): Tialpan, Mexico; Colegio de la Frontera Sur (ECOSUR): Chetumal, Mexico; Gobierno del Estado de Quintana Roo: Chetumal, Mexico; Programa de Pequeñas Donaciones-México (PPD): Merida, Mexico, 2011; ISBN 978-607-7607-45-8. [Google Scholar]
- Comisión Nacional de Áreas Naturales Protegidas (CONANP). Estudio Previo Justificativo Para La Declaratoria de La Reserva de La Biosfera Caribe Mexicano; CONANP: Quintana Roo, Mexico, 2016. [Google Scholar]
- Rodríguez-Martínez, R.E.; Medina-Valmaseda, A.E.; Blanchon, P.; Monroy-Velázquez, L.V.; Almazán-Becerril, A.; Delgado-Pech, B.; Vásquez-Yeomans, L.; Francisco, V.; García-Rivas, M.C. Faunal Mortality Associated with Massive Beaching and Decomposition of Pelagic Sargassum. Mar. Pollut. Bull. 2019, 146, 201–205. [Google Scholar] [CrossRef] [PubMed]
- UNIBIO. CNCR/Colección Nacional de Crustaceos; UNAM: Ciudad de México, Mexico, 2021. [Google Scholar] [CrossRef]
- BOLDSYSTEMS. Public Data Portal. Available online: Https://Www.Boldsystems.Org/Index.Php/Public_BINSearch?Searchtype=records (accessed on 1 February 2022).
- Elías-Gutiérrez, M.; Valdez-Moreno, M.; Topan, J.; Young, M.R.; Cohuo-Colli, J.A. Improved Protocols to Accelerate the Assembly of DNA Barcode Reference Libraries for Freshwater Zooplankton. Ecol. Evol. 2018, 8, 3002–3018. [Google Scholar] [CrossRef]
- Rathbun, M.J. The Grapsoid Crabs of America. Bull. U. S. Natl. Mus. 1918, i–xxii, 1–461. [Google Scholar] [CrossRef]
- Rathbun, M.J. The Spider Crabs of America. Bull. U. S. Natl. Mus. 1925, i–xx, 1–613. [Google Scholar] [CrossRef]
- Rathbun, M.J. The Cancroid Crabs of America of the Families Euryalidae, Portunidae, Atelecyclidae, Cancridae, and Xanthidae. Bull. U. S. Natl. Mus. 1930, 152, 1–609. [Google Scholar] [CrossRef]
- Williams, A.B. Marine Decapod Crustaceans of the Carolinas. Fish. Bull. Fish Wildl. Serv. 1965, 65, 1–298. [Google Scholar]
- Williams, A.B. The Mud Crab, Panopeus Herbstii, s.l. Partition into Six Species (Decapoda: Xanthidae). Fish. Bull. 1983, 81, 863–882. [Google Scholar]
- Williams, A.B. Shrimps, Lobsters, and Crabs of the Atlantic Coast of the Eastern United States, Maine to Florida. Estuaries 1984, 8, 77. [Google Scholar] [CrossRef]
- Abele, L.G.; Kim, W. An Illustrated Marine Decapod Crustacea] of Florida. State Fla. Dep. Environ. Regul. 1986, 8, 784. [Google Scholar]
- WoRMS Editorial Board. World Register of Marine Species. Available online: https://www.marinespecies.org (accessed on 1 March 2022). [CrossRef]
- Ivanova, N.V.; Dewaard, J.R.; Hebert, P.D.N. An Inexpensive, Automation-Friendly Protocol for Recovering High-Quality DNA. Mol. Ecol. Notes 2006, 6, 998–1002. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. Lond. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Prosser, S.; Martínez-Arce, A.; Elías-Gutiérrez, M. A New Set of Primers for COI Amplification from Freshwater Microcrustaceans. Mol. Ecol. Resour. 2013, 13, 1151–1155. [Google Scholar] [CrossRef]
- Sarmiento-Camacho, S.; Valdez-Moreno, M. DNA Barcode Identification of Commercial Fish Sold in Mexican Markets. Genome 2018, 61, 457–466. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Castillo, L.; Velázquez, D.; Iturbe, A.; López, D. La planeación urbana-regional en Costa Maya. In ANÁLISIS TERRITORIAL DEL TURISMO, Región Costa Maya, 1st ed.; Plaza Y Valdés: Ciudad de México, México, 2010; ISBN 978-607-402-295-7. [Google Scholar] [CrossRef]
- Vargas-Castillo, R.; Vargas-Zamora, J.A. Crustaceans (Decapoda & Stomatopoda) from Golfo Dulce (Pacific, Costa Rica) in the Collection of the Museum of Zoology, University of Costa Rica. UNED Res. J. Cuad. Investig. UNED 2020, 12, e2736. [Google Scholar]
- Thoma, B.P.; Guinot, D.; Felder, D.L. Evolutionary Relationships among American Mud Crabs (Crustacea: Decapoda: Brachyura: Xanthoidea) Inferred from Nuclear and Mitochondrial Markers, with Comments on Adult Morphology: Evolution of American Xanthoid Crabs. Zool. J. Linn. Soc. 2014, 170, 86–109. [Google Scholar] [CrossRef]
- Poupin, J. Les crustacés Décapodes des Petites Antilles, avec de nouvelles observations pour St. Martin, La Guadeloupe et La Martinique, 1st ed.; Patrimoines Naturels: Paris, France, 2018; pp. 1–264. [Google Scholar]
- Giraldes, B.W.; Smyth, D.M. Recognizing Panulirus meripurpuratus sp. nov. (Decapoda: Palinuridae) in Brazil—Systematic and biogeographic overview of Panulirus species in the Atlantic Ocean. Zootaxa 2016, 4107, 353–366. [Google Scholar] [CrossRef]
- Salas, S.; Bello, V.; Cabrera, M.A.; Rivas, R.; Santa María, A. Programa Maestro del Sistema Producto de la Pesquería de Langosta en Yucatán; CONAPESCA: Mérida, Mexico, 2012. [Google Scholar]
- Mendonça, L.M.C.; Guimarães, C.R.P.; Santos, R.C.; Alves, D.F.R.; Barros-Alves, S.P.; Silva, S.L.R.; Hirose, G.L. Decapod crustaceans from the continental shelf of Sergipe, northeastern Brazil. Zootaxa 2019, 4712, 301–344. [Google Scholar] [CrossRef] [PubMed]
- Holthuis, L.B. FAO Species Catalogue. Marine Lobsters of the World. An Annotated and Illustrated Catalogue of Marine Lobsters Known to Date; FAO Fisheries Synopsis: Rome, Italy, 1991; Volume 13. [Google Scholar]
- Poupin, J.; Corbari, L.A. Preliminary assessment of the deep-sea Decapoda collected during the KARUBENTHOS 2015 Expedition to Guadeloupe Island. Zootaxa 2016, 4190, 001–107. [Google Scholar] [CrossRef] [PubMed]
- Freitas, R.; Wirtz, P. First record of the sculptured mitten lobster Parribacus antarcticus (Crustacea, Decapoda, Scyllaridae) from the Cabo Verde Islands (Eastern Atlantic). Arquipelago-Life Mar. Sci. 2018, 36, 15–18. [Google Scholar] [CrossRef]
- Yang, M.; Liu, H.; He, Y. Complete mitochondrial genome of sculptured slipper lobster Parribacus antarcticus (Lund, 1793). Mitochondrial DNA Part B 2019, 4, 2297–2298. [Google Scholar] [CrossRef]
- Manzanilla-Domínguez, H.; Gasca, R.; Suárez-Morales, E. Notes on the distribution of phyllosoma larvae in an Oceanic Atoll-like Reef System in the Western Caribbean. Crustaceana 2005, 78, 505–512. [Google Scholar]
- Kropp, R.K.; Manning, R.B. The Atlantic gall crabs, Family Cryptochiridae (Crustacea: Decapoda: Brachyura). Smithson. Contrib. Zool. 1987, 462, 1–21. [Google Scholar] [CrossRef]
- Almeida, A.O.; Bezerra, L.E.A.; Souza, G.B.G.; Boehs, G. Shallow-water anomuran and brachyuran crabs (Crustacea: Decapoda) from southern Bahia, Brazil. Lat. Am. J. Aquat. Res. 2010, 38, 329–376. [Google Scholar]
- Diez García, Y.L.; Capote, A.J. List of marine crabs (Decapoda: Anomura and Brachyura) of shallow littoral of Santiago de Cuba, Cuba. Check List 2015, 11, 1–22. [Google Scholar] [CrossRef]
- Mantelatto, F.L.; Tamburus, A.F.; Magalhães, T.; Buranelli, R.C.; Terossi, M.; Negri, M.; Castilho, A.L.; Costa, R.C.; Zara, F.J. Checklist of decapod crustaceans from the coast of the São Paulo State (Brazil) supported by integrative molecular and morphological data: III. Infraorder Brachyura Latreille, 1802. Zootaxa 2020, 4872, 1–108. [Google Scholar] [CrossRef]
- Castro, P.; Ng, P.K.L. Revision of the family Euryplacidae Stimpson, 1871 (Crustacea: Decapoda: Brachyura: Goneplacoidea). Zootaxa 2010, 2375, 1–130. [Google Scholar] [CrossRef]
- Keith, D.E. Shallow-water and terrestrial brachyuran crabs of Roatan and the Swan Islands, Honduras. Sarsia 1985, 70, 251–278. [Google Scholar] [CrossRef]
- Colombara, A.M.; Quinn, D.; Chadwick, N.E. Habitat segregation and population structure of Caribbean Sea anemones and associated crustaceans on coral reefs at Akumal Bay, Mexico. Bull. Mar. Sci. 2017, 93, 1025–1047. [Google Scholar] [CrossRef]
- Wagner, H.P. The genera Mithrax Latreille, 1818 and Mithraculus White, 1847 (Crustacea: Brachyura: Majidae) in the Western Atlantic Ocean. Zool. Verh. 1990, 264, 1–65. [Google Scholar]
- Spivak, E.D.; Farias, N.E.; Ocampo, E.H.; Lovrich, G.A.; Luppi, T.A. Annotated catalogue and bibliography of marine and estuarine shrimps, lobsters, crabs and their allies (Crustacea: Decapoda) of Argentina and Uruguay (Southwestern Atlantic Ocean). Com. Téc. Mix. Frente Marít. 2019, 26, 1–178. [Google Scholar]
- Abele, L.G. A Review of the grapsid crab genus Sesarma (Crustacea: Decapoda: Grapsidae) in America, with the description of a new genus. Smithson. Contrib. Zool. 1992, 527, 1–60. [Google Scholar] [CrossRef]
- Simakova, U.V.; Zalota, A.K.; Spiridonov, V.A. Genetic analysis of population structure of alien North American mud crab Rhithropanopeus harrisii (Gould, 1841) in the Black Sea–Caspian Region. Russ. J. Biol. Invasions 2017, 8, 168–177. [Google Scholar] [CrossRef]
- Felder, D.L.; Palacios Theil, E. Three new symbiotic crabs of the genus Glassella Campos & Wicksten, 1997 from Atlantic and Gulf of Mexico Coasts of Florida, USA (Decapoda: Brachyura: Pinnotheridae). J. Crustac. Biol. 2020, 40, 899–917. [Google Scholar] [CrossRef]
- Garth, J.S. The Brachyura of the “Askoy” Expedition: With remarks on carcinological collecting in the Panama Bight. Bull. Am. Mus. Nat. Hist. 1948, 92, 1–66. [Google Scholar]
- Hendrickx, M. Checklist of brachyuran crabs (Crustacea: Decapoda) from the Eastern Tropical Pacific. Bull. L’Institut R. Sci. Nat. Belg. Biol. 1995, 65, 125–150. [Google Scholar]
- Rodrigues, I.B.; Cardoso, I.A.; Serejo, C.S. Catalogue and illustrated key of Achelous De Haan, 1833 and Portunus Weber, 1795 (Brachyura: Portunidae: Portuninae) species occurring in Brazilian waters. Nauplius 2017, 25, 2017005. [Google Scholar] [CrossRef]
- Williams, A.B. The swimming crabs of the genus Callinectes (Decapoda: Portunidae). Fish. Bull. 1974, 72, 685–798. [Google Scholar]
- Ortiz-León, H.J.; Cordero, E.S.; de Jesús-Navarrete, A. Distribución espacial y temporal del cangrejo Callinectes sapidus (Decapoda: Portunidae) en la Bahía de Chetumal, Quintana Roo, México. Rev. Biol. Trop. 2007, 55, 235–245. [Google Scholar] [CrossRef][Green Version]
- Rathbun, M.J. The Brachyura of the Biological Expedition to the Florida Keys and the Bahamas in 1893. Bull. Lab. Nat. Hist. State Univ. Iowa 1898, 4, 250–294. [Google Scholar]
- Felder, D.L. A new crab of the genus Nanoplax from the Gulf of Mexico, and assignment of Micropanope pusilla to a new genus (Crustacea, Brachyura, Pseudorhombilidae). Zootaxa 2020, 4810, 531–545. [Google Scholar] [CrossRef] [PubMed]
- Chace, F.A.; Hobbs, H.H. Bredin-Archbold-Smithsonian Biological Survey of Dominica: The Freshwater and Terrestrial Decapod Crustaceans of the West Indies with Special Reference to Dominica. Bull. U. S. Natl. Mus. 1969, 292, 1–258. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).